Abstract
Common polymorphisms in complement alternative pathway (AP) proteins C3 (C3R102G), factor B (fBR32Q), and factor H (fHV62I) are associated with age-related macular degeneration (AMD) and other pathologies. Our published work showed that fBR32Q influences C3 convertase formation, whereas fHV62I affects factor I cofactor activity. Here we show how C3R102G (C3S/F) influences AP activity. In hemolysis assays, C3102G activated AP more efficiently (EC50 C3102G: 157 nM; C3102R: 191 nM; P < 0.0001). fB binding kinetics and convertase stability were identical, but native and recombinant fH bound more strongly to C3b102R (KD C3b102R: 1.0 μM; C3b102G: 1.4 μM; P < 0.0001). Accelerated decay was unaltered, but fH cofactor activity was reduced for C3b102G, favoring AP amplification. Combining disease “risk” variants (C3102G, fB32R, and fH62V) in add-back assays yielded sixfold higher hemolytic activity compared with “protective” variants (C3102R, fB32Q, and fH62I; P < 0.0001). These data introduce the concept of a functional complotype (combination of polymorphisms) defining complement activity in an individual, thereby influencing susceptibility to AP-driven disease.
Keywords: inflammation, infection
Complement plays crucial roles in clearance of pathogens and immune complexes (1), with alternative pathway (AP) “tickover” in plasma providing a rapid response (2). Assembly of the AP C3-cleaving enzyme (convertase) involves Mg2+-dependent binding of factor B (fB) to C3b, forming the labile proenzyme C3bB; factor D (fD) then cleaves fB to yield active convertase (C3bBb). Convertase-generated C3b forms more C3bBb, providing exponential AP amplification. C3b clustered around the convertase creates a C5-cleaving enzyme (C3bBbC3b), triggering formation of the cytolytic membrane attack complex (MAC).
Nascent C3b binds pathogens and host cells indiscriminately. To prevent damage to self, multiple regulatory proteins limit complement activation by inactivating C3b/C4b, dissociating the C3/C5 convertases, or inhibiting MAC formation. Decay accelerating factor (DAF) (CD55) dissociates convertases, whereas membrane cofactor protein (MCP) (CD46) is an essential cofactor for factor I (fI) cleavage of C3b (3). In plasma, AP amplification is controlled by factor H (fH), which rapidly dissociates the enzymatic Bb domain from the C3 convertase and catalyses fI cleavage of C3b (4).
Maintenance of complement homeostasis involves equilibrium between activation and control. AP dysregulation is associated with many diseases (5), either due to loss-of-function/expression mutations in regulators or gain-of-function mutations in components; both scenarios cause uncontrolled complement activation and inflammation (6–10). Common polymorphisms in AP components (C3 and fB) and regulators (fH) also link to disease; the fH polymorphism fHY402H (rs1061170) is risk for age-related macular degeneration (AMD) (11–14), whereas fH62I (rs800292) and fB32Q (rs641153) polymorphisms are protective (11, 15, 16). A common polymorphism in C3, originally identified from electrophoretic mobility and dubbed C3S/F for slow/fast migration (17) (C3R102G, rs2230199; allele frequency: C3102R [C3S]: 0.79, C3102G [C3F]: 0.21), was recently linked with AMD (odds ratio 2.6) (18, 19). Each polymorphism also links to other disorders. fH62I is protective in dense deposit disease (DDD) and atypical hemolytic uremic syndrome (aHUS) (15), whereas fB32Q is risk for infections (20) and overrepresented in autoimmunity (21, 22). C3R102G is associated with IgA nephropathy (23), systemic vasculitis (24), kidney allograft dysfunction (25), and DDD (26).
Understanding mechanisms underlying these genetic associations is essential to prove causality, eliminate association through linkage, and aid understanding of disease etiology. The fH402H risk variant had no direct effect on fH AP regulation but influenced binding to sialylated surfaces (27, 28). The AMD-protective variant, fB32Q, formed AP convertase less efficiently, whereas the protective variant fH62I bound C3b more strongly and was a better cofactor for fI inactivation (29). These latter polymorphisms directly influenced AP activity, explaining their link to disease. Effects were additive, “risk” combinations (fB32R/fH62V) caused twofold increased AP activation compared with “protective” variants (fB32Q/fH62I) (29). To define mechanisms underlying disease associations of the common C3R102G (C3S/F) polymorphism, proteins were purified from homozygote donors and tested for ligand binding, convertase formation, and regulation. The AMD risk variant (C3b102G) bound fH less well compared with C3b102R, causing decreased fI cofactor activity, extended convertase lifetime, and enhanced AP amplification. By combining risk and protective AP variants, a “complotype” was revealed in which C3, fH, and fB variants collaborate to set levels of AP activity in plasma, thereby influencing risk in complement-dependent diseases.
Results
Differential Activity of C3R102G Variants in Hemolysis Assays.
