Abstract
General transcription factor IIH (TFIIH) is a complex RNA polymerase II basal transcription factor comprising 10 different polypeptides that display activities involved in transcription and DNA repair processes. Although biochemical studies have uncovered TFIIH importance, little is known about how the mRNAs that code for TFIIH subunits are regulated. Here it is shown that mRNAs encoding seven of the TFIIH subunits (p34, p44, p52, p62, XPB, CDK7, and p8) are regulated at the posttranscriptional level in a Dicer-dependent manner. Indeed, abolition of the miRNA pathway induces abnormal accumulation, stabilization, and translational activation of these seven mRNAs. Herein, miR-27a was identified as a key regulator of p44 mRNA. Moreover, miR-27a was shown to destabilize the p44 subunit of the TFIIH complex during the G2-M phase, thereby modulating the transcriptional shutdown observed during this transition. This work is unique in providing a demonstration of global transcriptional regulation through the action of a single miRNA.
Keywords: G2-M transition, cell-cycle, mitosis
To maintain cellular homeostasis, cells have evolved a myriad of specialized pathways, among which basal transcription and DNA repair play crucial roles. Both mechanisms are mainly regulated by the multisubunit complex termed TFIIH (general transcription factor IIH) formed by two modules, the core and the CAK [cyclin-dependent kinase (CDK)-activated kinase], each displaying distinguishable activities and differential protein composition (1–3). Whereas the core is constituted by seven polypeptides (p34, p44, p52, p62, XPB, XPD, and p8) and is indispensable for transcription and DNA repair (4), the CAK contains only three proteins (CDK7, CycH, and MAT1) and is required to modulate transcription by phosphorylating the carboxyl terminal domain of RNA polymerase II (5). Moreover, the CAK module regulates cell-cycle progression by phosphorylating CDKs during G2-M transition in an autonomous fashion (6).
Mutations in different TFIIH subunits lead to severe clinical disorders that range from mental retardation to cancer predisposition. This predisposition is manifested in patients displaying three clinically well-characterized syndromes—Xeroderma Pigmentosum, Cockaine syndrome, and Trichothiodystrophy—highlighting the importance of maintaining TFIIH integrity (7–9). Lessons learned from mutations encountered on patients helped to unravel which activities of TFIIH were modified in different clinical contexts. However, it has not been possible either to link specific mutations with a particular syndrome or with clinical severity (10–12). Nevertheless, all three clinical anomalies are taught, in principle, to be caused by diminished transcriptional and DNA repair activities often associated with reduced TFIIH stability. In vitro-obtained data show that naturally occurring mutations in XPD or p44 that alter the XPD/p44 or p44/p62 interaction may affect the composition of TFIIH by decreasing levels of XPD and CAK subunits associated with the core. On the other hand, mutations in XPB and p52 may prevent XPB anchoring to the core, ultimately leading to reduced amounts of core TFIIH (13).
The undoubted role of TFIIH as a transcription complex and the fact that transcription needs to be severely down-regulated during G2-M to resume cell-cycle progression, led to hypothesize that TFIIH might have a role in this regulatory event. Indeed, several reports have attempted to address whether TFIIH regulates the transcriptional shutdown observed during this transition (14, 15). In that regard, it was shown in Drosophila that XPD expression is down-regulated during G2-M with a concomitant increase in TFIIH-independent CAK activity, thus promoting mitotic progression (16). Because the absence of any core TFIIH subunit will irremediably lead to a transcriptional defect, we thus hypothesize that modulation of the TFIIH-coding mRNA at the expression/translation levels could modulate the transcriptional landscape of the cell through cell-cycle progression, specifically regulating the G2-M phase.
Herein we describe that 7 of 10 mRNAs that code for TFIIH subunits are regulated at the posttranscriptional level by the miRNA pathway. Importantly, we demonstrate that a cell cycle-dependent miRNA regulation of the p44 coding mRNA by miR-27a modulates the transcriptional shutdown observed during G2-M transition.
Results
TFIIH-Coding mRNAs Are Regulated at the Posttranscriptional Level in a Dicer-Dependent Manner.
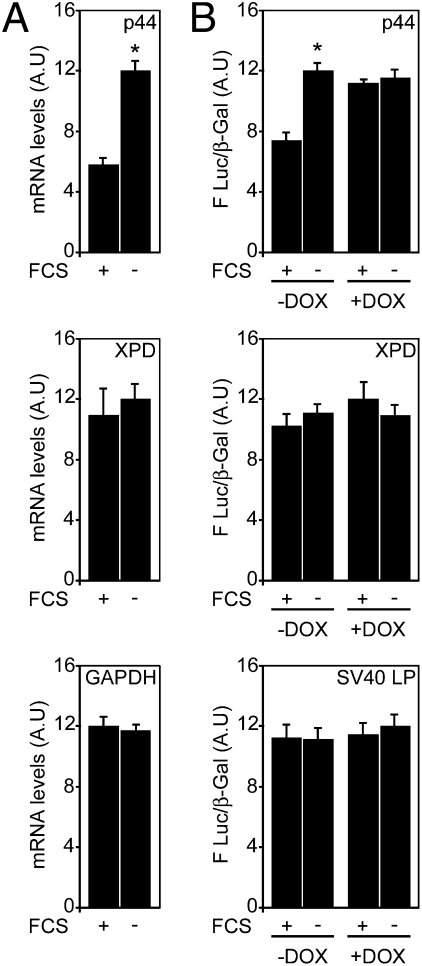
To address whether the levels of any TFIIH-coding mRNAs might be modulated, a screening was performed. For the screening, 293 cells were grown in normal (+FCS) or serum-depleted medium (−FCS) and mRNA levels of all subunits were measured by RT-qPCR. The level of two mRNAs remain unchanged (XPD, CycH), one was down-regulated (MAT1), and seven (p34, p44, p52, p62, XPB, CDK7, p8) were increased upon serum deprivation (Fig. 1A and Figs. S1 and S2).
