Abstract
As a key regulator of the unfolded protein response, the transcription factor XBP1 activates genes in protein secretory pathways and is required for the development of certain secretory cells. To elucidate the function of XBP1 in pancreatic β-cells, we generated β-cell-specific XBP1 mutant mice. Xbp1f/f;RIP-cre mice displayed modest hyperglycemia and glucose intolerance resulting from decreased insulin secretion from β-cells. Ablation of XBP1 markedly decreased the number of insulin granules in β-cells, impaired proinsulin processing, increased the serum proinsulin:insulin ratio, blunted glucose-stimulated insulin secretion, and inhibited cell proliferation. Notably, XBP1 deficiency not only compromised the endoplasmic reticulum stress response in β-cells but also caused constitutive hyperactivation of its upstream activator, IRE1α, which could degrade a subset of mRNAs encoding proinsulin-processing enzymes. Hence, the combined effects of XBP1 deficiency on the canonical unfolded protein response and its negative feedback activation of IRE1α caused β-cell dysfunction in XBP1 mutant mice. These results demonstrate that IRE1α has dual and opposing roles in β-cells, and that a precisely regulated feedback circuit involving IRE1α and its product XBP1s is required to achieve optimal insulin secretion and glucose control.
Keywords: diabetes, secretory granule, endoribonuclease
XBP1 is a transcription factor that plays a crucial role in the development of professional secretory cells (1, 2). As a key component of the unfolded protein response (UPR), XBP1 is activated by disturbances in endoplasmic reticulum (ER) protein-folding homeostasis (3, 4). In response to upstream signals, XBP1 mRNA undergoes an unconventional splicing by the ER transmembrane endoribonuclease IRE1 which ultimately generates the potent transcriptional transactivator XBP1s (1, 3). XBP1s promotes ER biogenesis and activates the expression of ER chaperone genes that are required for the folding and trafficking of secretory cargo proteins (5–7). Consistent with its critical role in facilitating protein secretion, XBP1 deficiency impairs the development of professional secretory cells such as plasma B cells, pancreatic acinar cells, and intestinal Paneth cells (8–11).
Insulin synthesis and secretion from pancreatic β-cells are dynamically regulated by environmental changes that may impose burdens on the islet cell. It has been suggested that the UPR plays a protective role in the adaptation of the β-cell to increased insulin production (12, 13). UPR pathways mediated by IRE1/XBP1 and ATF6 have been implicated in insulin secretion and β-cell survival, but their in vivo functions in β-cells are not fully understood (14–18).
Here we show that β-cell proliferation, proinsulin processing, and insulin secretion are impaired in mutant mice lacking XBP1 selectively in β-cells. Whereas impaired direct transcriptional activation of secretory pathway genes by loss of XBP1 partly accounted for abnormalities in the mutant mice, we provide evidence for an indirect mechanism involving IRE1α-mediated degradation of proinsulin-processing gene mRNAs as well. These studies demonstrate that a precisely regulated feedback circuit involving IRE1α and its product XBP1s is required to achieve optimal glucose control. This rheostat relies on achieving optimal levels of XBP1s protein to fine-tune the insulin secretory function of β-cells.
Results
Xbp1 Deletion in β-Cells Causes Hyperglycemia and Glucose Intolerance.
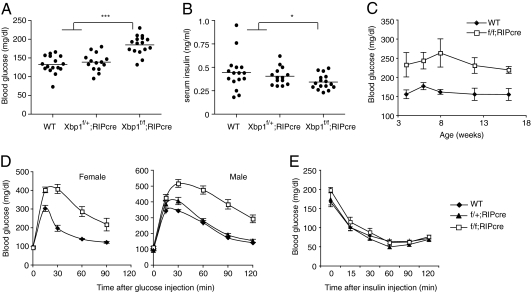
Deletion of Xbp1 in β-cells was achieved by crossing Xbp1flox mice (19) with RIP-cre transgenic mice that express cre recombinase under the control of the rat insulin gene promoter. Xbp1f/f;RIP-cre mice displayed slightly elevated blood glucose levels compared with littermate WT and heterozygous Xbp1f/+;RIP-cre mice (Fig. 1A). In accordance with increased blood glucose levels, serum insulin levels were slightly lower in Xbp1f/f;RIP-cre mice (Fig. 1B), indicating that XBP1-deficient β-cells secrete insufficient amounts of insulin to maintain normal glucose levels. Blood glucose and insulin levels were not altered in heterozygous Xbp1f/+;RIP-cre mice (Fig. 1 A and B). The modest hyperglycemic phenotype of Xbp1f/f;RIP-cre mice was maintained up to 16 wk of age (Fig. 1C). Glucose homeostasis was further assessed by glucose tolerance tests (GTT), which demonstrated impaired glucose clearance in both male and female Xbp1f/f;RIP-cre mice (Fig. 1D). Glucose tolerance in Xbp1f/+;RIP-cre mice was indistinguishable from WT, indicating that glucose homeostasis was not significantly affected by Xbp1 heterozygosity or by cre expression in β-cells. Notably, administration of insulin to Xbp1f/f;RIP-cre mice effectively lowered blood glucose, indicating that insulin signaling and sensitivity in peripheral tissues were not altered by the absence of XBP1 in β-cells (Fig. 1E).