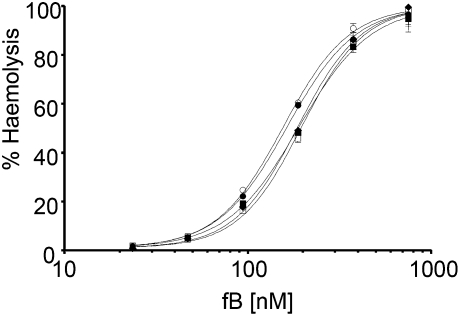
C3, from plasma of healthy individuals homozygous for C3102R (n = 3) or C3102G (n = 2) and free of hydrolyzed C3 and aggregates, was added back to normal human serum depleted of C3 (NHSΔC3) and used to deposit C3b and AP convertases on antibody-coated sheep erythrocytes (ShEA); lysis was developed using C3-supplemented methylamine-treated NHS (NHS-MA). NHSΔC3 supplemented with C3102R required 1.3-fold more fB to achieve lysis equivalence with C3102G-supplemented NHSΔC3, demonstrating that plasma containing C3102G (AMD risk) displayed higher AP activity (Fig. 1). EC50 (fB) values were internally consistent for different C3102G (158 nM and 156 nM), and C3102R (188 nM, 197 nM, and 187 nM) donors, and were significantly different (two-tailed, unpaired t test; P < 0.0001).
Fig. 1.
Hemolytic activity of the C3 variants C3102G and C3102R. C3 was purified from plasma of two donors homozygous for C3102G (circles) and three donors homozygous for C3102R (squares/diamond) and was added to C3-depleted serum to deposit C3b on ShEA. C3 convertase was formed and lysis was developed using NHS-MA supplemented with C3 of each variant. Data points represent mean ± SD of triplicate measurements. Curves were fitted using nonlinear regression to calculate the EC50.
Formation and Decay of C3b102G and C3b102R AP Convertases.
To explore molecular mechanisms underlying differential hemolytic activity, formation and natural decay of the AP C3 convertase was analyzed using surface plasmon resonance (SPR) (Biacore). AP C3 convertase formed on Biacore chips replicates natural convertase: it is labile, cleaves C3, deposits nascent C3b, and is regulated by decay accelerators (30, 31). Either fB alone, or fB and fD together, were flowed over each C3b variant. Neither proenzyme (C3bB; Fig. S1; KD C3b102G: 0.85 ± 0.10 μM (n = 5); KD C3b102R: 0.71 ± 0.15 μM (n = 5); two-tailed, unpaired t test; P = 0.11) nor convertase (C3bBb; Fig. S2; KD C3b102G: 67 ± 7 nM, n = 10; KD C3b102R: KD: 71 ± 8 nM, n = 18; two-tailed, unpaired t test; P = 0.23) formation differed significantly between variants. Both enzymes decayed at the same rate with calculated half-lives at 25 °C (ln2/kd) of 369 ± 35 s for C3b102G Bb and 385 ± 33 s for C3b102RBb (two-tailed, unpaired t test; P = 0.06). In multiple experiments using C3b102G and C3b102R from different donors, run on different days and chip surfaces, results were consistent. The fB32R variant was used in all analyses.
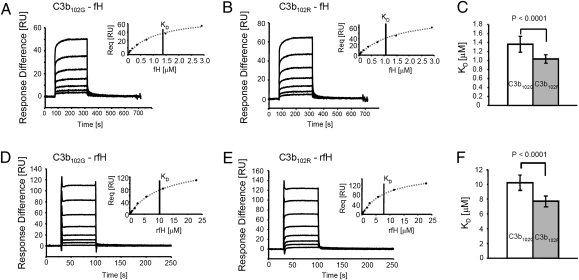
Binding of Convertase Regulators to C3bR102G Variants.
After convertase decay, residual C3b is a ligand for cofactors, fH and MCP, which catalyze cleavage/inactivation by fI. C3b interaction with regulators was assessed using SPR. Each variant C3b was coupled to the Biacore chip, and native fH flowed across (Fig. 2 A and B). Binding affinity (KD) of fH for C3b102G at steady-state equilibrium (Fig. 2 A and B, Inset), was 1.3-fold lower than for C3b102R (KD = 1.36 ± 0.17 μM, n = 17; KD = 1.03 ± 0.09 μM, n = 16; P < 0.0001; Fig. 2C). Analyses were reproduced with native fH from three different donors and repeated at least three times on different chips and days and with protein from two C3b102G and three C3b102R donors. The fH62V variant was used throughout these analyses. Binding of fH to immobilized C3(H2O)102R and C3(H2O)102G (hydrolyzed C3) was measured in identical experiments; measured affinities were reduced compared with C3b and were not significantly different for the variants [C3(H2O)102R, KD = 8.85 ± 0.64 μM; C3(H2O)102G, KD = 9.02 ± 0.13 μM; P = 0.727; Fig. S3].
Fig. 2.
Binding of native fH and recombinant fH to C3bR102G variants. Native fH (A and B) or rfH1-4 (D and E) were flowed over immobilized C3b102G (A and D) or C3b102R (B and E) at concentrations between 45 nM and 2.9 μM (fH) and 80 nM–23 μM (rfH1–4) and equilibrium binding response was measured. Affinity (KD) was determined by steady-state analysis (Inset). Multiple analyses on different days with different donors confirmed consistent and significant differences in native fH (C) or rfH1-4 (F) binding to the C3bR102G variants.
C3b binding analyses were repeated using recombinant fH comprising short consensus repeats (SCRs) 1-4 (rfH1–4) to investigate whether the observed affinity difference was due to differential binding of SCRs 1–4, containing fH decay and cofactor activities (32, 33). rfH1–4 bound C3b102G with 1.3-fold lower affinity than C3b102R (KD = 10.2 ± 1.0 μM, n = 5 versus KD = 7.7 ± 0.7 μM, n = 6; P < 0.0001; Fig. 2 E and F), replicating findings with native fH. Binding affinities of soluble recombinant MCP (sMCP) and soluble recombinant DAF (sDAF) for immobilized C3b variants were measured precisely as described for fH. There was no difference in C3b variant binding affinities of either sMCP (KD C3b102G = 1.48 ± 0.24 μM, n = 11; KD C3b102R = 1.43 ± 0.23 μM, n = 6; P = 0.68; Fig. S4 A–C) or sDAF (KD C3b102G 12.2 ± 1.7 μM, n = 19; C3b102R KD = 12.8 ± 1.2 μM, n = 10; P = 0.39; Fig. S4 D–F).