Fig. 1.
The mRNA that codes for the p44 subunit of TFIIH is posttranscriptionally regulated. (A) 293 cells were subjected or not to serum deprivation (+FCS and −FCS). mRNA levels were measured by RT-qPCR. XPD subunit and GAPDH are shown as controls. Results are presented as arbitrary units (AU) and are expressed as the mean of three experiments performed in triplicate ± SD of mRNA accumulation. The higher value obtained for each subunit was arbitrarily normalized to 12. *P < 0.05 as determined by Student's t test. (B) 293-Dicer cells were transfected with 3′UTR reporter plasmids for p44 or XPD and subjected or not to serum deprivation (+FCS and −FCS). Dicer dependency was assessed by addition of doxycyclin (+DOX) that triggers the expression of a shRNA targeting Dicer mRNA. Results are expressed as arbitrary units representing the mean ± SD of Luc activity/β-galactosidase activity; n = 3 experiments performed in triplicate. *P < 0.05 as determined by Student's t test.
The stability of mRNA is often regulated by elements located in their 3′ untranslated regions (3′UTR) (17–19). In that regard, miRNAs have emerged as key regulators of mRNA translation and decay. These 21- to 24-nucleotide long single-stranded RNAs are generated from sequential processing of primary-miRNA transcripts by Drosha and Dicer (20–22) and integrated into the RNA-induced silencing complex (RISC). Once into the RISC, miRNAs bind the 3′UTR of target mRNAs to suppress translation or direct their degradation (23–25). To address whether the regulation of the mRNA levels of the different TFIIH subunits was dependent on their 3′UTR sequences, luciferase (Luc) reporter plasmids bearing the 3′UTR of each of the seven TFIIH subunits were constructed and transfected into 293-Dicer cells genetically modified to develop a conditional knock-down of Dicer (293-Dicer) in the presence of doxycyclin (DOX) (Fig. S3) (26). We observed that the luciferase activity of the seven reporters increased upon serum deprivation in cells that were not treated with DOX (Fig. 1B and Fig. S4). Conversely, results obtained upon DOX addition (+DOX) indicate that the miRNA machinery is required to maintain the expression levels under normal growth conditions, whereas this posttranscriptional control is abolished in serum-deprived cells (Fig. 1B and Fig. S4).
miR-27a Is a Regulator of p44 mRNA.
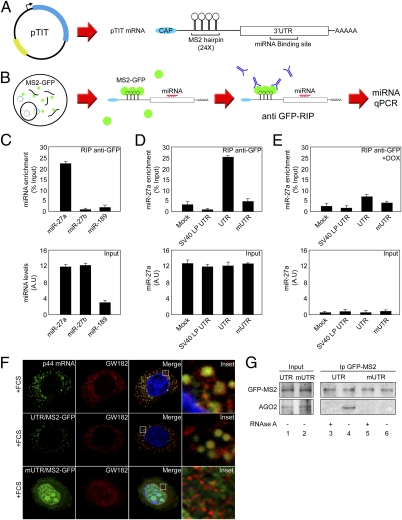
Our results indicate that the posttranscriptional fate of several TFIIH subunits is regulated by their 3′UTR and is controlled by a Dicer-dependent mechanism. Prediction algorithms of miRNA target sites (miRGator) confirmed that these seven TFIIH-coding mRNAs are potentially targeted by miRNAs (Table S1). Because the p44 subunit plays a role in RNA pol II promoter-escape modulating transcription (27) and its coding mRNA presents only three predicted miRNA binding sites (miR-27a, miR-27b, miR-189), we focused our study on p44 mRNA. To experimentally identify miRNAs regulating p44 mRNA translation/stability, we used a method that allows the identification of miRNAs binding to a particular 3′UTR in vivo (28) and further validated the interaction by intracellular localization/interaction with the RISC complex. The method is based on the ability of the bacteriophage MS2-coat protein (MS2) to bind with an MS2-RNA repetitive sequence (29, 30). A reporter system (pTIT) containing the MS2-RNA repetition (24×) followed by p44 3′UTR under the control of an SV40 promoter was generated (Fig. 2A) and transfected into a stable cell line derived from 293-Dicer cells expressing the MS2-GFP fusion protein. Transfected cells express the MS2-RNA repetition followed by p44-3′UTR (Fig. 2A), which interacts with the MS2-GFP protein produced by the host cell (Fig. 2B). Immunopurification of the MS2-GFP/MS2-RNA complex by GFP-immunoprecipitation allowed the identification of the coimmunoprecipitated miRNAs interacting with p44-3′UTR (RT-qPCR). In normal growth conditions, only miR-27a—but neither miR-27b nor miR-189—interacts with p44 WT 3′UTR (UTR) (Fig. 2C and Fig. S5). This binding was abolished in reporters bearing either an unrelated 3′UTR (SV40 LP) or a p44 3′UTR mutated in the miR-27a binding site (mUTR) (Fig. S5), confirming the specificity of the miR-27a-p44 3′UTR interaction (Fig. 2D). Moreover, in the absence of miRNAs no interaction was detected (Fig. 2E). Furthermore, endogenous p44 mRNA as well as the reporter containing the WT p44 3′UTR (UTR) colocalize with integral components of p-bodies, including GW182, DCP-1, and AGO-2 (31, 32) (Fig. 2F, Top and Middle, and Fig. S6), whereas the mutated reporter (mUTR) is excluded (Fig. 2F, Bottom). These data indicate that the subcellular localization of the endogenous p44 mRNA is under the control of its 3′UTR through its interaction with miR-27a.
