Abstract
The phosphorylation of H2Ax on its S139 site, γH2Ax, is important during DNA double-strand repair and is considered necessary for assembly of repair complexes, but its functional role after other kinds of DNA damage is less clear. We have measured the survival of isogenic mouse cell lines with the H2Ax gene knocked out, and replaced with wild-type or mutant (S139A) H2Ax genes, exposed to a range of agents with varied mechanisms of DNA damage. Knockout and mutant cells were sensitive to γ-rays, etoposide, temozolamide, and endogenously generated reactive oxygen species, each of which can include double-strand breaks among their spectra of DNA lesions. The absence or mutation of H2Ax had no influence on sensitivity to cisplatin or mitomycin C. Although UV light induced the highest levels of γH2Ax, mutation of S139 had no influence on UV sensitivity or the UV DNA damage response. Complete loss of H2Ax reduced the survival of cells exposed to UV light and reduced pChk1 induction, suggesting that sites other than S139 may impact the ATR-pChk1 pathway. The relative intensity of γH2Ax measured in Western blots in wild-type cells did not correlate with the functional importance of γH2Ax. The use of γH2Ax as a general biomarker of DNA damage is therefore potentially misleading because it is not an unambiguous indicator of double-strand breaks, and a significant fraction of DNA repair, especially involving nucleotide excision or crosslink repair, can occur without functional involvement of γH2Ax.
Keywords: alkylation, rotenone, caffeine, poly(ADP-ribose) polymerase, veliparib
Phosphorylation of the minor histone H2Ax (γH2Ax) has been a prominent feature of recent research on the mechanism of DNA double-strand break (DSB) repair (1, 2). Phosphorylation is carried out by members of the phosphatidylinositol 3-kinase–related kinase (PI3KK) family that includes ATM, ATR, and DNA PKcs. The ATM kinase is activated either by upstream binding of the Mre11/Rad50/Nbs1 complex to DNA breaks (3) or by reactive oxygen (4) and is required for downloading many components of the repair mechanisms (1, 2). Mice lacking H2Ax are prone to cancer, and fibroblasts derived from these mice are sensitive to X-rays and fail to repair DSBs (5). The formation of γH2Ax is therefore functionally important for executing the DSB DNA damage response (DDR).
Less clear is the functional role of γH2Ax during nucleotide excision repair (NER) and arrest of DNA replication following large adduct damage, such as UV light-induced photoproducts and DNA crosslinks where DSBs are presumably not initially formed (6–11). Several distinct distributions of γH2Ax occur in UV damaged cells (12–17). A low level nuclear-wide formation of γH2Ax occurs as a consequence of NER (12, 18, 19). A focal distribution of γH2Ax occurs at arrested replication forks coincident with PCNA and Mre11 (14–17). High-intensity nuclear-wide γH2Ax occurs in association with S-phase apoptosis (17). Despite these close associations of γH2Ax formation with the UV-DDR, none of these experiments directly address the functional importance of γH2Ax.
We established an isogenic set of mouse cell lines in which H2Ax is deleted and replaced with a wild-type gene or mutant H2Ax (S139A) gene that could not be phosphorylated on the major phosphorylation site, serine 139 (2). These cell lines (knockout, wild-type, and mutant) were used to determine the functional importance of γH2Ax for cell survival following exposure to γ-rays, endogenous sources of reactive oxygen (ROS), etoposide, alkylation, UV light, cisplatin, and crosslink damage. We conclude that γH2Ax is only important during repair of DSBs or ROS, and has very limited functional importance during the DDR after UV, cisplatin, and crosslink damage where DSBs are not the major lesion, and that the quantity of γH2Ax generated has little relation to its functional importance. Our observations imply that other sites in H2Ax may play a role in the UV-DDR, but that S139 has a negligible role.
Results
We first confirmed that only the cells expressing the wild-type gene, but not knockout and mutant genes, would phosphorylate the S139 site after γ-rays and UV irradiation (Fig. 1). The FLAG tag on the vector confirmed the presence of the transfected wild-type and mutant genes (Fig. 1A). Expression of the DSB binding protein 53BP1 that localizes to DSBs was unaffected by loss of γH2Ax (Fig. 1B). We then determined the survival of cells after exposure to a range of DNA damaging agents (Figs. 2and 3, and Table 1). The H2Ax knockout and mutant cells were sensitive to killing by γ-rays, as expected (Fig. 2A). The γH2Ax knockout and mutant cells were sensitive to etoposide (Fig. 2B) (20–22), to ROS generated from the mitochondria by rotenone exposure (Fig. 2C) (23), and to the alkylating agent temozolamide (Fig. 3A) (24, 25).