Fig. 1.
β-cell-specific deletion of Xbp1 causes hyperglycemia and glucose intolerance in mice. (A) Blood glucose and (B) serum insulin levels were measured after fasting mice for 6 h. Each dot represents an individual male mouse (7-wk-old). (C) Fed-state blood glucose levels of WT and β-cell-specific Xbp1 knockout (Xbp1f/f;RIP-cre) mice were measured along the time course. n = 6–8 male mice per group. Values represent means ± SEM. (D) GTTs were performed on 7- to 8-wk-old mice. n = 6–8 mice per group. (E) ITT was performed on 16-wk-old male mice. n = 5–7 mice per group. *P < 0.05; ***P < 0.001.
Akita mice carrying the C96Y mutation in the Ins2 gene have been widely used as an animal model for ER stress in pancreatic β-cells (20, 21). The C96Y mutation disrupts a disulfide bond in the insulin molecule, leading to the degradation of the misfolded proinsulin protein and the compromise of ER function (20, 22). To determine whether XBP1 had any protective role in β-cells against the ER stress caused by the misfolded insulin, we generated Xbp1f/f;Ins2+/Akita;RIP-cre mice that are deficient for Xbp1 and produce C96Y mutant insulin. As expected, Xbp1-sufficient Akita mice (Xbp1f/f;Ins2+/Akita) developed progressive hyperglycemia both in males and females (Fig. S1). Notably, blood glucose level was markedly increased in XBP1-deficient Akita mice (Xbp1f/f;Ins2+/Akita;RIP-cre) compared with the control XBP1-sufficient mice, suggesting that XBP1 plays a protective role in preserving protein-folding homeostasis in stressed β-cells.
Loss of β-Cells and Decreased Pancreatic Insulin Content in XBP1-Deficient Mice.
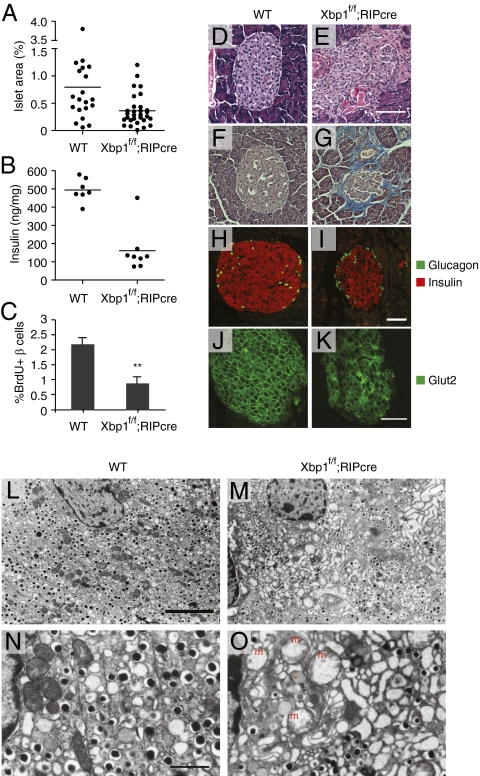
Hyperglycemia and glucose intolerance in Xbp1f/f;RIP-cre mice in the absence of a detectable alteration in systemic insulin sensitivity strongly suggested a defect in insulin production from XBP1-deficient pancreatic β-cells. Histological analysis of pancreatic sections revealed that islet area was diminished in Xbp1f/f;RIP-cre mice by ∼50% (Fig. 2A). Pancreatic insulin content was also decreased in Xbp1f/f;RIP-cre mice, consistent with β-cell insufficiency (Fig. 2B). TUNEL and active caspase 3 staining revealed no difference in β-cell apoptosis between WT and Xbp1f/f;RIP-cre mice. In contrast, BrdU incorporation into β-cells was significantly decreased in Xbp1f/f;RIP-cre mice (Fig. 2C), suggesting that impaired proliferation contributed to the reduced islet mass in Xbp1f/f;RIP-cre mice. Xbp1f/f;RIP-cre islets frequently displayed morphological abnormalities, including disorganization of islet structure, indistinct separation of islets from exocrine pancreatic tissue (Fig. 2 D and E), and peri-islet fibrosis (Fig. 2 F and G). Immunofluorescent staining of pancreatic sections revealed decreased abundance of insulin-producing β-cells and diminished insulin fluorescence intensity per cell in Xbp1f/f;RIP-cre mice (Fig. 2 H and I and Fig. S2). In addition, XBP1-deficient β-cells displayed a diffuse localization pattern of the Glut2 glucose transporter protein both in the cytosol and in the plasma membrane compared with the localized plasma membrane immunofluorescence in the WT control, suggesting that XBP1 is required for normal Glut2 trafficking to the plasma membrane (Fig. 2 J and K).