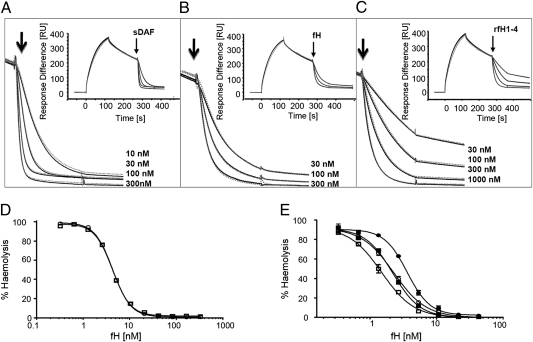
To test effects of C3R102G polymorphism on convertase accelerated decay, different concentrations of sDAF, native fH and rfH1–4 were flowed over C3b102GBb or C3b102R Bb-coated surfaces to catalyze rapid decay of Bb (30, 31) (Fig. 3 A–C). The convertases showed identical susceptibility to accelerated decay by each regulator. Cofactor activity for fI cleavage was not tractable using Biacore because mass change from C3b to iC3b (C3f release) was not measurable.
Fig. 3.
Regulatory activities of native fH, rfH1–4, and sDAF on surface-bound convertase formed from C3R102G variants. (A–C) Identical amounts (1,500 RU) of C3b102R (gray dotted line) and C3b102G (black solid line) were deposited on the chip surfaces using thioester coupling. Convertase was formed by flowing (20 μL/min) fB and fD in the presence of Mg2+ and allowed to decay naturally for 150 s. At the arrow, either (A) sDAF (10, 30, 100, and 300 nM; from Upper to Lower curve), (B) fH (30, 100, and 300 nM), or (C) rfH1–4 (30, 100, 300, and 1,000 nM) was injected (90 s) and accelerated decay of the convertase was monitored. Inset shows whole sensorgram; expanded decay curves are illustrated. (D) Decay accelerating activity of fH was quantitated by incubating C3b-coated ShEA with fB and fD to form convertase and then treating with varying concentrations of fH to decay convertase before developing lysis. (E) Cofactor activity was analyzed by treating C3b-coated ShEA with fH and fI before forming convertase and developing lysis. In D and E, curves were fitted using nonlinear regression to calculate the IH50. Data points represent mean ± SD of three determinations. C3b102G-ShEA are shown as circles, C3b10R-ShEA as squares, fH62V as filled symbols, and fH62I as open symbols.
Hemolysis Assays to Investigate Decay and Cofactor Activities of fH on Variant Convertases.
fH and rfH1–4 bound less well to C3b102G compared with C3b102R, but reduced binding affinity did not cause decreased decay acceleration. To confirm this finding and test whether affinity differences affected fI cofactor activity, hemolysis assays were performed using C3 variants spiked into NHS depleted of C3, fH, and fB (NHSΔC3BH) to deposit C3b on ShEA. AP convertase was then assembled using fB and fD and incubated with different fH concentrations to promote convertase decay before developing lysis (Fig. 3D). No difference in rate of accelerated decay of the variant convertases was seen, in agreement with Biacore data (IH50 for C3102G: 4.4 ± 0.2 nM; C3102R: 4.0 ± 0.5 nM; P = 0.31).
Cofactor activity was measured by treating C3b-coated ShEA with fI and different concentrations of fH to inactivate C3b. Convertase was formed on residual C3b and lysis developed. The C3102R variant showed 1.5-fold higher sensitivity to inactivation by fI compared with C3102G when fH62I was used as cofactor (IH50: 1.5 ± 0.2 nM for C3b102R; 2.3 ± 0.1 nM for C3b102G; two-tailed unpaired t test; P < 0.001) and 1.7-fold higher sensitivity to inactivation catalyzed by fH62V (IH50: 3.6 ± 0.1 nM for C3b102G; 2.2 ± 0.1 nM for C3b102R; two-tailed unpaired t test; P < 0.0001; Fig. 3E). Strikingly, combining two protective variants (C3102R and fH62I) caused a 2.4-fold reduction in ShEA lysis compared with risk combination (C3102G and fH62V; two-tailed unpaired t test; P < 0.0001; Fig. 3E).
Different Combinations of Polymorphic Variants of C3, fH, and fB Regulate AP Activity in Plasma.