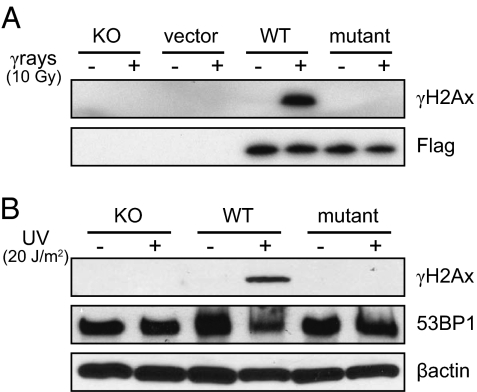
Fig. 1.
Expression of γH2Ax formation is restricted to cells transfected with wild-type H2Ax. (A) γ-Rays (10 Gy, 30 min); (B) UV (20 J⋅m−2, 4 h) cells. The Flag epitope indicates the presence of the H2Ax vector in both wild-type and mutant cell lines. 53BP1 is expressed in each cell type as this is a DSB binding protein that is constitutive but relocates to DSBs, and is used as an independent marker.
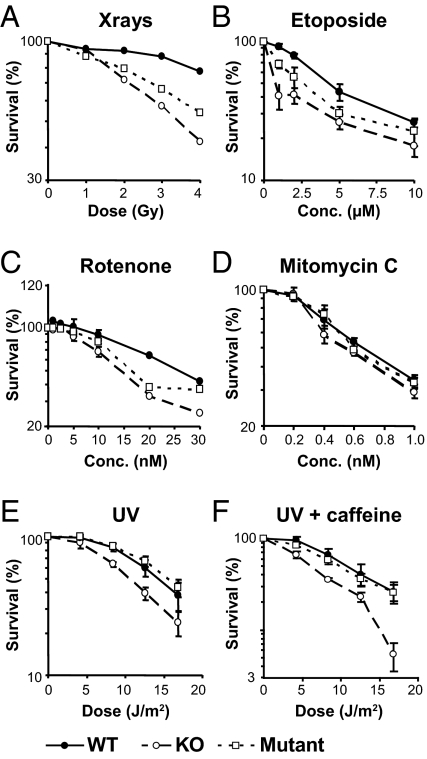
Fig. 2.
Survival of knockout, mutant, and wild-type cells after exposure to γ-rays, UV light, and various chemicals (●, wild-type; □, mutant; ○, knockout) (A) γ-Rays; (B) etoposide; (C) rotenone; (D) mitomycin C; (E) UV grown in medium without additions; (F) UV grown with caffeine (1 mM). All agents were assayed at least in triplicate (see legend to Fig. 3), except γ-rays that were only assayed once to confirm that the knockout and mutant cells showed the known relevance of γH2Ax for ionizing radiation.
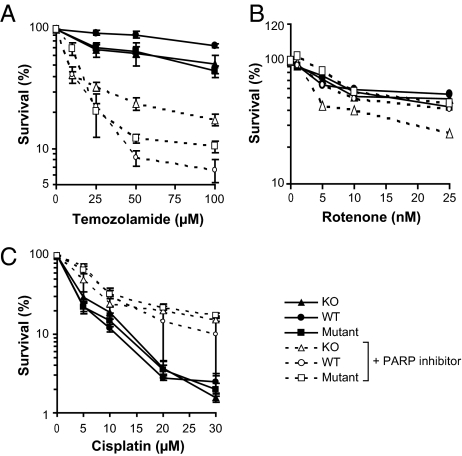
Fig. 3.
Survival curves for cells exposed to (A) temozolamide, (B) rotenone, and (C) cisplatin in the presence or absence of the PARP-1 inhibitor veliparib (50 μM). Note the dose range for rotenone is much lower than used in Fig 2 and does not detect the sensitivity of knockout cells. Circles, wild type H2Ax; triangles, H2Ax knockout; squares, mutant. Solid symbols and lines are cells grown without inhibitor; open symbols and dashed lines are cells grown in veliparib.
Table 1.