Fig. 2.
Histopathological analysis of islets of β-cell-specific Xbp1 KO mice. (A) Islet area relative to total pancreas. One random pancreas section per mouse was examined. Each dot represents an individual mouse of the indicated genotype of 4–6 mo of age. (B) Pancreatic insulin contents of 12- to 16-wk-old male mice. (C) BrdU-positive β-cells were counted after BrdU/insulin double staining. n = 4–5 mice per group. **P < 0.01. Pancreatic sections from WT and Xbp1f/f;RIP-cre mice were stained with (D and E) hematoxylin and eosin, (F and G) trichrome blue, (H and I) insulin and glucagon, and (J and K) Glut2 antibodies followed by fluorescence-conjugated secondary antibodies. (Scale bars, 100 μm.) (L–O) Islet sections were examined by TEM. [Scale bar, 5 μm (L and M); 1 μm (N and O).] The asterisk indicates an electron lucent granule. m, swollen mitochondria.
Ultrastructural analysis by transmission electron microscopy (TEM) revealed that XBP1-deficient β-cells contained fewer insulin granules than WT β-cells (Fig. 2 L and M). Further, ER distension and mitochondrial swelling were evident in most β-cells but not in α-cells of Xbp1f/f;RIP-cre mice (Fig. 2 N and O and Fig. S3). Electron lucent insulin granules were readily observed in XBP1-deficient β-cells (Fig. 2O and Fig. S3), similar to what we had previously observed in β-cells of newborn XBP1 germ-line knockout mice rescued by transgenic expression of XBP1 in the liver (10). These data suggest that XBP1 is essential for the biogenesis of insulin secretory granules and the maintenance of ER and mitochondrial integrity in β-cells.
XBP1 Is Required for Glucose-Stimulated Insulin Secretion and Proinsulin Processing.
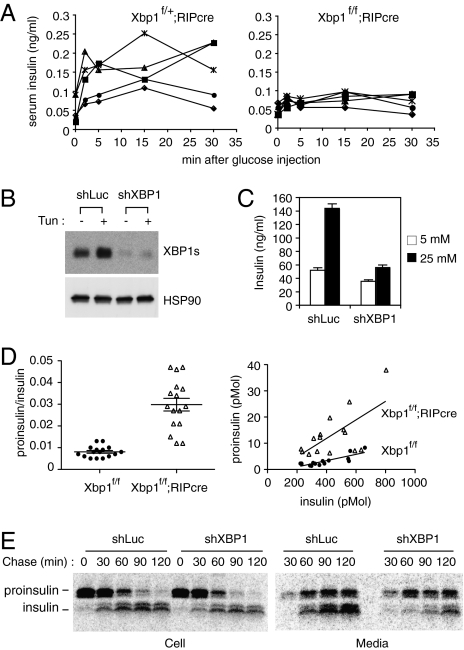
To further evaluate the role of XBP1 in insulin secretion from β-cells, we measured insulin secretion from control and Xbp1f/f;RIP-cre mice following a bolus glucose administration (Fig. 3A). Heterozygous Xbp1f/+;RIP-cre mice were used as controls to avoid potential nonspecific effects of cre expression in β-cells. Glucose administration markedly increased serum insulin levels in control mice within 2 min, and these levels were maintained for up to 30 min (Fig. 3A). In contrast, glucose-stimulated insulin secretion was severely reduced in Xbp1f/f;RIP-cre mice, indicating that XBP1 plays an important role in this process.
Fig. 3.
Impaired insulin secretion and proinsulin maturation in XBP1-deficient β-cells. (A) Glucose-stimulated insulin secretion assays. Serum insulin levels were measured in control heterozygous Xbp1f/+;RIP-cre and Xbp1f/f;RIP-cre mice at indicated time points after a bolus glucose injection. Each line represents an individual mouse. (B) Min6 cells stably expressing shRNAs targeting control luciferase or XBP1 mRNAs were tested for XBP1s expression by Western blot. (C) Cells were pretreated with 2.5 mM glucose for 2 h and then cultured in 5 mM or 25 mM glucose media for 2 h. Culture media were collected to measure insulin content by ELISA. (D) Serum proinsulin levels relative to total insulin in WT and Xbp1f/f;RIP-cre mice were determined. Data from both males and females were combined, because sex difference was not significant. The graphs display proinsulin:insulin ratio (Left) or proinsulin and insulin concentrations (pmol per liter) (Right). Each dot represents an individual mouse (4- to 6-mo-old). (E) Min6 cells expressing control or XBP1 shRNA were pulse-labeled for 30 min with [35S]Met/Cys and then cultured in chase media for the indicated time. Cells and culture supernatants were harvested for immunoprecipitation of radiolabeled insulin and proinsulin, which were revealed by SDS/PAGE followed by autoradiography.