Fig. 2.
Identification of miRNAs that bind to p44 3′UTR in vivo. (A) Schematic representation of pTIT reporter system and its expressed mRNA. (B) Scheme representing miRNA identification workflow using pTIT reporter system. (C) 293-Dicer cells expressing the MS2-GFP fusion protein were transfected with pTIT-UTR, subjected to RIP with an anti-GFP antibody, and miRNA detection was assessed by RT-qPCR. Results are represented as fold-enrichment. miRNA input (27a, 27b, and 189) are depicted as control for RIP assay. (D) 293-Dicer cells expressing MS2-GFP were mock-treated or transfected with pTIT wt 3′UTR (UTR), pTIT seed region mutant 3′UTR (mUTR) or pTIT unrelated 3′UTR (SV40LP UTR) and subjected to RIP. miRNA detection was performed by RT-qPCR. Results are represented as miR-27a enrichment. miR-27a input levels are depicted as control. (E) 293-Dicer cells expressing MS2-GFP were treated with DOX, transfected as in D, and subjected to RIP. miRNA detection was performed by RT-qPCR. Results are represented as miR-27a enrichment. miR-27a input levels are depicted. (F) HeLA cells were processed for RNA-FISH against p44 mRNA (green channel) or cotransfected with an MS2-GFP expressing plasmid with pTIT-UTR or pTIT-mUTR. MS2-GFP labels pTIT produced mRNAs (green channel) and p-bodies; localization is analyzed by GW182 staining (red channel). Merged and enlarged (Inset) images are depicted. (G) 293-Dicer cells expressing the MS2-GFP were transfected with pTIT-UTR or pTIT-mUTR and used for immunoprecipitation with anti-GFP antibody. Precipitates were immunoblotted with anti-GFP (Upper) to reveal the MS2-GFP fusion protein and with anti-AGO2 (Lower). Lysates were subjected or not to RNAse A treatment (± RNase A) to analyze the RNA dependency of the interaction.
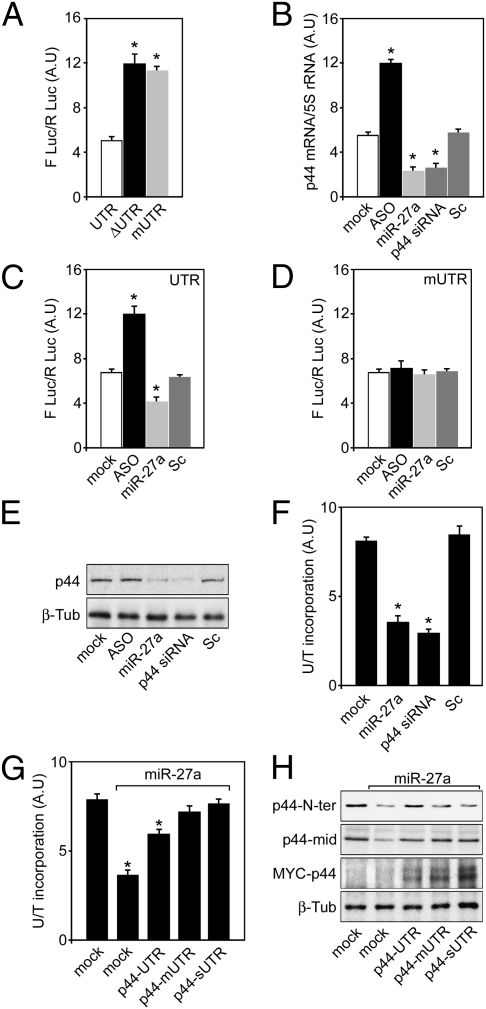
To further characterize miR-27a/p44 3′UTR interaction, MS2-GFP obtained from cells transfected with pTIT-UTR or pTIT-mUTR was immunoprecipitated. Whereas AGO2 is recruited to the reporter bearing the p44 WT-3′UTR (Fig. 2G, lane 4), this was abolished in p44 mUTR-reporters (Fig. 2G, lane 6) and when RNase A was added to the immunocomplexes, indicating an RNA-dependent mechanism (Fig. 2G, compare lanes 3 and 5). To confirm the specificity of the miR-27a-p44 3′UTR interaction, three luciferase reporters containing either the p44 WT-3′UTR (UTR), a p44 3′UTR deletion mutant that lacks the first 53 nt eliminating the miR-27a binding site (ΔUTR) or a reporter mutated within the miR-27a seed region (mUTR) were constructed (Fig. S5). Reporters were transfected in 293-Dicer cells and Luc activity was measured. The abolition miR-27a binding site (Fig. 3A, lane 2) and its mutation in the seed region (Fig. 3A, lane 3) resulted in increased Luc activity, reflecting a higher translation efficiency of these reporters compared with the WT 3′UTR (Fig. 3A, lane 1). This finding confirms the specificity of the miR-27a-p44 3′UTR interaction and emphasizes the requirement of a functional target sequence in the 3′UTR to achieve full translational control of p44.