Agents used and main mechanisms of action
| Agent | (LD50) in H2Ax WT | Main mechanism of action | Main repair pathways* |
| Cs137 (γ-rays) | 10 Gy | MLDSs† | NHEJ, HR, SSB repair, BER |
| Etoposide | 10 μM | Topoisomerase inhibitor (DSBs in S phase) | NHEJ, HR |
| Rotenone | 25 nM | Mitochondrial complex I inhibitor (MLDSs†) | NHEJ, HR, SSB repair, BER |
| Cisplatin | 5 μM | G-G intrastrand crosslinks | NER |
| UV light | 16 J⋅m−2 | T = T, [6-4] photoproducts | NER |
| UV light (caffeine) | 11 J⋅m−2 | T = T, [6-4] photoproducts, Inhibition of ATR/pChk1 | NER |
| Mitomycin C | 0.5 μM | DNA-DNA interstrand crosslinks | Crosslink repair |
| Temozolamide | 200 μM | Alkylation (N7, O6), DSBs‡ | BER, AGMT, DSB repair‡ |
*The pathways listed are those mainly responsible for the classes of damage produced by each agent. NHEJ, nonhomologous endjoining; HR, homologous recombination; SSB repair, single strand break repair; NER, nucleotide excision repair; BER, base excision repair; AGMT, O6alkyl transferase.
†MLDS, multiple locally damaged sites that include SSBs, DSBs, apurinic (AP) sites, and base damage caused by external ionizing (γ) radiation or endogenously generated ROS from mitochondrial poisoning.
‡Recent work has shown that a fraction of the AP sites generated during BER can react in nucleosomes to produce DSBs (25).
Survival of knockout or mutant cells exposed to mitomycin C (Fig. 2D) or cisplatin (Fig. 3C) was not different from wild-type. Survival of mutant cells following UV damage, either grown with or without caffeine that inhibits the ATR-Chk1 pathway (26, 27), was also not different from wild-type. The survival of knockout cells was, however, reduced after UV to similar extents with or without caffeine (Fig. 2 E and F). UV mainly induced γH2Ax during the S phase in the wild-type mouse cells, and was increased by growth in caffeine, as previously reported for human cells (Fig. S1) (17).
To analyze the role of base excision repair (BER), we determined cell survival following ROS and temozolamide exposure in the presence of the inhibitor of poly(ADP ribose) polymerase (PARP-1) veliparib (Fig. 3 A and B). PARP-1 is an important component of BER (28). Inhibition of PARP-1 increased cell killing by temozolamide, consistent with the role of BER in repairing alkylation damage (Fig. 3A). The LD50 in the presence of veliparib was reduced to 15 to 20 μM, a 10-fold decrease (Table 1), in contrast to a reduction by only a factor of 2 when H2Ax was knocked out (Fig. 3A). A PARP-1–dependent BER is therefore a major pathway in cells damaged by temozolamide, and γH2Ax is required for repair of a smaller fraction of the total damage (25). Inhibiting PARP-1 slightly sensitized knockout but not mutant cells to low doses of rotenone (Fig. 3B) (2, 29). Cells exposed to cisplatin were not sensitized by veliparib, but showed an increased survival (Fig. 3C).
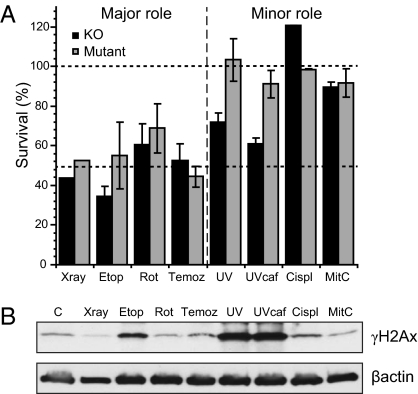
The survival curves were used to calculate the relative dependence on H2Ax for each DNA damaging agent based on the ratios of the doses required to reduce survival by 50% (LD50) in knockout or mutant cells compared with wild-type (Fig. 4A). Two classes of agents could be discerned: those for which H2Ax played a major role in survival and those for which H2Ax played a minor or insignificant role (Fig. 4A). The total quantity of γH2Ax induced 4 h after the start of exposure to LD50 doses in Western blots for cells that contained wild-type H2Ax showed no consistent correlation with the importance of H2Ax for survival (Fig. 4B). Within the group of agents for which H2Ax played a major role in cell survival (Fig. 4A), detectable levels of γH2Ax could only detected at this dose level for etoposide (Fig. 4B). The γ-ray–induced foci contributed little to net increases in total γH2Ax. Within the group of agents for which H2Ax played only a minor role (Fig. 4A), higher levels of γH2Ax were detected after UV than for any agent in which H2Ax had a major role, and low levels after cisplatin and mitomycin C (Fig. 4B). The increased cell killing by UV in knockout cells suggests that other phosphorylation sites (e.g., Y142) or ubiquitylation sites are more important than S139 for UV sensitivity (Fig. 4A) (30).