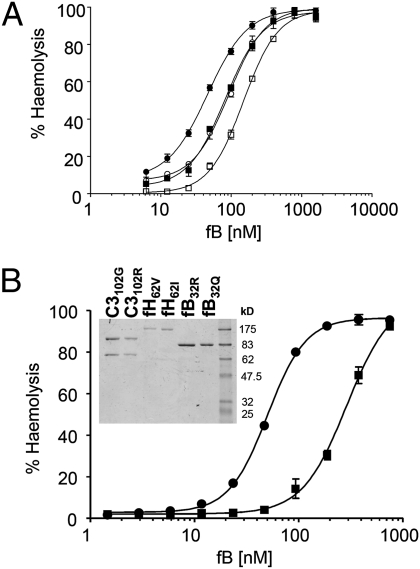
Data above show that different combinations of fH and C3 variants yield different plasma AP activities. We previously showed that fB32Q forms C3 convertase less efficiently than fB32R, causing decreased AP amplification (34). To test whether C3 and fB variants collaborate to influence AP activity, NHS depleted of C3 and fB (NHSΔC3B) spiked with C3 variants was used to deposit variant C3b on ShEA. Convertase was then formed by incubating with fD and either fB32R or fB32Q and lysis developed (Fig. 4A). Complementary effects of C3 (filled/open symbols) and fB (circles/squares) variants were apparent; protective combination requiring 3.2-fold more fB to achieve lysis comparable with risk combination (EC50: C3102G, fB32R: 46.5 ± 2.8 nM; C3102R, fB32Q: 149.1 ± 1 nM; P < 0.0001).
Fig. 4.
Polymorphic variations in C3, fB, and fH collaborate to control AP hemolytic activity. (A) C3b was deposited on ShEA using NHSΔC3B spiked with either variant of C3 (C3102R, squares; C3102G, circles); convertase was formed using fD and either fB32R (filled symbols) or fB32Q (open symbols); lysis was developed with NHSΔBH. (B) C3 variants were spiked into NHSΔC3BH and used to deposit C3b on cells, which were then treated with fI and a specific variant of fH. Convertase was formed by adding different concentrations of a specific fB variant with fD, and lysis was developed. The complotype predicted to yield highest AP activity (C3102G, fB32R, fH62V) is shown by circles, whereas that predicted to yield lowest AP activity (C3102R, fB32Q, fH62I) is represented by squares. Data points represent mean ± SD of three determinations (error bars depicted for each point); nonlinear regression was used to calculate the EC50. Inset shows SDS/PAGE of purified C3, fB, and fH variants.
Complementarity of the fHV62I polymorphism was investigated by comparing the variant set promoting most AP amplification (C3102G/fB32R/fH62V) with that causing least amplification (C3102R/fB32Q/fH62I). C3 variants spiked into NHSΔC3BH were used to deposit C3b on ShEA (depletion of fH eliminated C3b cleavage by fI), followed by incubation with equivalent concentrations of either fH variant and fI, and finally varying amounts of fB variants and fD to form convertase. Lysis was developed and hemolysis measured (Fig. 4B). The C3102G/fB32R/fH62V combination had sixfold greater AP hemolytic activity compared with C3102R/fB32Q/fH62I at equivalent concentrations and in identical depleted serum (EC50 50 ± 1 nM versus 289 ± 14 nM, P < 0.0001). This finding was confirmed in whole serum AP hemolysis by adding back pure C3, fB, and fH variants to hydrazine-treated NHS depleted of fB and fH (NHS-R3ΔBH). AP serum hemolytic activity (AH50) of serum repleted with C3102G/fB32R/fH62V was 206 units/mL compared with 108 units/mL when repleted with C3102R/fB32Q/fH62I.
Discussion
Common polymorphisms in complement proteins alter risk for diverse diseases (11, 15, 16, 19). Polymorphisms in AP proteins C3, fB, and fH strongly influence risk for AMD with quoted odds ratios for homozygotes of 3.51–7.4 for fHY402H (11, 14), 2.6 for C3R102G (19), 0.36 for fBR32Q (16), and 0.54 for fHV62I (11); each also influences risk for other diseases and/or infections, implying that small changes in AP proteins can dramatically affect health. fHY402H likely alters capacity of fH to bind surfaces (27, 28), but the reasons why other AP polymorphisms influence disease susceptibility were unknown until our demonstration that fBR32Q and fHV62I directly impacted on AP activation and amplification (29, 34). Association of the common C3 polymorphism (C3S/F; C3R102G) with disease has been recognized for decades, but the molecular basis was enigmatic. We report here that the C3R102G polymorphism affects AP activation by influencing efficiency of regulation by fH and that different combinations of common variations in C3, fB, and fH dramatically alter AP activity, up to sixfold at the extremes when pure proteins were used in the assay. When variants were added back to whole serum, the C3102R, fB32Q, fH62I combination of variants had 50% alternative pathway activity (AH50) compared with the most active variants. These data reveal an AP complotype providing clear explanation for association of AP protein polymorphisms with disease.
Given the long association of C3R102G with disease, it is surprising that the functional basis had remained unknown; however, our studies of polymorphisms in fB and fH had demonstrated that functional consequences of the C3R102G polymorphism were likely to be subtle (29, 34). Meticulous attention was therefore given to protein purity and integrity, with replication using protein from several homozygous individuals. Pure protein added back to C3-depleted serum was used to deposit C3b and assemble AP convertase on ShEA, before developing lysis. AP hemolytic activity driven by the C3102R convertase was consistently less than by C3102G convertase (Fig. 1). This was not due to differences in kinetics of convertase formation or decay, but to altered susceptibility to regulation. Specifically, fH bound C3b102G with lower affinity compared with C3b102R (Fig. 2 A–C). This difference was replicated using recombinant fH SCRs 1–4, locating differential binding in these amino-terminal domains where decay and cofactor activities reside (Fig. 2 D–F) (32, 33). Decay accelerating activity of fH was not affected by this change in affinity (Fig. 3 B–D), but fI cofactor activity for C3b102G inactivation was reduced, decreasing the rate of C3b inactivation and increasing overall AP activity (Fig. 3E). The effect was specific for fH as binding affinities and regulatory control by DAF and MCP were unaffected by the C3bR102G polymorphism. Binding affinity of fH for hydrolyzed C3 was weak and showed no difference for the C3(H20)R102G variants; thus, the polymorphism affects primarily the amplification convertase rather than the initiation convertase and tickover loop.