Fig. 3.
miR-27a regulates basal transcription by targeting p44 mRNA. (A) 293-Dicer cells were transfected with Firefly luciferase 3′UTR reporters bearing p44 3′UTR (UTR), p44 Δ3′UTR (ΔUTR), or p44 m3′UTR (mUTR) and Firefly luciferase activity was measured. Results are plotted as arbitrary units (A.U) representing the mean ± SD of Firefly luciferase activity/Renilla luciferase activity; n = 3 experiments performed in triplicate. *P < 0.05 as determined by Student's t test. (B) 293-Dicer cells were transfected with an antisense for miR-27a (ASO), a mimic for miR-27a (miR-27a), a siRNA against p44 mRNA (p44 siRNA), or a scrambled oligonucleotide (Sc) and the p44 mRNA was measured by RT-qPCR. Results are expressed as arbitrary units representing the mean ± SD of p44 mRNA/5S rRNA; n = 3 experiments performed in triplicate. *P < 0.05 as determined by Student's t test. (C) 293-Dicer cells were transfected with a p44 3′UTR Firefly luciferase reporter and either ASO, miR-27a, or Sc respectively and Firefly luciferase activity was determined. Results are expressed as arbitrary units representing the mean ± SD of Firefly luciferase/Renilla luciferase activity; n = 3 experiments performed in triplicate. *P < 0.05 as determined by Student's t test. (D) 293-Dicer cells were transfected with a p44 3′mUTR Firefly luciferase reporter and either ASO, miR-27a, or Sc respectively and Firefly luciferase activity was determined. Results are expressed as in C. (E) 293-Dicer cells were transfected with miR-27a antisense (ASO), miR-27a mimic (miR-27a), p44 siRNA, or scrambled (Sc) oligonucleotides. Cell extracts were processed for Western-blot for p44. β-Tubulin is shown as control. (F) 293-Dicer cells were transfected with ASO, miR-27a, p44 siRNA or Sc and general transcription was assessed by 3H-Uridine incorporation to RNA. Radiolabeled RNA was normalized to 14C-Thymidine incorporated to DNA before the 3H-Uridine pulse. Results are expressed as arbitrary units representing the mean ± SD of 3H-Uridine/14C-Thymidine; n = 3 experiments performed in triplicate. *P < 0.05 as determined by Student's t test. (G) 293-Dicer cells were cotransfected with miR-27a and either (MYC) p44-UTR, p44-mUTR or p44-sUTR and general transcription was assessed as in F. Results are expressed as arbitrary units representing the mean ± SD of 3H-Uridine/14C-Thymidine; n = 3 experiments performed in triplicate. *P < 0.05 as determined by Student's t test. (H) 293-Dicer cells were cotransfected with miR-27a and either (MYC) p44-UTR, p44-mUTR, or p44-sUTR and subjected to Western blot against p44 (N-terminal and central region antibodies) and MYC-p44 (MYC epitope). Note that the N-terminal p44 antibody recognizes only endogenous p44 (N-terminal MYC epitope masking), whereas the antibody that recognizes a central region of p44 detects both species. β-Tubulin is shown as control.
The next goal was to investigate the functional consequences of miR-27a-p44 mRNA interaction. Therefore, an antisense oligonucleotide that blocks miR-27a (ASO) was used and endogenous p44 mRNA levels were measured (Fig. 3B). Our results confirmed that inactivating miR-27a with ASO results in the accumulation of endogenous p44 mRNA (Fig. 3B, lane 2). Conversely, transfection of a miR-27a mimic reduces p44 mRNA levels (miR-27a) (Fig. 3B, lane 3). Interestingly, a siRNA directed against p44 mRNA (Fig. 3B, lane 4) reduces p44 levels to the same extent than miR-27a (Fig. 3B, lanes 3 and 4). Neither mock (Fig. 3B, lane 1) nor scrambled transfected oligonucleotides (Fig. 3B, lane 5) display any difference. Moreover, cotransfection of p44-3′UTR Luc reporter with miR-27a ASO increases translation of the reporter (Fig. 3C, lane 2), whereas cotransfection with a mimic reduces p44 translational rate (miR-27a) (Fig. 3C lane 3). No differences were observed in mock- (Fig. 3C, lane 1) or scrambled-treated samples (Fig. 3C, lane 4). Moreover, none of these treatments display any difference relative to mock- or scrambled-treated cells when the reporter carrying the p44 3′UTR was mutated at the miR-27a binding site (mUTR) (Fig. 3D). Western blot against the endogenous p44 subunit was performed in cells treated with ASO, miR-27a, p44 siRNA, or Sc. As expected, cells transfected with miR-27a or p44 siRNA display reduced amounts of p44 respect to mock- or Sc-treated cells. Taken together, our results demonstrate that miR-27a mediates the destabilization of p44 mRNA and controls its translation and stability. Intriguingly, ASO treatment resulted in a very mild increase of p44 protein, suggesting that the regulation of this subunit by the endogenous miR-27a should take place in a minority of the cell population (Fig. 3E).
miR-27a Modulates Basal Transcription by Targeting the p44 Coding mRNA.