Fig. 4.
Relative sensitivity of H2Ax mouse knockout (black histograms) or mutant (gray histograms) following exposure to γ-rays, etoposide, rotenone, temozolamide, UV, cisplatin, and mitomycin C. (A) The doses required to kill 50% of the population (LD50) were recorded from multiple repeats of the survival curves in Figs. 2 and 4 [γ-rays n = 1 (Fig. 2A); Etop n-3; Rot n = 3; Temoz n = 6; UV n = 6; UV plus caffeine n = 6; Cispt n = 3; MitC n = 3] and means and SEs calculated. Lower dashed line indicates the relative sensitivity when the damage is predominantly double strand breaks, as for γ-rays and etoposide. The upper dashed line at 100% indicates the absence of any sensitization from inactivation of γH2Ax. (B) Western analysis of the levels of γH2Ax induced in cells with wild-type H2Ax 4 h after the start of exposure to an LD50 dose of each designated agent or 30 min in the case of γ-rays. Abbreviations: Etop, etoposide; Rot, rotenone; Temoz, temozolamide; Cispl,cisplatin; Mit C, mitomycin C.
The reduced survival of knockout cells to UV (Fig. 4A) could represent either a small modulation of many branches of the UV DDR, or a major inhibition of one branch. We therefore examined [6-4] photoproduct excision, apoptosis, cell cycle arrest, and Rad51 induction and found no major effects resulting from loss or mutation of H2Ax (Figs. S2–S4). The recovery of RNA synthesis after UV proceeded to similar extents in wild-type, knockout and mutant cells, whereas RNA synthesis in Cs-b cells continued to decline because of defects in transcription-coupled repair (Fig. S2C). The induction of pChk1 by UV exposure was reduced by caffeine to a greater extent in knockout cells, than in wild-type or mutant cells (Fig. S3) suggesting a role for H2Ax but not the S139 site in the ATR-pChk1 pathway (27).
Discussion
Histones are extensively modified by phosphorylation and ubiquitylation in response to DNA damage via the PI3KK class of kinases (1, 2, 31, 32). ATM and ATR phosphorylate over 700 protein targets during the DNA damage response (33), although not all targets are likely to be equally important. The minor histone H2Ax is an intensively studied target phosphorylated by ATM, ATR, and other kinases on several sites near the C-terminal end of the protein. Phosphorylation of serine 139 has been shown to play a major role in recruiting repair and recombination proteins to facilitate homologous and nonhomologous DSB repair (1). In yeast, analogous modifications increase sensitivity to X-rays, ROS, and UV light (32). The functional role of H2Ax modifications in mammalian cells is, however, less clear in response to agents that do not form DSBs directly. Because a large class of DNA damaging agents, including UV light, chemotherapy agents, and carcinogens make covalent modifications to DNA that only result in DNA breakage indirectly during repair or at blocked forks, we investigated the functional role of H2Ax in cells exposed to a range of these DNA damaging agents.
The survival of cells exposed to various agents indicates that H2Ax has a major role during repair of damage that includes DSBs but only a minor role, if any role at all, during BER, NER, or crosslink repair (Figs. 2–4 and Table 1). The levels of γH2Ax induced in normal cells, however, bore little relationship to the relative importance of H2Ax for cell survival (Fig. 4B) and each category of agent had some unique features. Survival of H2Ax knockout and mutant cells was decreased following γ-rays, etoposide, temozolamide, and rotenone-induced ROS (Figs. 2 and 3). Etoposide is an inhibitor of topoisomerase II and causes DSBs during the S phase (20–22). We have previously shown that after etoposide exposure, γH2Ax coincides with 53BP1, another marker of DSBs in human cells (17). Temozolamide is an alkylating agent used in treating advanced melanoma that forms adducts to N7 and O6 of guanine, requiring BER and O6-alkyltransferase for repair (24). BER transiently generates abasic sites that can be converted to DSBs in chromatin (25). The reduced survival of H2Ax knockout and mutant cells exposed to rotenone indicates that poisoning of mitochondrial complex I (23) generates sufficient ROS to reach the nucleus, activate ATM kinase, and cause DNA damage that requires H2Ax for survival. The damage from ROS and γ-rays is likely to be a mixture of single-strand breaks, DSBs, and oxidative damage to DNA bases (Table 1). The sensitivity of cells exposed to rotenone and temozolamide to inhibition of PARP-1 shows that BER is involved in repair of alkylation damage to a greater extent than for ROS (Fig. 3 A and B). The reduced survival of knockout cells compared with mutant H2Ax to ROS (Fig. 3B) also indicates that sites other than S139 can be important (2, 29).