The observation that a reduction in binding affinity between C3b and fH adversely affected cofactor but not decay accelerating activity implies that cofactor activity is more dependent on tight binding, an interpretation supported by our published analyses of fHV62I, where lower binding affinity for C3b in fH62V reduced cofactor but not decay activities compared with fH62I (29). C3R102G polymorphism maps to a positively charged area at the interface between the MG1 domain and TED (thioester-containing domain) (Fig. S5) (35). fH SCR3 binds MG2 and SCR4 contacts TED in close proximity to MG1. Decreasing overall positive charge in C3b by substituting Arg with Gly at position 102 likely alters interdomain associations around the fH binding site, influencing affinity and cofactor activity. In contrast, binding of fB involves different C3b domains; hence, convertase assembly and decay are unaffected by the polymorphism (Fig. S5) (36, 37).
The C3R102G functional effect, modest in isolation, when placed in the context of other AP variants, translates to a large functional effect. C3R102G and fHV62I polymorphisms both influence cofactor activity, and effects were additive (Fig. 3E), as were effects of C3R102G and fBR32Q variants (Fig. 4A). Comparison of the protective set (C3102R, fB32Q, fH62I) with risk (C3102G, fB32R, fH62V) revealed a sixfold difference in hemolytic activity, graphically illustrating that different combinations of complement polymorphic variants yield dramatic differences in complement activity (Fig. 4B). We demonstrate that different combinations of activators and regulators (the complotype) set the level of systemic complement activity in an individual, thereby determining risk for various pathologies involving complement. These findings provide functional underpinning to genetic analyses describing altered disease risk depending on the set of complement genes inherited (38, 39). Maintenance of complement homeostasis involves a balance between activation and regulation. Inheritance of a highly active complement system (e.g., homozygous C3102G, fB32R, fH62V), whereas beneficial in fighting infection by enhanced opsonization, may predispose to pathology in inflammatory diseases such as AMD.
The extreme risk and protective complotypes illustrated here will be rare but they represent the extremities of a continuum in AP activity that define disease risk. Biomarker studies reported complement activation products in AMD plasma, indicative of low-grade, systemic AP activation (39–41). Inheritance of a more active complotype may be detrimental only in the elderly, where chronic inflammation has been sustained long-term, whereas inheritance of a less active complotype, although protective against inflammation, may increase susceptibility to infection, a negative selective pressure in populations where infection is prevalent. This is illustrated in a study of a cohort of 347 Dutch immigrants to Surinam in the 19th century that lost 60% of their number soon after arrival due to typhoid and yellow fever. Survivors of these epidemics showed higher frequency of the C3102G allele compared with the parent population (P = 0.00001), providing evidence that strong selective pressure in the face of infection selects this C3 variant in a population (42).
We demonstrate here that sets of common polymorphisms in just three complement proteins dramatically alter AP activity; however, the complotype extends beyond these proteins and the AP. Protein levels will influence systemic complement activity, as will polymorphisms in other complement proteins. For example, fI- and fH-related proteins are associated with AMD (43, 44), whereas polymorphisms in C5/TRAF1 are associated with rheumatoid arthritis and systemic lupus erythematosus (45, 46). Complement dysregulation often manifests in inflammatory diseases when there are “multiple hits,” exemplified in aHUS where two or three hits in the complement cascade are required to trigger pathology (47). Frequently, one or more of these hits is a mutation in a complement protein causing overactivation of components or decreased function of regulators; however, our data show that the complotype can also tip the balance toward dysregulation and pathology. An individual's complotype will therefore influence susceptibility to, or severity of, diseases involving complement dysregulation; knowledge of the complotype will not only aid disease prediction but also dictate avenues of therapy aimed at restoring equilibrium in the complement cascade.
Materials and Methods
Preparation of Complement Components and Activation Fragments.
C3, fB, and fH were purified from plasma of consented volunteers, genotyped and homozygous for the relevant protein; hydrolyzed C3 was generated by hydroxylamine treatment of pure C3 (SI Materials and Methods). C3 variants were purified using classical chromatography (Na2SO4 cut; anion and cation exchange), described in SI Materials and Methods). On the day of analysis, C3 samples were dialyzed into 50 mM Na-phosphate, pH 6, fractionated on Mono S (GE Healthcare) to remove hydrolyzed C3, and gel filtered (Superdex200 10/300; GE Healthcare) to remove aggregates and buffer exchange [Hepes-buffered saline (HBS): 10 mm Hepes, 150 mM NaCl], complement fixation diluent (CFD) (Oxoid), AP buffer (5 mM sodium barbitone pH 7.4, 150 mM NaCl, 7 mM MgCl2, 10 mM EGTA pH 7.5) as appropriate, and used immediately without concentration. Factor B and fH were affinity purified from EDTA plasma on anti-Bb (mAb JC1, in-house) or anti-fH (mAb 35H9, in-house) HiTrap columns (GE Healthcare). Eluted protein was gel filtered to remove aggregates and minor contaminants and buffer exchange. Factor D and fI were from Comptech. C3b was prepared by fluid phase AP activation using purified proteins as described (31) and gel filtered to remove aggregates. Recombinant fH (rfH1–4) was isolated from Escherichia coli inclusions and refolded as described in SI Materials and Methods.