We then investigated whether the regulation of p44 by miR-27a had a functional consequence on transcriptional activity. The introduction of miR-27a mimic led to a 60% reduction of total 3H-Uridine incorporation into RNA compared with control cells (Fig. 3F), indicating that reduced amounts of p44 are sufficient to induce a transcriptional shutdown, underscoring the biological relevance of our findings. We previously showed that p44 mUTR and sUTR are insensitive to the presence/activity of miR-27a, whereas the WT 3′UTR is regulated by this miRNA (Figs. 2 and 3). To confirm that miR-27a action over p44 mRNA was responsible for the transcriptional shutdown observed, rescue experiments were performed using an N-terminal MYC epitope tagged-p44 overexpressing vector where p44 coding mRNA was controlled by UTR, mUTR, or synthetic sUTR. The capacity of these vectors to rescue the transcriptional shutdown observed upon miR-27a transfection was assessed. For that, the transcriptional activity in 293-Dicer cells cotransfected with miR-27a and either with p44-UTR, p44-mUTR, or p44-sUTR was measured. Whereas overexpression of p44-UTR only partially rescued the transcriptional shutdown induced by miR-27a (Fig. 3H), overexpression of p44-mUTR and p44-sUTR substantially rescue transcription (Fig. 3G), indicating that only p44 mRNA that cannot be targeted by miR-27a restores transcription to normal levels, demonstrating that p44 is the key target of miR-27a.
miR-27a Interacts and Destabilizes p44 Coding mRNA During G2-M.
During the G2-M phase, transcription is severely down-regulated. Interestingly, miR-27a has been recently implicated in the control of G2-M transition in transformed cells (33). Our observation that miR-27a dramatically reduces transcription by destabilizing p44 prompted us to analyze whether miR-27a-dependent destabilization of p44 plays a role in the transcriptional shutdown associated to G2-M. For that analysis, 293-Dicer cells were synchronized in G2-M by nocodazole treatment and miR-27a and p44 mRNA/protein levels were measured at different time points upon nocodazole release (Fig. 4 A–C, and Figs. S7 and S8A). After 2 h, cells entering mitosis showed a high level of miR-27a that correlated with a low level of p44 mRNA and p44 protein. At later time points, miR-27a levels decayed and p44 mRNA and protein levels concomitantly increased. To confirm the direct interaction of p44 3′UTR with miR-27a during G2-M, 293-Dicer cells expressing the MS2-GFP were transfected with pTIT-UTR reporter (Fig. 3 A and B) and synchronized with nocodazole (Fig. S7). After release, the MS2-GFP was immunoprecipitated and the levels of MS2-p44 UTR mRNA interacting with miR-27a (RT-qPCR) were measured. The miR-27a level was maximum in immunoprecipitates obtained from cells harvested at the moment of release (Fig. 4D, 0 h), whereas the amount of MS2-p44 UTR polyadenylated mRNA was the minimum observed (Fig. 4E, 0 h). Moreover, the levels of interacting miR-27a detected diminished through cell-cycle progression and MS2-p44 UTR polyadenylated mRNA concomitantly increased (Fig. 4 D and E). This finding suggested that after nocodazole release, an MS2-p44 UTR derived molecule that cannot be detected by oligo dT priming (Fig. 4E, 0 h) interacts with miR-27a (Fig. 4D, 0 h). Thus, total MS2-p44 UTR RNA was determined by performing reverse transcription using random primers and qPCR. Results showed that deadenylated p44 RNA was predominant at the moment of release (Fig. 4F, 0 h) and diminishes during cell-cycle progression (Fig. 4F). Taken together, these data demonstrate that during G2-M phase transition p44 3′UTR interacts with miR-27a, and suggests that p44 mRNA degradation might occur through a deadenylation process.
Fig. 4.
miR-27a targets p44 3′UTR during G2-M transition and induces transcriptional shutdown. (A) 293-Dicer cells were synchronized with nocodazole in G2-M and released from drug treatment. Total RNA was collected at indicated time points and miR-27a was measured by RT-qPCR. Data were normalized to U49 snoRNA. (B) 293-Dicer cells were treated as described in A, total RNA was collected at indicated time points and p44 mRNA was determined by RT-qPCR. Obtained values were normalized as in A. (C) 293-Dicer cells were treated as in A. Samples were collected at indicated time points and processed for Western blot against p44 and β-tubulin. (D) 293-Dicer cells expressing MS2-GFP were transfected with pTIT-UTR, synchronized as in A and released from drug treatment. Samples obtained at indicated time points were subjected to RIP and miR-27a detection was performed by RT-qPCR. Results are represented as arbitrary units (A.U). (E) 293-Dicer cells expressing MS2-GFP were treated as described in D and level of MS2 p44 3′UTR polyadenylated mRNA was determined by RT-qPCR. (F) 293-Dicer cells expressing the MS2-GFP fusion protein were treated as described in D and the level of total MS2 p44 3′UTR RNA was assessed by RT-qPCR. (G) U49 snoRNA was measured as a control in the input. Results are expressed as arbitrary units. (H) 293-Dicer cells were synchronized with aphidicolin in G1-S boundary, released from aphidicolin block and transfected either with ASO or Sc oligonucleotides. Twelve hours after release, cells were harvested and p44 content was analyzed by Western blot. Cycling cells were treated in parallel as control. Actin was included as loading control. (I) 293-Dicer cells were transfected with miR-27a antisense (ASO) or scrambled (Sc) oligonucleotides and pulsed with BrU, fixed, and stained with propidium iodide. G2-M population was determined by FACS analysis. Upper and lower limits of BrU incorporation in the population of cells form mock-treated samples was determined as the decile 95% (red punctuated line). (J) Percentage of cells in G2-M of the 293-Dicer cells analyzed in H are depicted as a table. Results are represented as the percentage of cells in G2-M ± SD of n = 2 experiments performed in duplicate.