In contrast to the major role of H2Ax in response to damage that may include DSBs, there was little sensitivity to UV, cisplatin, or mitomycin C (Figs. 2 D–F, 3C, and 4A). The S139A mutation conferred no UV sensitivity (Figs. 2E and 4A), despite UV being the agent that showed the highest levels of γH2Ax (Fig. 4B). Complete loss of H2Ax did confer an intermediate level of sensitivity, indicating a role for sites other than S139 (Fig. 4A) (2, 29). We further examined whether the reduced survival of H2Ax knockout cells to UV light was because of a small impact throughout the UV-DDR, or a specific effect on one pathway, but found no major impacts (Figs. S2–S4). These results are consistent with our previous demonstration that few DSBs are generated by UV damage during replication arrest or apoptosis, based on the low frequency of coincident foci containing both γH2Ax and 53BP1 (17).
Several lines of evidence suggest a minor role for H2Ax during the ATR-pCHK1 recovery pathway that depends on other sites than S139. Knockout but not mutant cells showed a reduced UV LD50 (Fig. 4A) and a reduced induction of pChk1 by UV when ATR was inhibited by caffeine (Fig. S3). This finding suggests a minor role for sites other than S139 that is revealed when major pathways are compromised. This role could also involve DSBs if the fraction of arrested replication forks that break in the absence of the ATR-pChk1 recovery pathway is increased (34). Whether replication fork breakage is a passive process or enzymatically induced is at present unknown, but similar studies in cells exposed to cross-linking agents implicated the 5′ NER nuclease (ERCC1/XPF) or Fanconi's anemia nucleases in such breakage (35).
Given the demonstration of extensive S139 H2Ax phosphorylation in UV damaged cells (Figs. 1B and 4B, and Fig. S1), it remains enigmatic that H2Ax may simply be a passive phosphorylation substrate without any function. Abnormal DNA structures associated with damage, repair, and replication arrest could expose the C-terminal tail of H2Ax to adventitious phosphorylation by the PI3KK family irrespective of whether every phosphorylation is subsequently important (33). Only the additional presence of DSBs would then allow repair factors transiently bound to γH2Ax to remain and execute repair. Conversely, when cells are exposed to agents where there is little independent evidence for the presence of DSBs, the formation and quantity of γH2Ax provides no evidence for the presence of DSBs. For example, exposure of cells to an inhibitor of histone deacetylase produced γH2Ax that could merely represent phosphorylation associated with altered histone conformation and not DSBs (36, 37). Observation of extensive γH2Ax formation during germ cell maturation also cannot be used as evidence for DSBs in the absence of other independent measurements, and therefore needs to be reevaluated (38).
There are several implications of this study. (i) Although γH2Ax is a biomarker of DNA damage, indicating the activation of damage-dependent kinases, it is only a functional component of a restricted subset of repair processes. (ii) Neither the amount nor the distribution of γH2Ax as foci or nuclear-wide staining is a necessary indication of its functional role. (iii) The function of γH2Ax may be restricted to DSBs, but conversely, its formation does not always indicate the presence of DSBs. (iv) The use of γH2Ax as a general biomarker of DNA damage and repair is potentially misleading because it is not a consistent quantifiable measure (39).
Materials and Methods
Cell Lines.
The mouse cell line lacking H2Ax (knockout) was derived from a mouse knockout strain (5) and generously provided by A. Nussenzweig (National Cancer Institute, Bethesda, MD) and derivatives were grown in 25 μg/mL blasticidin. Cockayne B (Cs-b) mutant mouse embryo fibroblasts were kindly donated by V. Bohr (National Institute on Aging, Baltimore) and used as a positive control for defective transcription-coupled repair (Fig. S2). All cells were cultured in Eagle's minimal essential media with Earle's balanced salt solution, supplemented with 10% FBS, 10 μg/mL streptomycin, 10 units/mL penicillin, and 0.292 g/L glutamine.
Vector Construction.