Hemolysis Assays.
NHS depleted of C3 (NHSΔC3), C3 and fB (NHSΔC3B), fB and fH (NHSΔBH), or all three components (NHSΔC3BH or NHS-R3ΔBH) was prepared as described previously and in SI Materials and Methods(29). NHS-MA was prepared as described (48). Hemolysis assays were performed as described with modifications as specified (29). Curves were fitted using nonlinear regression and EC50/IH50 calculated (GraphPad Prism software); statistical significance was evaluated using Microsoft Excel software (two-tailed unpaired t test).
Preparation of cells.
C3b was deposited on antibody-coated sheep erythrocytes (ShEA) using either 5% (vol/vol) NHSΔC3, 10% (vol/vol) NHSΔC3B (presence of fH), or 10% (vol/vol) NHSΔC3BH (absence of fH). Purified C3 was added back to depleted serum at either 420 nM (5% serum, Fig. 1), 790 nM (10% serum; decay assay Fig. 3D), or 260 nM (5% serum; cofactor assay Fig. 3E). OmCI (0.6 μM; gift from Varleigh, London, United Kingdom) was used to block terminal pathway activation as described previously (29, 49). ShEA-C3b cells were resuspended in AP buffer at 2% (vol/vol).
Comparison of C3 from different homozygote donors.
ShEA-C3b were generated using NHSΔC3 with C3 add-back; C3bBb was formed by incubating 1 volume of cells with 1 volume of 16 nM fD mixed with fB (24–789 nM) for 15 min at 37 °C. Further convertase formation was prevented and lysis developed by adding 1 volume of NHS-MA (4% vol/vol in PBS, 20 mM EDTA) containing 530 nM C3. Lysis was calculated by measuring hemoglobin release, control incubations were ShEA incubated in buffer only (0%) or in 0.001% Triton X (100%). Percentage lysis = 100 × (A410 test sample – A410 0% control)/(A410 100% control – A410 0% control).
fH decay and cofactor assays.
These assays were performed as previously described (29) other than serum used to deposit C3b was depleted of donor C3 and had purified C3 added-back (SI Materials and Methods). The ability of fH to decay preformed convertase on ShEA-C3b, or catalyze inactivation of C3b to iC3b by fI before convertase formation, was assessed by extent of hemolysis.
Complotype assays.
Combined effect of C3 and fB variants was assessed using the assay described above. C3b was deposited on ShEA using NHSΔC3B with C3 add-back; different concentrations (6 nM–1.5 μM) of either fB32Q or fB32R were used to form convertase with either C3b102R or C3b102G. Combined effect of C3 and fH variants was assessed using cofactor assays with either fH62I or fH62V. Combined effect of all three variant proteins (C3R102G, fBR32Q, and fHV62I) was assessed using NHSΔC3BH with C3 add-back to deposit C3b on ShEA, incubation in AP buffer with fI as described for the cofactor assay, and incubation with 3.4 nM fH (chosen to yield incomplete cleavage of C3b in the assay). Cells were resuspended in AP buffer and lysis developed as described for decay assay, except that varying concentrations of fB variants (1.5–750 nM) were used to form convertase. AH50 was calculated as previously described using variants added back to NHS-R3ΔBH (SI Materials and Methods).
Surface Plasmon Resonance.
All analyses used a Biacore T100 (GE Healthcare), run in HBS, 0.01% surfactant P20 as described and in SI Materials and Methods (34). To assess enzyme kinetics, fB alone (6–788 nM; proenzyme) or fB (3–397 nM) in the presence of 40 nM fD (convertase) was flowed across immobilized C3b in HBS, 0.01% surfactant P20, 1 mM MgCl2; proenzyme was regenerated with EDTA and convertase with sDAF (0.3 μM) or rfH1–4 (1 μM). Regulator binding to C3b [or C3(H20) where stated] was analyzed by flowing varying concentrations of fH [45 nM–2.9 μM for C3b; 0.2 μM–28 μM for C3(H20), rfH1–4 (89 nM–23 μM), sMCP (100 nM–20 μM), or sDAF (0.78–50 μM); for fH interactions, the surface was regenerated with 10 mM sodium acetate pH 4, 1 M NaCl.
Decay of AP C3 convertase was assessed by forming equal amounts of convertase (788 nM fB, 20 nM fD) on each variant C3b surface (1,500 RU) for 150 s. fH, rfH1–4, and sDAF in the range 10–1,000 nM were injected for 90 s, and accelerated decay was monitored. Reference curves for each concentration of regulator binding the C3b surface were subtracted to adjust for regulator interaction with C3b alone.
Supplementary Material
Acknowledgments
We thank Dr. David Cole (Cardiff University, Cardiff, United Kingdom) for advice on refolding rfH1–4 and Prof. Susan Lea (Oxford University, Oxford, United Kingdom) for sMCP and sDAF. This work was supported by the Medical Research Council, United Kingdom (G0701298 to C.L.H. and B.P.M.), the Spanish Ministerio de Educación y Cultura (SAF2008-00226 to S.R.deC.) and the Ciber de Enfermedades Raras and the Fundación Renal Iñigo Alvarez de Toledo (S.R.deC.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1019338108/-/DCSupplemental.