To examine the role of XBP1 in insulin secretion at the cellular level in an alternative system, we silenced XBP1 mRNA in Min6 insulinoma cells by stably expressing an XBP1-specific shRNA (Fig. 3B). It was notable that Min6 cells expressed significant amounts of XBP1s protein at baseline even in the absence of chemical ER stress inducers. Stimulation of control Min6 cells with high glucose increased insulin secretion by approximately threefold (Fig. 3C). Consistent with the defective insulin secretion observed in Xbp1f/f;RIP-cre mice in response to a bolus glucose injection, glucose-stimulated insulin secretion was markedly blunted in XBP1 knockdown Min6 cells (Fig. 3C), indicating that XBP1 is required for optimal insulin secretion from β-cells.
Insulin is synthesized as a precursor form that undergoes a series of maturation steps that involve proteolytic cleavage and disulfide-bond formation (23, 24). Given the importance of XBP1 in cellular secretory function in many cell types (9, 10, 25, 26), we asked whether XBP1 also plays a role in proinsulin maturation in β-cells and measured the ratio of serum proinsulin to total insulin levels. WT mice displayed very low proinsulin:insulin ratios, indicating efficient insulin maturation in normal β-cells (Fig. 3D). In contrast, serum proinsulin:insulin ratios were increased by approximately fourfold in Xbp1f/f;RIP-cre mice. The increase in proinsulin concentration mainly accounted for the increase in proinsulin:insulin ratios in Xbp1f/f;RIP-cre mice (Fig. 3D Right). Taken together, these results demonstrate that XBP1 deficiency impairs proinsulin processing and/or folding of mature insulin, resulting in increased secretion of proinsulin at the expense of mature insulin.
Proinsulin processing and secretion were also monitored in Min6 insulinoma cells that expressed control luciferase or XBP1-specific shRNAs in pulse-chase experiments (Fig. 3E). In both control and XBP1 knockdown cells, radiolabeled proinsulin was gradually converted to mature insulin, which was then secreted into the media together with unprocessed proinsulin (Fig. 3E). Interestingly, XBP1 knockdown or overexpression of dominant-negative XBP1 markedly diminished the accumulation of radiolabeled mature insulin but not proinsulin in the culture supernatant, increasing the ratio of secreted proinsulin to insulin (Fig. 3E and Fig. S4A).
We hypothesized that XBP1 deficiency perturbed protein-folding homeostasis in the ER, leading to inefficient insulin processing in β-cells. To test this hypothesis, we added pharmacological drugs that interfere with ER protein folding after pulse labeling and examined proinsulin processing and secretion (Fig. S4B). The addition of tunicamycin or thapsigargin to inhibit protein glycosylation in the ER or deplete ER calcium stores during chase periods resulted in a marked increase of both intracellular and secreted proinsulin. In contrast, the amount of secreted mature insulin was decreased by tunicamycin or thapsigargin treatment (Fig. S4B), suggesting that perturbed ER function impaired efficient proinsulin processing, leading to an increase in proinsulin:insulin ratio in the culture media.
XBP1 Is Required for the Expression of Genes in the Insulin Secretory Pathway.
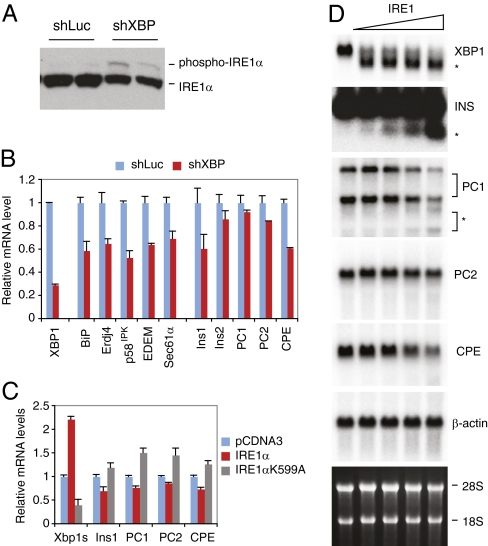
We have demonstrated that XBP1 deficiency results in feedback activation of IRE1α in certain cell types (9, 11, 19). Interestingly, recent studies demonstrated that IRE1α can cleave certain mRNAs for degradation, a process termed “IRE1-mediated degradation” (17, 27, 28). Notably, insulin mRNA was shown to be cleaved for degradation by IRE1α at multiple sites (17, 27). IRE1α phosphorylation was significantly induced in Min6 cells transfected with XBP1 shRNA, demonstrating that the feedback regulation of IRE1α by XBP1 also occurs in β-cells (Fig. 4A). Thus, the deletion of the Xbp1 gene in β-cells results in both XBP1 deficiency and IRE1α hyperactivation.
Fig. 4.
Feedback activation of IRE1α by XBP1 deficiency, and IRE1α-mediated mRNA cleavage. (A) Western blot and (B) Quantitative RT-PCR analysis of Min6 cells expressing luciferase or XBP1 shRNAs. (C) 293T cells were cotransfected with EGFP, Ins1, PC1, PC2, and CPE plasmids together with WT or K599A mutant IRE1α. Cells were harvested 24 h after transfection for quantitative RT-PCR analysis. EGFP mRNA levels were used for normalization. Values represent fold changes relative to vector controls. (D) Total Min6 RNA was incubated with increasing amounts of recombinant IRE1α and then separated on an agarose gel. Northern blot was performed using the indicated 32P-labeled probes. The asterisks indicate cleavage products.