Human H2Ax cDNA was cloned into a gateway pENTR vector (Invitrogen) with a Flag-tag at the N terminus. A nonphosphorylatable variant of H2Ax with serine139 replaced by alanine139 (S139A) was generated using PCR-directed mutagenesis. Constructs were sequenced, and transferred into a Gateway-modified pWZL-blasticidin retrovirus destination vector. To obtain retrovirus stocks, Phoenix A cells were transfected with six micrograms of pWZL, pWZL-H2Ax or pWZL-H2Ax-S139A vectors by lipofection (Fugene; Roche). pWZL amphotropic viruses were collected 2 d after transfection and used to infect the H2Ax knockout cell line. Cultures arose from polyclonal expansion of infected cells grown in blasticidin (25 μg/mL; Invitrogen) medium for 1 wk. The cell line corrected by wild-type H2Ax was designated as “wild-type”; the cell line transfected with the S139A mutant was designated “mutant” for this article.
Cell Survival.
Cell survival was determined from cells irradiated or exposed continuously to DNA damaging agents by measuring colorimetric activation of MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; Sigma-Aldrich] at 5 to 7 d. The doses for 50% survival (LD50) were calculated from multiple independent individual survival curves (see captions for number) and the ratios of LD50 values for knockout and mutant cell lines relative to wild-type were calculated, from which the mean and SEMs were calculated.
Exposures.
Cells were irradiated with UV light (254 nm, 1.3 J⋅m−2⋅s) or Cs137 γ-rays (2.4 Gy⋅min−1). Cells were exposed continuously to a range of concentrations of various agents listed in Table 1 (SI Materials and Methods). Cells were harvested at 4 h for γH2Ax analysis, 20 h for apoptosis, or 5 to 7 d for survival.
Photoproduct Analysis.
Levels of genomic [6-4] photoproducts ([6-4]PP) in UV irradiated cells were measured using a slot blot immunoassay using monoclonal antibodies to [6-4]PP (64M-2) and CPDs (MBL, Inc.). The recovery of RNA synthesis was determined by labeling cells with 0.02 mCi/mL [methyl-14C]-thymidine for 24 h, then washed with PBS, irradiated with UV, and allowed to incubate in medium for 0, 4, 8, or 24 h following which they were incubated for 0.5 h with 1 mCi/mL [5,6-3H]-uridine and scintillation counted for 3H and 14C. Results are plotted as the 3H/14C ratio, normalized to unirradiated cells.
Western Blots.
Western blots were used to determine protein concentrations as described previously (17) using PARP-1 (p85) (Trevigen), β-actin (Upstate), pCHK1(p317), γH2Ax (S139), and 53BP1 (Cell Signaling) antibodies.
Flow Cytometry.
Flow cytometry was carried out using the H2Ax phosphorylation assay kit (Upstate Biotechnology) and analyzed using a FACS Caliber Flow Cytometer (Becton Dickinson) equipped with Cell Quest. Data were analyzed by using both Cell Quest and FloJo software (Tree Star).
Apoptosis.
Cell death was quantified on the basis of cell detachment from the culture surface, and confirmed by detection of the PARP-1 85-kDa apoptotic marker in floating cells (Trevigen) (17, 40). The percentage of apoptotic cells released from the substrate was calculated as the number of detached cells divided by the total number of attached plus detached cells 20 h after doses of 10 J⋅m−2.
Immunofluorescence.
γH2Ax and Rad 51 were measured using immunofluorescence microscopy as described previously (17). Images were acquired at room temperature with a Zeiss Axioplan 2 microscope equipped with a 100× Zeiss Plan-Neofluar 1.3 Oil objective lens and a Zeiss Axiocam color camera under the control of Axiovision 4.2 software. Images used for comparison between different treatments or cell lines were acquired with the same instrument settings and exposure time and were processed equally without recourse to any image processing software. Quantification was carried out using ImageJ software.
Supplementary Material
Acknowledgments
This work was supported by grants from the University of California Cancer Research Coordinating Committee and the Lily Drake Cancer Research Fund at the University of San Francisco, and generous donations from the Mt. Zion Health Fund, the Xeroderma Pigmentosum Society of New York State, the Xeroderma Pigmentosum Family Support Group of California, and the Luke O'Brien Foundation (Cockayne syndrome). Work by T.K.D. and D.H.O. was supported by National Cancer Institute Grant CA105958 and a Veterans Affairs Award (to D.H.O.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1105866108/-/DCSupplemental.