References
- 1.Walport MJ. Complement. First of two parts. N Engl J Med. 2001;344:1058–1066. doi: 10.1056/NEJM200104053441406. [DOI] [PubMed] [Google Scholar]
- 2.Lachmann PJ, Hughes-Jones NC. Initiation of complement activation. Springer Semin Immunopathol. 1984;7:143–162. doi: 10.1007/BF01893018. [DOI] [PubMed] [Google Scholar]
- 3.Morgan BP, Meri S. Membrane proteins that protect against complement lysis. Springer Semin Immunopathol. 1994;15:369–396. doi: 10.1007/BF01837366. [DOI] [PubMed] [Google Scholar]
- 4.Fearon DT. Regulation by membrane sialic acid of beta1H-dependent decay-dissociation of amplification C3 convertase of the alternative complement pathway. Proc Natl Acad Sci USA. 1978;75:1971–1975. doi: 10.1073/pnas.75.4.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holers VM. The spectrum of complement alternative pathway-mediated diseases. Immunol Rev. 2008;223:300–316. doi: 10.1111/j.1600-065X.2008.00641.x. [DOI] [PubMed] [Google Scholar]
- 6.Rodríguez de Córdoba S, Esparza-Gordillo J, Goicoechea de Jorge E, Lopez-Trascasa M, Sánchez-Corral P. The human complement factor H: Functional roles, genetic variations and disease associations. Mol Immunol. 2004;41:355–367. doi: 10.1016/j.molimm.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Atkinson JP, Liszewski MK, Richards A, Kavanagh D, Moulton EA. Hemolytic uremic syndrome: An example of insufficient complement regulation on self-tissue. Ann N Y Acad Sci. 2005;1056:144–152. doi: 10.1196/annals.1352.032. [DOI] [PubMed] [Google Scholar]
- 8.Zipfel PF, Heinen S, Józsi M, Skerka C. Complement and diseases: Defective alternative pathway control results in kidney and eye diseases. Mol Immunol. 2006;43:97–106. doi: 10.1016/j.molimm.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 9.Goicoechea de Jorge E, et al. Gain-of-function mutations in complement factor B are associated with atypical hemolytic uremic syndrome. Proc Natl Acad Sci USA. 2007;104(1):240–245. doi: 10.1073/pnas.0603420103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodríguez de Córdoba S, Harris CL, Morgan BP, Llorca O. Lessons from functional and structural analyses of disease-associated genetic variants in the complement alternative pathway. Biochim Biophys Acta. 2011;1812:12–22. doi: 10.1016/j.bbadis.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 11.Hageman GS, et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci USA. 2005;102(20):7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haines JL, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308(5720):419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 13.Edwards AO, et al. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308(5720):421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 14.Klein RJ, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308(5720):385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pickering MC, et al. Spontaneous hemolytic uremic syndrome triggered by complement factor H lacking surface recognition domains. J Exp Med. 2007;204(6):1249–1256. doi: 10.1084/jem.20070301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gold B, et al. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat Genet. 2006;38(4):458–462. doi: 10.1038/ng1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Botto M, Fong KY, So AK, Koch C, Walport MJ. Molecular basis of polymorphisms of human complement component C3. J Exp Med. 1990;172:1011–1017. doi: 10.1084/jem.172.4.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spencer KL, et al. C3 R102G polymorphism increases risk of age-related macular degeneration. Hum Mol Genet. 2008;17:1821–1824. doi: 10.1093/hmg/ddn075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yates JR, et al. Complement C3 variant and the risk of age-related macular degeneration. N Engl J Med. 2007;357(6):553–561. doi: 10.1056/NEJMoa072618. [DOI] [PubMed] [Google Scholar]
- 20.Messias-Reason IJ, Urbanetz L, Pereira da Cunha C. Complement C3 F and BF S allotypes are risk factors for Chagas disease cardiomyopathy. Tissue Antigens. 2003;62:308–312. doi: 10.1034/j.1399-0039.2003.00101.x. [DOI] [PubMed] [Google Scholar]
- 21.Papiha SS, Duggan-Keen M, Roberts DF. Factor B (BF) allotypes and multiple sclerosis in north-east England. Hum Hered. 1991;41:397–402. doi: 10.1159/000154033. [DOI] [PubMed] [Google Scholar]
- 22.Hägglöf B, et al. Studies of HLA, factor B (Bf), complement C2 and C4 haplotypes in type 1 diabetic and control families from northern Sweden. Hum Hered. 1986;36:201–212. doi: 10.1159/000153627. [DOI] [PubMed] [Google Scholar]
- 23.Rambausek M, et al. Genetic polymorphism of C3 and Bf in IgA nephropathy. Nephrol Dial Transplant. 1987;2:208–211. [PubMed] [Google Scholar]
- 24.Finn JE, et al. Molecular analysis of C3 allotypes in patients with systemic vasculitis. Nephrol Dial Transplant. 1994;9:1564–1567. [PubMed] [Google Scholar]
- 25.Andrews PA, Finn JE, Mathieson PW, Sacks SH. Molecular analysis of C3 allotypes related to transplant outcome in human renal allografts. Transplantation. 1995;60:1342–1346. [PubMed] [Google Scholar]
- 26.Finn JE, Mathieson PW. Molecular analysis of C3 allotypes in patients with nephritic factor. Clin Exp Immunol. 1993;91:410–414. doi: 10.1111/j.1365-2249.1993.tb05917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prosser BE, et al. Structural basis for complement factor H linked age-related macular degeneration. J Exp Med. 2007;204:2277–2283. doi: 10.1084/jem.20071069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herbert AP, et al. Structure shows that a glycosaminoglycan and protein recognition site in factor H is perturbed by age-related macular degeneration-linked single nucleotide polymorphism. J Biol Chem. 2007;282:18960–18968. doi: 10.1074/jbc.M609636200. [DOI] [PubMed] [Google Scholar]