To investigate the effects of both the direct transcriptional activation of target genes by XBP1 and the indirect effect of XBP1 loss on IRE1α hyperactivation in β-cells, we examined the expression of classical UPR genes as well as genes involved in insulin secretory pathways in Min6 cells transiently transfected with XBP1 siRNA (Fig. S5A) or in stable cell lines expressing XBP1 shRNA (Fig. 4B). Unfortunately, isolation of primary islets from Xbp1f/f;RIP-cre mice was unsuccessful, yielding only small amounts (<10% of WT control) of minute islets associated with residual exocrine tissue (Fig. S5B), likely due to the structural abnormalities of the mutant islets (Fig. 2C), hence precluding gene expression analysis in primary cells. XBP1 mRNA levels were only slightly decreased in the KO islets compared with the WT control, likely due to the presence of a very small number of XBP1-deficient β-cells and the consequent enrichment of non-β-cells, or the contamination of WT β-cells that failed to undergo cre recombination. XBP1 knockdown down-regulated ER chaperone genes such as BiP (Hspa5), ERdj4 (Dnajb9), and p58IPK (Dnajc3), which have been documented as direct XBP1 targets (5), and decreased ER/Golgi dimensions, consistent with the role of XBP1 in ER membrane biogenesis (Fig. 4B and Fig. S5 A and C) (6, 7). Interestingly, Ins1 and a number of genes involved in the insulin secretory pathway, such as PC1, PC2, CPE, and synaptophysin (Syp), were also down-regulated in XBP1 knockdown cells (Fig. 4B and Fig. S5A). Decreased expression of CPE was also observed in β-cells of Xbp1f/f;RIP-cre mice by immunostaining (Fig. S5D), suggesting that the in vitro system recapitulated in vivo events. Surprisingly, we failed to demonstrate specific binding of XBP1 to PC1, PC2, CPE, or Ins1 promoters in chromatin immunoprecipitation (ChIP) assays (Fig. S5E), suggesting that XBP1 did not directly regulate these genes. In addition, IRE1α siRNA did not reduce PC1, PC2, or CPE mRNA levels, despite efficiently decreased XBP1s and its direct targets, BiP and ERdj4 (Fig. S4F). We tested whether XBP1 indirectly regulated these via mRNA degradation by activated IRE1α, as demonstrated previously for Ins1 mRNA (17, 27). Indeed, cotransfection of IRE1α with plasmids expressing Ins1, PC1, PC2, or CPE significantly reduced the mRNA levels of these genes compared with an inactive mutant IRE1α (Fig. 4C). To investigate the specific cleavage of these mRNAs by IRE1α, we incubated the total RNA from Min6 cells with recombinant IRE1α protein and examined the cleavage of each mRNA species by Northern blot (Fig. 4D). IRE1α efficiently cleaved XBP1 mRNA. IRE1α also cleaved insulin, PC1, PC2, and CPE mRNAs, albeit at low efficiency, decreasing the full-length mRNAs and/or generating small amounts of cleaved mRNAs (Fig. 4D).
Discussion
Here we report the occurrence of hyperglycemia and islet cell loss in adult mice lacking XBP1 selectively in β-cells of the pancreas. Biochemical analysis revealed two mechanisms by which XBP1 deficiency led to this phenotype. XBP1 deficiency not only compromised the canonical ER stress response in β-cells but also caused constitutive hyperactivation of its upstream activator IRE1α, allowing degradation of a subset of mRNAs encoding insulin secretory pathway genes. It remains to be determined to what extent IRE1α hyperactivation versus loss of XBP1s transactivation contributes to the impaired proliferation and insulin secretory function of XBP1-deficient β-cells.
Pancreatic β-cells require the UPR signaling pathways regulated by IRE1/XBP1 and PERK/eIF2α for proliferation, survival, and insulin secretion. Whereas XBP1 inactivation impaired the postnatal insulin secretory function of β-cells, PERK appears to be essential at the embryonic or neonatal stage of development for the viability of β-cells, but not at the adult stage (29). Further, PERK deletion or eIF2α mutation did not cause any measurable activation of the other UPR branches in β-cells (13, 29), suggesting that PERK/eIF2α might have a unique function in the β-cell which is distinct from its function in relieving ER stress. Nonetheless, regulated eIF2α phosphorylation is critical for β-cell survival and insulin secretion in adults, as an inducible mutation at the eIF2α phosphorylation target site caused dramatic loss of β-cells within a few weeks, highlighting the importance of other eIF2α kinases in β-cells (13). Notably, inactivation of XBP1 (this report) or blockade of eIF2α phosphorylation caused similar cellular structural abnormalities, such as ER distension and mitochondrial swelling, and reduced insulin contents, suggesting overlapping functions regulated by these two UPR signaling pathways (13). XBP1 also displays a remarkable similarity to ATF6α, another key UPR transcription factor, in DNA binding specificity, and can form heterodimers with ATF6α, suggesting that they might work cooperatively (5, 30). Although ablation of ATF6α did not cause any overt abnormalities in mice (30, 31), it will be interesting to determine whether XBP1 and ATF6α have compensatory roles in β-cells and other secretory cells in vivo.