References
- 1.Lowndes NF, Toh GW-L. DNA repair: The importance of phosphorylating histone H2AX. Curr Biol. 2005;15:R99–R102. doi: 10.1016/j.cub.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 2.Pinto DM, Flaus A. Structure and function of histone H2AX. Subcell Biochem. 2010;50:55–78. doi: 10.1007/978-90-481-3471-7_4. [DOI] [PubMed] [Google Scholar]
- 3.Jazayeri A, Balestrini A, Garner E, Haber JE, Costanzo V. Mre11-Rad50-Nbs1-dependent processing of DNA breaks generates oligonucleotides that stimulate ATM activity. EMBO J. 2008;27:1953–1962. doi: 10.1038/emboj.2008.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo Z, Kozlov S, Lavin MF, Person MD, Paull TT. ATM activation by oxidative stress. Science. 2010;330:517–521. doi: 10.1126/science.1192912. [DOI] [PubMed] [Google Scholar]
- 5.Celeste A, et al. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnes L, et al. GammaH2AX, an accurate marker that analyzes UV genotoxic effects on human keratinocytes and on human skin. Photochem Photobiol. 2010;86:933–941. doi: 10.1111/j.1751-1097.2010.00744.x. [DOI] [PubMed] [Google Scholar]
- 7.Zhao H, Traganos F, Darzynkiewicz Z. Kinetics of the UV-induced DNA damage response in relation to cell cycle phase. Correlation with DNA replication. Cytometry A. 2010;77:285–293. doi: 10.1002/cyto.a.20839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yajima H, Lee KJ, Zhang S, Kobayashi J, Chen BP. DNA double-strand break formation upon UV-induced replication stress activates ATM and DNA-PKcs kinases. J Mol Biol. 2009;385:800–810. doi: 10.1016/j.jmb.2008.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stiff T, et al. ATR-dependent phosphorylation and activation of ATM in response to UV treatment or replication fork stalling. EMBO J. 2006;25:5775–5782. doi: 10.1038/sj.emboj.7601446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halicka HD, et al. Histone H2AX phosphorylation after cell irradiation with UV-B: Relationship to cell cycle phase and induction of apoptosis. Cell Cycle. 2005;4:339–345. [PubMed] [Google Scholar]
- 11.Ward IM, Minn K, Chen J. UV-induced ataxia-telangiectasia-mutated and Rad3-related (ATR) activation requires replication stress. J Biol Chem. 2004;279:9677–9680. doi: 10.1074/jbc.C300554200. [DOI] [PubMed] [Google Scholar]
- 12.Marti TM, Hefner E, Feeney L, Natale V, Cleaver JE. H2AX phosphorylation within the G1 phase after UV irradiation depends on nucleotide excision repair and not DNA double-strand breaks. Proc Natl Acad Sci USA. 2006;103:9891–9896. doi: 10.1073/pnas.0603779103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu C, et al. Cell apoptosis: Requirement of H2AX in DNA ladder formation, but not for the activation of caspase-3. Mol Cell. 2006;23:121–132. doi: 10.1016/j.molcel.2006.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Limoli CL, Giedzinski E, Morgan WF, Cleaver JE. Polymerase eta deficiency in the xeroderma pigmentosum variant uncovers an overlap between the S phase checkpoint and double strand break repair. Proc Natl Acad Sci USA. 2000;97:7939–7946. doi: 10.1073/pnas.130182897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Limoli CL, Giedzinski E, Bonner WM, Cleaver JE. UV-induced replication arrest in the xeroderma pigmentosum variant leads to DNA double-strand breaks, gamma-H2AX formation, and Mre11 relocalization. Proc Natl Acad Sci USA. 2002;99:233–238. doi: 10.1073/pnas.231611798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Limoli CL, Giedzinski E, Cleaver JE. Alternative recombination pathways in UV-irradiated XP variant cells. Oncogene. 2005;24:3708–3714. doi: 10.1038/sj.onc.1208515. [DOI] [PubMed] [Google Scholar]
- 17.de Feraudy S, Revet I, Bezrookove V, Feeney L, Cleaver JE. A minority of foci or pan-nuclear apoptotic staining of gammaH2AX in the S phase after UV damage contain DNA double-strand breaks. Proc Natl Acad Sci USA. 2010;107:6870–6875. doi: 10.1073/pnas.1002175107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanasoge S, Ljungman M. H2AX phosphorylation after UV irradiation is triggered by DNA repair intermediates and is mediated by the ATR kinase. Carcinogenesis. 2007;28:2298–2304. doi: 10.1093/carcin/bgm157. [DOI] [PubMed] [Google Scholar]
- 19.Matsumoto M, et al. Perturbed gap-filling synthesis in nucleotide excision repair causes histone H2AX phosphorylation in human quiescent cells. J Cell Sci. 2007;120:1104–1112. doi: 10.1242/jcs.03391. [DOI] [PubMed] [Google Scholar]