- 29.Tortajada A, et al. The disease-protective complement factor H allotypic variant Ile62 shows increased binding affinity for C3b and enhanced cofactor activity. Hum Mol Genet. 2009;18:3452–3461. doi: 10.1093/hmg/ddp289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harris CL, Pettigrew DM, Lea SM, Morgan BP. Decay-accelerating factor must bind both components of the complement alternative pathway C3 convertase to mediate efficient decay. J Immunol. 2007;178:352–359. doi: 10.4049/jimmunol.178.1.352. [DOI] [PubMed] [Google Scholar]
- 31.Harris CL, Abbott RJ, Smith RA, Morgan BP, Lea SM. Molecular dissection of interactions between components of the alternative pathway of complement and decay accelerating factor (CD55) J Biol Chem. 2005;280(4):2569–2578. doi: 10.1074/jbc.M410179200. [DOI] [PubMed] [Google Scholar]
- 32.Gordon DL, Kaufman RM, Blackmore TK, Kwong J, Lublin DM. Identification of complement regulatory domains in human factor H. J Immunol. 1995;155:348–356. [PubMed] [Google Scholar]
- 33.Kühn S, Zipfel PF. Mapping of the domains required for decay acceleration activity of the human factor H-like protein 1 and factor H. Eur J Immunol. 1996;26:2383–2387. doi: 10.1002/eji.1830261017. [DOI] [PubMed] [Google Scholar]
- 34.Montes T, Tortajada A, Morgan BP, Rodríguez de Córdoba S, Harris CL. Functional basis of protection against age-related macular degeneration conferred by a common polymorphism in complement factor B. Proc Natl Acad Sci USA. 2009;106:4366–4371. doi: 10.1073/pnas.0812584106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu J, et al. Structure of complement fragment C3b-factor H and implications for host protection by complement regulators. Nat Immunol. 2009;10:728–733. doi: 10.1038/ni.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Torreira E, Tortajada A, Montes T, Rodríguez de Córdoba S, Llorca O. 3D structure of the C3bB complex provides insights into the activation and regulation of the complement alternative pathway convertase. Proc Natl Acad Sci USA. 2009;106:882–887. doi: 10.1073/pnas.0810860106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Janssen BJ, et al. Insights into complement convertase formation based on the structure of the factor B-cobra venom factor complex. EMBO J. 2009;28:2469–2478. doi: 10.1038/emboj.2009.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sobrin L, et al. Genetic profile for five common variants associated with age-related macular degeneration in densely affected families: A novel analytic approach. Eur J Hum Genet. 2010;18:496–501. doi: 10.1038/ejhg.2009.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hecker LA, et al. Genetic control of the alternative pathway of complement in humans and age-related macular degeneration. Hum Mol Genet. 2010;19:209–215. doi: 10.1093/hmg/ddp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scholl HP, et al. Systemic complement activation in age-related macular degeneration. PLoS ONE. 2008;3:e2593. doi: 10.1371/journal.pone.0002593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reynolds R, et al. Plasma complement components and activation fragments: Associations with age-related macular degeneration genotypes and phenotypes. Invest Ophthalmol Vis Sci. 2009;50:5818–5827. doi: 10.1167/iovs.09-3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Vries RR, Meera Khan P, Bernini LF, van Loghem E, van Rood JJ. Genetic control of survival in epidemics. J Immunogenet. 1979;6:271–287. doi: 10.1111/j.1744-313x.1979.tb00684.x. [DOI] [PubMed] [Google Scholar]
- 43.Fagerness JA, et al. Variation near complement factor I is associated with risk of advanced AMD. Eur J Hum Genet. 2009;17:100–104. doi: 10.1038/ejhg.2008.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zipfel PF, et al. Deletion of complement factor H-related genes CFHR1 and CFHR3 is associated with atypical hemolytic uremic syndrome. PLoS Genet. 2007;3:e41. doi: 10.1371/journal.pgen.0030041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kurreeman FA, et al. European Consortium on Rheumatoid Arthritis Families Replication of the tumor necrosis factor receptor-associated factor 1/complement component 5 region as a susceptibility locus for rheumatoid arthritis in a European family-based study. Arthritis Rheum. 2008;58:2670–2674. doi: 10.1002/art.23793. [DOI] [PubMed] [Google Scholar]
- 46.Nishimoto K, et al. Association study of TRAF1-C5 polymorphisms with susceptibility to rheumatoid arthritis and systemic lupus erythematosus in Japanese. Ann Rheum Dis. 2010;69:368–373. doi: 10.1136/ard.2008.104315. [DOI] [PubMed] [Google Scholar]
- 47.Esparza-Gordillo J, et al. Predisposition to atypical hemolytic uremic syndrome involves the concurrence of different susceptibility alleles in the regulators of complement activation gene cluster in 1q32. Hum Mol Genet. 2005;14(5):703–712. doi: 10.1093/hmg/ddi066. [DOI] [PubMed] [Google Scholar]
- 48.Harris CL. Functional assays for complement regulators. Methods Mol Biol. 2000;150:83–101. doi: 10.1385/1-59259-056-X:83. [DOI] [PubMed] [Google Scholar]
- 49.Hepburn NJ, et al. In vivo characterization and therapeutic efficacy of a C5-specific inhibitor from the soft tick Ornithodoros moubata. J Biol Chem. 2007;282:8292–8299. doi: 10.1074/jbc.M609858200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.