Although both IRE1α and XBP1 deficiencies ablate the active transcription factor XBP1s, there are also fundamental differences between these two. First, XBP1 deficiency can cause feedback activation of IRE1α (9, 11, 19), which could result in the hyperactivation of IRE1α’s “XBP1-independent” functions such as insulin mRNA degradation (17, 27). Second, IRE1α deficiency does not ablate XBP1u protein encoded by the unspliced XBP1 mRNA. Although XBP1u is unstable and has no transactivation ability (32, 33), it might have distinct, yet-to-be explored functions. In line with this possibility, recent studies demonstrated that the phenotypes of IRE1α and XBP1 KO mice are not identical, suggesting the presence of unique functions specific to each gene (18). However, it is unclear to what extent these XBP1-independent functions of IRE1α contribute to the viability and insulin secretory function of β-cells. It is important to note that the cleavage of proinsulin-processing enzyme mRNAs requires higher IRE1α activity than XBP1 splicing (Fig. 4E), suggesting that the former would occur only when IRE1α is hyperactivated. The physiological and pathological conditions that activate IRE1α leading to IRE1-mediated mRNA degradation need to be defined, but it is conceivable that IRE1α would modulate β-cell function depending on the severity of the ER stress signal received (27).
The proinsulin:insulin ratio is increased in type 2 diabetes and in populations at high risk for developing the disease (34–38). One explanation put forth is that the increased secretory demand on β-cells under these conditions might result in the secretion of incompletely processed proinsulin (39, 40). One scenario is that the increased secretory demand in the setting of chronic hyperglycemia induces ER stress, as recently demonstrated by signs of UPR activation in β-cells of diabetic humans and mice (41, 42). Hence, it is conceivable that disturbance of ER folding capacity and/or XBP1 function could predispose individuals to diabetes, especially in the setting of insulin resistance, which would markedly increase the demand on β-cell secretory function (43).
The regulatory mechanisms of insulin secretion in β-cells are also conserved in other endocrine and neuroendocrine cells that secrete peptide hormones and neuropeptides (44). CPE and prohormone convertases also function in the biogenesis of secretory granules in neurons and the secretion of neuropeptides (45). XBP1 KO mice did not display any overt neurological defects (46), although it has been reported that axonal growth of developing neurons requires XBP1 (47). The possible role of XBP1 in the regulation of secretory pathways in other cell types, including neurons, awaits further investigation.
Materials and Methods
GTT, insulin tolerance test (ITT), and Glucose-Stimulated Insulin Secretion Assays.
For GTT and glucose-stimulated insulin secretion assays, mice were fasted for 16 h with free access to water and then injected i.p. with glucose at 2 g/kg of body weight. Blood glucose concentrations were measured using an Ascensia Breeze glucometer (Bayer). Plasma insulin concentrations were determined using an Ultra Sensitive Rat Insulin ELISA Kit (Crystal Chem). For ITT, mice were fasted for 6 h and injected i.p. with 0.75 units/kg body weight of human insulin (Eli Lilly).
Pancreatic Insulin Content and Plasma Proinsulin Assay.
Insulin was extracted from pancreata by homogenizing the tissue in acidified ethanol. After removing insoluble materials, insulin levels in pancreas extracts were measured by ELISA as above. Serum proinsulin levels were measured using an ELISA kit (ALPCO Diagnostics).
Histological Analysis.
Details of histological analysis are provided in SI Materials and Methods. For immunohistochemistry, we used guinea pig anti-insulin (4011-01; Linco), rabbit anti-glucagon (AB932; Chemicon), mouse anti-CPE (610758; BD Biosciences), rabbit anti-Glut2 (07-1402; Millipore), and mouse anti-BrdU (Bu20a; Biolegend). For the β-cell proliferation assay, BrdU (Sigma) was administered to 4-wk-old male mice i.p. at 50 mg/kg 24 h before sacrifice.
Cell Culture and Pulse-Chase Experiments.
Min6 cells were cultured in DMEM/high glucose supplemented with 15% FBS, nonessential amino acids, 50 μM β-mercaptoethanol, and penicillin/streptomycin. After starvation in methionine/cysteine-free medium for 30 min, cells were labeled with [35S]methionine/cysteine (PerkinElmer) for 30 min, as described previously (19). Cells and culture supernatants were harvested after chase in cold media and insulin was immunoprecipitated using an anti-insulin monoclonal antibody (I1208; Sigma). Tricine gels (10%) (Invitrogen) were used to resolve 35S-labeled insulin species. Densitometry was performed using a PhosphorImager (Bio-Rad).