miR-27a/p44 Mediated Transcriptional Shut Down Is Necessary for Cell-Cycle Progression.
As previously shown (Fig. 3E), only slight differences were observed in ASO-treated cycling cells, indicating a narrow regulatory window for endogenous miR-27a to operate over p44. Results displayed in Fig. 4 A to G suggested that the effect of endogenous miR-27a in regulating p44 occurs mainly during G2-M, which corresponds with ∼22% of cells in a cycling population (Fig. S9A). To confirm that miR-27a regulates p44 through G2-M transition, 293-Dicer cells were synchronized with aphidicolin in G1-S boundary, released, and transfected either with ASO or Sc oligonucleotides. Twelve hours after release, cells were harvested (∼70% of the cells were in G2-M) (Fig. S9B) and p44 content was analyzed by Western blot. In these G2-M enriched samples, an evident increment of p44 protein was observed in ASO treated cells compared with mock- or Sc-treated controls (Fig. 4H and Fig. S8B). Taken together, these results demonstrate that miR-27a regulates p44 protein level at the G2-M transition.
To analyze the involvement of endogenous miR-27a in regulating global transcription during G2-M, a cycling population of 293-Dicer cells was transfected with ASO or Sc oligonucleotides, pulsed with BrU to determine its incorporation to nascent RNA by immunodetection, and stained with propidium iodide for cell-cycle analysis. The percentage of cells at each phase of the cell cycle was determined by FACS. The population of cells at G2-M phase was selected and the intensity of fluorescence corresponding to BrU incorporation was measured. Results show that impeding endogenous miR-27a action with ASO causes an increment in p44 protein levels (Fig. 4H) and a higher transcriptional activity (Fig. 4I). Indeed, 43% of the cell population in ASO-treated cells display higher levels of BrU incorporation relative to mock- or Sc-treated cells (Fig. 4I). Moreover, an abnormal accumulation of cells in G2-M (Fig. 4J) is observed upon ASO treatment. All in all, our data indicate that miR-27a mediated p44 down-regulation is required for normal cell-cycle progression. Whether the posttranscriptional regulation of the other six TFIIH subunits shown to be modulated by the miRNA pathway (Figs. S1 and S3) is also participating in this process remains to be elucidated.
Discussion
In eukaryotic cells, mitosis is accompanied by a global repression of transcription caused by inactivation of the transcriptional machinery (14, 15). In this regard, abrogation of TFIIH transcriptional activity achieved by inhibition of the CAK-associated RNA pol II carboxyl terminal domain phosphorylation occurs during the G2-M phase. Although it has been generally accepted that the CAK module should dissociate from the core to achieve mitotic inhibition, the mechanism that modulates TFIIH disassembly has remained elusive. Herein we described that several TFIIH subunits are regulated at the posttranscriptional level by the miRNA pathway. Particularly, we demonstrate that miR-27a directly binds and regulates the stability of the mRNA that codes for the p44 during G2-M transition, providing a control mechanism relying on the relative abundance of core subunits in G2-M boundaries. In our model, as in Drosophila (16), down-regulation of a particular subunit could result into core-CAK dissociation, thus reducing global transcriptional activity. As an alternative, we provide evidence indicating that miR-27a could tether the abundance of core TFIIH by modulating the availability of p44, ultimately regulating transcription. As we have learned from the clinic, many mutations lead to the loss of core TFIIH, ultimately resulting in transcription/DNA repair defects (7–12). Moreover, it has been shown that in a recombinant reconstituted TFIIH in vitro transcriptional system, the lack of any core subunit impairs transcription, thus supporting a model were the absence of any core protein could modulate the transcriptional landscape of the cell (34). Therefore, it is fair to postulate that in patients displaying Xeroderma Pigmentosum, Cockaine syndrome, and Trichothiodystrophy symptoms where no mutations have been allocated, regulatory miRNAs/transcription factors modulating the expression levels of TFIIH subunits could provide an explanation. Furthermore, many features of previously mentioned syndromes (i.e., chromosomal instability, cancer) can be explained by problems associated to chromosome segregation and mitosis, which might result from improper p44 down-regulation. In that regard, the overexpression of miR-27a in several cancer types has been proposed to promote oncogenic features, supporting that improper transcriptional shutdown/inefficient DNA repair might lead to improper resolution of mitosis and cancer (32, 35–37).