- 20.Muslimović A, Nyström S, Gao Y, Hammarsten O. Numerical analysis of etoposide induced DNA breaks. PLoS ONE. 2009;4:e5859. doi: 10.1371/journal.pone.0005859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donà F, et al. Loss of histone H2AX increases sensitivity of immortalized mouse fibroblasts to the topoisomerase II inhibitor etoposide. Int J Oncol. 2008;33:613–621. [PubMed] [Google Scholar]
- 22.Smart DJ, et al. Assessment of DNA double-strand breaks and gammaH2AX induced by the topoisomerase II poisons etoposide and mitoxantrone. Mutat Res. 2008;641:43–47. doi: 10.1016/j.mrfmmm.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: A dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newlands ES, Stevens MF, Wedge SR, Wheelhouse RT, Brock C. Temozolomide: A review of its discovery, chemical properties, pre-clinical development and clinical trials. Cancer Treat Rev. 1997;23:35–61. doi: 10.1016/s0305-7372(97)90019-0. [DOI] [PubMed] [Google Scholar]
- 25.Sczepanski JT, Wong RS, McKnight JN, Bowman GD, Greenberg MM. Rapid DNA-protein cross-linking and strand scission by an abasic site in a nucleosome core particle. Proc Natl Acad Sci USA. 2010;107:22475–22480. doi: 10.1073/pnas.1012860108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarkaria JN, et al. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 1999;59:4375–4382. [PubMed] [Google Scholar]
- 27.Heffernan TP, et al. ATR-Chk1 pathway inhibition promotes apoptosis after UV treatment in primary human keratinocytes: potential basis for the UV protective effects of caffeine. J Invest Dermatol. 2009;129:1805–1815. doi: 10.1038/jid.2008.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masaoka A, Horton JK, Beard WA, Wilson SH. DNA polymerase beta and PARP activities in base excision repair in living cells. DNA Repair (Amst) 2009;8:1290–1299. doi: 10.1016/j.dnarep.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cook PJ, et al. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature. 2009;458:591–596. doi: 10.1038/nature07849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bergink S, et al. DNA damage triggers nucleotide excision repair-dependent monoubiquitylation of histone H2A. Genes Dev. 2006;20:1343–1352. doi: 10.1101/gad.373706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore JD, Krebs JE. Histone modifications and DNA double-strand break repair. Biochem Cell Biol. 2004;82:446–452. doi: 10.1139/o04-034. [DOI] [PubMed] [Google Scholar]
- 32.Moore JD, Yazgan O, Ataian Y, Krebs JE. Diverse roles for histone H2A modifications in DNA damage response pathways in yeast. Genetics. 2007;176:15–25. doi: 10.1534/genetics.106.063792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 34.Squires S, et al. p53 prevents the accumulation of double-strand DNA breaks at stalled-replication forks induced by UV in human cells. Cell Cycle. 2004;3:1543–1557. doi: 10.4161/cc.3.12.1272. [DOI] [PubMed] [Google Scholar]
- 35.Kratz K, et al. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell. 2010;142:77–88. doi: 10.1016/j.cell.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 36.Lee JH, Choy ML, Ngo L, Foster SS, Marks PA. Histone deacetylase inhibitor induces DNA damage, which normal but not transformed cells can repair. Proc Natl Acad Sci USA. 2010;107:14639–14644. doi: 10.1073/pnas.1008522107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Namdar M, Perez G, Ngo L, Marks PA. Selective inhibition of histone deacetylase 6 (HDAC6) induces DNA damage and sensitizes transformed cells to anticancer agents. Proc Natl Acad Sci USA. 2010;107:20003–20008. doi: 10.1073/pnas.1013754107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wossidlo M, et al. Dynamic link of DNA demethylation, DNA strand breaks and repair in mouse zygotes. EMBO J. 2010;29:1877–1888. doi: 10.1038/emboj.2010.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watters GP, Smart DJ, Harvey JS, Austin CA. H2AX phosphorylation as a genotoxicity endpoint. Mutat Res. 2009;679:50–58. doi: 10.1016/j.mrgentox.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 40.Cleaver JE, et al. Polymerase eta and p53 jointly regulate cell survival, apoptosis and Mre11 recombination during S phase checkpoint arrest after UV irradiation. DNA Repair (Amst) 2002;1:41–57. doi: 10.1016/s1568-7864(01)00004-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.