RNA Isolation and Quantitative RT-PCR.
Total RNAs were isolated using TRIzol (Invitrogen) and treated with DNase I (Ambion). cDNA synthesis, SYBR Green-based real-time PCR, and ChIP assays were performed as described previously (19). Primer sequences are shown in Table S1.
RNA Cleavage Assay.
Detailed methods are provided in SI Materials and Methods. Total RNA isolated from Min6 cells was incubated with the cytoplasmic domain of human IRE1α and subjected to Northern blot analysis.
Supplementary Material
Acknowledgments
We thank Dr. R. Kaufman for WT and K599A mutant IRE1α constructs, D. Hu for assistance with histologic analyses, K. Sigrist for assistance with animal maintenance, C. Cahill for assistance with electron microscopy, Drs. R. N. Kulkarni, C. W. Liew, and U. Ozcan for technical assistance, and Dr. A. Shulman for critical reading of this manuscript. This study was supported by National Institutes of Health Grants AI32412 (to L.H.G.) and DK082448 (to L.H.G.) and by a grant from the American Heart Association (to A.-H.L.).
Footnotes
Conflict of interest statement: L.H.G. is a member of the board of directors of and holds equity in the Bristol Myers Squibb Corporation.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1105564108/-/DCSupplemental.
References
- 1.Schröder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 2.Lee AH, Glimcher LH. Intersection of the unfolded protein response and hepatic lipid metabolism. Cell Mol Life Sci. 2009;66:2835–2850. doi: 10.1007/s00018-009-0049-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 4.Kaufman RJ. Stress signaling from the lumen of the endoplasmic reticulum: Coordination of gene transcriptional and translational controls. Genes Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- 5.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaffer AL, et al. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21:81–93. doi: 10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 7.Sriburi R, Jackowski S, Mori K, Brewer JW. XBP1: A link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol. 2004;167:35–41. doi: 10.1083/jcb.200406136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iwakoshi NN, et al. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat Immunol. 2003;4:321–329. doi: 10.1038/ni907. [DOI] [PubMed] [Google Scholar]
- 9.Kaser A, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee AH, Chu GC, Iwakoshi NN, Glimcher LH. XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. EMBO J. 2005;24:4368–4380. doi: 10.1038/sj.emboj.7600903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Todd DJ, et al. XBP1 governs late events in plasma cell differentiation and is not required for antigen-specific memory B cell development. J Exp Med. 2009;206:2151–2159. doi: 10.1084/jem.20090738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harding HP, Ron D. Endoplasmic reticulum stress and the development of diabetes: A review. Diabetes. 2002;51(Suppl 3):S455–S461. doi: 10.2337/diabetes.51.2007.s455. [DOI] [PubMed] [Google Scholar]
- 13.Back SH, et al. Translation attenuation through eIF2α phosphorylation prevents oxidative stress and maintains the differentiated state in β cells. Cell Metab. 2009;10:13–26. doi: 10.1016/j.cmet.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lipson KL, et al. Regulation of insulin biosynthesis in pancreatic β cells by an endoplasmic reticulum-resident protein kinase IRE1. Cell Metab. 2006;4:245–254. doi: 10.1016/j.cmet.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 15.Elouil H, et al. Acute nutrient regulation of the unfolded protein response and integrated stress response in cultured rat pancreatic islets. Diabetologia. 2007;50:1442–1452. doi: 10.1007/s00125-007-0674-4. [DOI] [PubMed] [Google Scholar]
- 16.Matsuda T, et al. Ablation of C/EBPβ alleviates ER stress and pancreatic β cell failure through the GRP78 chaperone in mice. J Clin Invest. 2010;120:115–126. doi: 10.1172/JCI39721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lipson KL, Ghosh R, Urano F. The role of IRE1α in the degradation of insulin mRNA in pancreatic β-cells. PLoS One. 2008;3:e1648. doi: 10.1371/journal.pone.0001648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwawaki T, Akai R, Kohno K. IRE1α disruption causes histological abnormality of exocrine tissues, increase of blood glucose level, and decrease of serum immunoglobulin level. PLoS One. 2010;5:e13052. doi: 10.1371/journal.pone.0013052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, et al. A mutation in the insulin 2 gene induces diabetes with severe pancreatic β-cell dysfunction in the Mody mouse. J Clin Invest. 1999;103:27–37. doi: 10.1172/JCI4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oyadomari S, et al. Targeted disruption of the Chop gene delays endoplasmic reticulum stress-mediated diabetes. J Clin Invest. 2002;109:525–532. doi: 10.1172/JCI14550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nozaki J, et al. The endoplasmic reticulum stress response is stimulated through the continuous activation of transcription factors ATF6 and XBP1 in Ins2+/Akita pancreatic β cells. Genes Cells. 2004;9:261–270. doi: 10.1111/j.1356-9597.2004.00721.x. [DOI] [PubMed] [Google Scholar]