It is important to bear in mind that although we identified miR-27a as a regulatory molecule for TFIIH through p44 down-regulation, six additional TFIIH coding mRNAs are also regulated by the miRNA pathway. Thus, miRNAs targeting TFIIH-coding mRNAs could, in principle, modulate the availability of different subunits favoring the formation of differential TFIIH-derived subcomplexes. Supporting the notion that different subcomplexes coexist, it has been recently shown that the XPD subunit is part of a different complex, termed MMXD (38), involved in chromosomal segregation. In addition, it has been shown that members of the proteosome pathway interact with particular CAK components regulating its degradation (39). Therefore, to address whether different TFIIH-derived complexes exist naturally, to unravel their biological function, and to assess whether miRNA or proteosome pathways cooperate for the regulation of TFIIH in vivo are major goals to achieve.
Materials and Methods
Cell Culture and Dicer Knockdown
293-Dicer cells (Dicer kd-2b2) were grown and Dicer knockdown was induced as described in ref. 28) and in SI Materials and Methods. MS2-GFP stable cell line generation and RNA immunoprecipitation protocols are available in SI Materials and Methods.
miRNA Detection, Mimicry, Knockdown, siRNA, and Cell Synchronization.
Both miR-27a and miR-27b levels were determined by reverse transcription followed by qPCR with LNA-specific primers (EXIQON). Mimicry for miR-27a, siRNA (smartpool-p44 mRNA) and ASO (miR-27a) molecules were used at 10 nM final concentration (Dharmacon). Cell synchronization procedures are described in SI Materials and Methods.
Transcription Assays.
293-Dicer cells were plated at low density in the presence of 2.5 μL/mL of medium of 14C-Thymidine (50 μCi; Perkin-Elmer) and were grown for 3 d. After radioactive medium removal, cells were mock-treated or transfected according to experimental design. Sixteen hours after transfection, cells were pulsed with 10 μL/mL of 3H-Uridine (1 mCi; Perkin-Elmer) for 1 h and processed to determine radioactive incorporation to RNA/DNA.
BrU Pulse and FACS Analysis.
Transfected 293-Dicer cells were pulsed with 2.5 mM BrU (Sigma) for 30 min, methanol-fixed, and incubated overnight with anti-BrDU antibody (Santa Cruz) in blocking buffer at 4 °C (SI Materials and Methods). Samples were washed with ice-cold 10 mM PBS and incubated with a secondary antibody Alexa 488 conjugated (Molecular Probes) for 3 h at room temperature, washed again, and incubated in 10 mM PBS supplemented with propidium iodide at 50 μM final concentration overnight at 4 °C. Cell-cycle analysis and BrU incorporation were assessed by FACS (FACScan).
Supplementary Material
Acknowledgments
I thank H. Gronemeyer and colleagues from the Institut de Génétique et de Biologie Moléculaire et Cellulaire for helpful discussions and critical reading of the manuscript; M. Lieb for expert technical assistance in FACS analysis; and H. Gronemeyer and J. M. Egly for financial support. 293-Dicer cells were a generous gift from W. Filipowicz; AGO2-GFP and PA-Dcp1 expressing plasmids were a gift from A. Leung and P. Sharp. This study was supported by funds from Centre National de la Recherche Scientifique, Institut National de la Santé et de la Recherche Médicale, La Ligue Contre le Cancer, and the French National Research Agency. M.M.P. was a recipient of a fellowship from the Fondation pour la Recherche Medicale.
Footnotes
The author declares no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1014018108/-/DCSupplemental.
References
- 1.Bradsher J, Coin F, Egly JM. Distinct roles for the helicases of TFIIH in transcript initiation and promoter escape. J Biol Chem. 2000;275:2532–2538. doi: 10.1074/jbc.275.4.2532. [DOI] [PubMed] [Google Scholar]
- 2.de Laat WL, Jaspers NG, Hoeijmakers JH. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 3.Schultz P, et al. Molecular structure of human TFIIH. Cell. 2000;102:599–607. doi: 10.1016/s0092-8674(00)00082-9. [DOI] [PubMed] [Google Scholar]
- 4.Zurita M, Merino C. The transcriptional complexity of the TFIIH complex. Trends Genet. 2003;19:578–584. doi: 10.1016/j.tig.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Serizawa H, et al. Association of Cdk-activating kinase subunits with transcription factor TFIIH. Nature. 1995;374:280–282. doi: 10.1038/374280a0. [DOI] [PubMed] [Google Scholar]
- 6.Nigg EA. Cyclin-dependent kinase 7: At the cross-roads of transcription, DNA repair and cell cycle control? Curr Opin Cell Biol. 1996;8:312–317. doi: 10.1016/s0955-0674(96)80003-2. [DOI] [PubMed] [Google Scholar]
- 7.Botta E, et al. Reduced level of the repair/transcription factor TFIIH in trichothiodystrophy. Hum Mol Genet. 2002;11:2919–2928. doi: 10.1093/hmg/11.23.2919. [DOI] [PubMed] [Google Scholar]