- 23.Weiss MA. Proinsulin and the genetics of diabetes mellitus. J Biol Chem. 2009;284:19159–19163. doi: 10.1074/jbc.R109.009936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steiner DF, Bell GI, Rubenstein AH, Chan SJ. Chemistry and biosynthesis of the islet hormones; insulin, islet amyloid polypeptide (amylin), glucagon, somatostatin, and pancreatic polypeptide. In: DeGroot LJ, editor. Endocrinology. 5th Ed. Philadelphia: Saunders; 2005. pp. 925–960. [Google Scholar]
- 25.Reimold AM, et al. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- 26.Huh WJ, et al. XBP1 controls maturation of gastric zymogenic cells by induction of MIST1 and expansion of the rough endoplasmic reticulum. Gastroenterology. 2010;139:2038–2049. doi: 10.1053/j.gastro.2010.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han D, et al. IRE1α kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 2009;138:562–575. doi: 10.1016/j.cell.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hollien J, et al. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol. 2009;186:323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang W, et al. PERK EIF2AK3 control of pancreatic β cell differentiation and proliferation is required for postnatal glucose homeostasis. Cell Metab. 2006;4:491–497. doi: 10.1016/j.cmet.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 30.Yamamoto K, et al. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6α and XBP1. Dev Cell. 2007;13:365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 31.Wu J, et al. ATF6α optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13:351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 32.Lee AH, Iwakoshi NN, Anderson KC, Glimcher LH. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc Natl Acad Sci USA. 2003;100:9946–9951. doi: 10.1073/pnas.1334037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 34.Heaton DA, et al. Increased proinsulin levels as an early indicator of B-cell dysfunction in non-diabetic twins of type 1 (insulin-dependent) diabetic patients. Diabetologia. 1988;31:182–184. doi: 10.1007/BF00276853. [DOI] [PubMed] [Google Scholar]
- 35.Pradhan AD, et al. Insulin, proinsulin, proinsulin:insulin ratio, and the risk of developing type 2 diabetes mellitus in women. Am J Med. 2003;114:438–444. doi: 10.1016/s0002-9343(03)00061-5. [DOI] [PubMed] [Google Scholar]
- 36.Haffner SM, et al. Disproportionately increased proinsulin levels are associated with the insulin resistance syndrome. J Clin Endocrinol Metab. 1994;79:1806–1810. doi: 10.1210/jcem.79.6.7989488. [DOI] [PubMed] [Google Scholar]
- 37.Mykkänen L, et al. Serum proinsulin levels are disproportionately increased in elderly prediabetic subjects. Diabetologia. 1995;38:1176–1182. doi: 10.1007/BF00422366. [DOI] [PubMed] [Google Scholar]
- 38.Kahn SE, et al. Proinsulin as a marker for the development of NIDDM in Japanese-American men. Diabetes. 1995;44:173–179. doi: 10.2337/diab.44.2.173. [DOI] [PubMed] [Google Scholar]
- 39.Seaquist ER, et al. Hyperproinsulinemia is associated with increased β cell demand after hemipancreatectomy in humans. J Clin Invest. 1996;97:455–460. doi: 10.1172/JCI118435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alarcón C, Leahy JL, Schuppin GT, Rhodes CJ. Increased secretory demand rather than a defect in the proinsulin conversion mechanism causes hyperproinsulinemia in a glucose-infusion rat model of non-insulin-dependent diabetes mellitus. J Clin Invest. 1995;95:1032–1039. doi: 10.1172/JCI117748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marchetti P, et al. The endoplasmic reticulum in pancreatic β cells of type 2 diabetes patients. Diabetologia. 2007;50:2486–2494. doi: 10.1007/s00125-007-0816-8. [DOI] [PubMed] [Google Scholar]
- 42.Laybutt DR, et al. Endoplasmic reticulum stress contributes to β cell apoptosis in type 2 diabetes. Diabetologia. 2007;50:752–763. doi: 10.1007/s00125-006-0590-z. [DOI] [PubMed] [Google Scholar]
- 43.Hotamisligil GS. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 2010;140:900–917. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim T, Gondré-Lewis MC, Arnaoutova I, Loh YP. Dense-core secretory granule biogenesis. Physiology (Bethesda) 2006;21:124–133. doi: 10.1152/physiol.00043.2005. [DOI] [PubMed] [Google Scholar]
- 45.Cawley NX, et al. The carboxypeptidase E knockout mouse exhibits endocrinological and behavioral deficits. Endocrinology. 2004;145:5807–5819. doi: 10.1210/en.2004-0847. [DOI] [PubMed] [Google Scholar]
- 46.Hetz C, et al. Unfolded protein response transcription factor XBP-1 does not influence prion replication or pathogenesis. Proc Natl Acad Sci USA. 2008;105:757–762. doi: 10.1073/pnas.0711094105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hayashi A, et al. The role of brain-derived neurotrophic factor (BDNF)-induced XBP1 splicing during brain development. J Biol Chem. 2007;282:34525–34534. doi: 10.1074/jbc.M704300200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.