- 8.Kraemer KH, et al. Xeroderma pigmentosum, trichothiodystrophy and Cockayne syndrome: A complex genotype-phenotype relationship. Neuroscience. 2007;145:1388–1396. doi: 10.1016/j.neuroscience.2006.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lehmann AR. DNA repair-deficient diseases, xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. Biochimie. 2003;85:1101–1111. doi: 10.1016/j.biochi.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 10.Lehmann AR. XPD structure reveals its secrets. DNA Repair (Amst) 2008;7:1912–1915. doi: 10.1016/j.dnarep.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 11.Dubaele S, et al. Basal transcription defect discriminates between xeroderma pigmentosum and trichothiodystrophy in XPD patients. Mol Cell. 2003;11:1635–1646. doi: 10.1016/s1097-2765(03)00182-5. [DOI] [PubMed] [Google Scholar]
- 12.Winkler GS, et al. TFIIH with inactive XPD helicase functions in transcription initiation but is defective in DNA repair. J Biol Chem. 2000;275:4258–4266. doi: 10.1074/jbc.275.6.4258. [DOI] [PubMed] [Google Scholar]
- 13.Schärer OD. Hot topics in DNA repair: The molecular basis for different disease states caused by mutations in TFIIH and XPG. DNA Repair (Amst) 2008;7:339–344. doi: 10.1016/j.dnarep.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gottesfeld JM, Forbes DJ. Mitotic repression of the transcriptional machinery. Trends Biochem Sci. 1997;22:197–202. doi: 10.1016/s0968-0004(97)01045-1. [DOI] [PubMed] [Google Scholar]
- 15.Akoulitchev S, Reinberg D. The molecular mechanism of mitotic inhibition of TFIIH is mediated by phosphorylation of CDK7. Genes Dev. 1998;12:3541–3550. doi: 10.1101/gad.12.22.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J, Larochelle S, Li X, Suter B. Xpd/Ercc2 regulates CAK activity and mitotic progression. Nature. 2003;424:228–232. doi: 10.1038/nature01746. [DOI] [PubMed] [Google Scholar]
- 17.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol. 2007;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leung AK, Calabrese JM, Sharp PA. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc Natl Acad Sci USA. 2006;103:18125–18130. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomari Y, Zamore PD. MicroRNA biogenesis: Drosha can't cut it without a partner. Curr Biol. 2005;15:R61–R64. doi: 10.1016/j.cub.2004.12.057. [DOI] [PubMed] [Google Scholar]
- 21.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002;21:5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zamore PD. RNA interference: Listening to the sound of silence. Nat Struct Biol. 2001;8:746–750. doi: 10.1038/nsb0901-746. [DOI] [PubMed] [Google Scholar]
- 24.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 25.Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 2005;19:2837–2848. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmitter D, et al. Effects of Dicer and Argonaute down-regulation on mRNA levels in human HEK293 cells. Nucleic Acids Res. 2006;34:4801–4815. doi: 10.1093/nar/gkl646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tremeau-Bravard A, Perez C, Egly JM. A role of the C-terminal part of p44 in the promoter escape activity of transcription factor IIH. J Biol Chem. 2001;276:27693–27697. doi: 10.1074/jbc.M102457200. [DOI] [PubMed] [Google Scholar]
- 28.Kim HH, et al. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodriguez AJ, Shenoy SM, Singer RH, Condeelis J. Visualization of mRNA translation in living cells. J Cell Biol. 2006;175:67–76. doi: 10.1083/jcb.200512137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fusco D, et al. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr Biol. 2003;13:161–167. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: How many mechanisms? Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 32.Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mertens-Talcott SU, Chintharlapalli S, Li X, Safe S. The oncogenic microRNA-27a targets genes that regulate specificity protein transcription factors and the G2-M checkpoint in MDA-MB-231 breast cancer cells. Cancer Res. 2007;67:11001–11011. doi: 10.1158/0008-5472.CAN-07-2416. [DOI] [PubMed] [Google Scholar]
- 34.Tirode F, Busso D, Coin F, Egly JM. Reconstitution of the transcription factor TFIIH: Assignment of functions for the three enzymatic subunits, XPB, XPD, and cdk7. Mol Cell. 1999;3:87–95. doi: 10.1016/s1097-2765(00)80177-x. [DOI] [PubMed] [Google Scholar]
- 35.Ma Y, Yu S, Zhao W, Lu Z, Chen J. miR-27a regulates the growth, colony formation and migration of pancreatic cancer cells by targeting Sprouty2. Cancer Lett. 2010;298:150–158. doi: 10.1016/j.canlet.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 36.Li X, et al. MicroRNA-27a indirectly regulates estrogen receptor alpha expression and hormone responsiveness in MCF-7 breast cancer cells. Endocrinology. 2010;151:2462–2473. doi: 10.1210/en.2009-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun Q, et al. Hsa-mir-27a genetic variant contributes to gastric cancer susceptibility through affecting miR-27a and target gene expression. Cancer Sci. 101:2241–2247. doi: 10.1111/j.1349-7006.2010.01667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ito S, et al. MMXD, a TFIIH-independent XPD-MMS19 protein complex involved in chromosome segregation. Mol Cell. 2010;39:632–640. doi: 10.1016/j.molcel.2010.07.029. [DOI] [PubMed] [Google Scholar]
- 39.Sandrock B, Egly JM. A yeast four-hybrid system identifies Cdk-activating kinase as a regulator of the XPD helicase, a subunit of transcription factor IIH. J Biol Chem. 2001;276:35328–35333. doi: 10.1074/jbc.M105570200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.