Abstract
A class of peptides from the bombolitin family, not previously identified for nitroaromatic recognition, allows near-infrared fluorescent single-walled carbon nanotubes to transduce specific changes in their conformation. In response to the binding of specific nitroaromatic species, such peptide–nanotube complexes form a virtual “chaperone sensor,” which reports modulation of the peptide secondary structure via changes in single-walled carbon nanotubes, near-infrared photoluminescence. A split-channel microscope constructed to image quantized spectral wavelength shifts in real time, in response to nitroaromatic adsorption, results in the first single-nanotube imaging of solvatochromic events. The described indirect detection mechanism, as well as an additional exciton quenching-based optical nitroaromatic detection method, illustrate that functionalization of the carbon nanotube surface can result in completely unique sites for recognition, resolvable at the single-molecule level.
Keywords: bionanotechnology, explosives detection, pesticides, spectroscopy, optical sensors
Single-walled carbon nanotubes (SWNT) emit near-infrared (NIR) bandgap photoluminescence (PL) (1, 2), which is highly responsive to its physical and chemical environment (3–6). SWNT are unique among nanoscale sensor platforms in their ability to detect the adsorption of as few as a single molecule of an analyte (7, 8). For such a capability to be extended to specific classes of organic molecules, chemical approaches must be developed that allow for selective molecular recognition. Stepwise quenching of SWNT PL by single-molecule adsorption events to the nanotube surface (7) has been demonstrated (8, 9). Our previous findings extend this resolution to biologically important reactive oxygen species and demonstrate multiplexed detection of redox-active analytes by two optical modes, leading to species identification (9).
The current work investigates the selective optical detection of binding events by single-SWNT PL modulation, employing both intensity- and wavelength-based signal transduction. We find that specific noncovalently bound polymers can be harnessed to change the properties of the nanotube–polymer complex, resulting in complete modulation of the nanotube sensitivity to certain analytes. Resolution of an entire class of molecules can be achieved by the nanotube via reporting the conformational state of a peptide, for example. Nanotube emission undergoes solvatochromic shifts due to nitroaromatic compound-mediated secondary structure changes of the amphipathic bombolitin II oligopeptide. In this work, we probe solvatochromic interactions at the single-nanotube level by a strategy in which two spectrally adjacent optical channels measure anticorrelated, quantized fluctuations, signifying molecular binding events. In addition, we find that an oligonucleotide of single-stranded DNA with the sequence (AT)15 [ss(AT)15] oligonucleotide imparts optical selectivity of SWNT for trinitrotoluene (TNT) via intensity modulation. Although nanotubes do not normally detect this analyte, the electronic and steric effects of this encapsulating sequence allow single-molecule detection by reversible excitonic quenching.
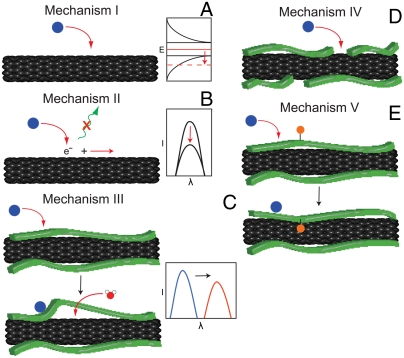
We assert that five distinct mechanisms for the selective modulation of SWNT PL have been identified to date, depending upon the analyte, SWNT surface chemistry, and solvent (Fig. 1). Adsorption to SWNT of an analyte with a favorable reduction potential may shift the nanotube Fermi level, resulting in bleaching of the absorption transition and concomitant excitonic loss (3, 10) (mechanism 1, Fig. 1A). Examples of this mechanism include hydrogen peroxide binding (8, 9, 11), pH effects (12), and several redox-active molecules discussed in the literature (13). A second mechanism, exciton quenching, involves the adsorption of a moiety that perturbs the exciton itself, preventing recombination and/or increasing competitive nonradiative decay, effectively quenching the PL (3) (mechanism 2, Fig. 1B). Examples from the literature include certain dye molecules (13). Solvatochromic shifting is typically caused by the direct changing of the solvent environment or by perturbing an encapsulating polymer, thereby changing the accessibility of the SWNT surface to specific moieties on the polymer or the solvent (14) (mechanism 3, Fig. 1C). A demonstrated example is ion-induced conformational changes to DNA-encapsulated SWNT (15, 16). Selectivity can be imparted to the above mechanisms by engineering polymer-encapsulated nanotube complexes which block the binding of interferents to SWNT, hence imparting steric selectivity (mechanism 4, Fig. 1D). An analyte-specific switchable polymer containing a quenching ligand, for example, is an additional strategy for engineering selectivity (mechanism 5, Fig. 1E), which was recently demonstrated with a polyvinyl alcohol derivative functionalized with glucose-binding protein (17). For mechanisms involving solvatochromism, no previous examples exist which demonstrate single-molecular resolution.
Fig. 1.
Mechanisms of SWNT PL modulation. (A) Mechanism I, Fermi level shifting via redox-active analyte adsorption to the nanotube surface. (B) Mechanism II, quenching induced by exciton disruption in response to analyte binding. (C) Mechanism III, solvatochromic shifting due to perturbation of a SWNT-bound polymer. (D) Mechanism IV, selectivity of analyte binding mediated by polymer wrapping. (E) Mechanism V, analyte-activated polymer switching resulting in intensity and/or wavelength modulation.
Results and Discussion
A Peptide–SWNT Chaperone Sensor.
A mixture of SWNT encapsulated by bombolitin II, a variant of a bumblebee venom-derived amphiphilic peptide, was screened against a library of 42 analytes. The (7,5) nanotube species exhibits quenching due to several redox-active compounds (discussed later) as well as red shifts relative to the control peak wavelength, with slight concomitant intensity variation, in response to several nitroaromatic and nitro-group-containing nonaromatic compounds (Fig. S1 and Table S1). Picric acid, cyclotrimethylenetrinitramine (RDX), 2,4-dinitrophenol, and 4-nitro-3(trifluoromethyl)phenol (TFM) induce spectral shifts without significant signal attenuation. Other shifting analytes induce large-intensity diminutions.
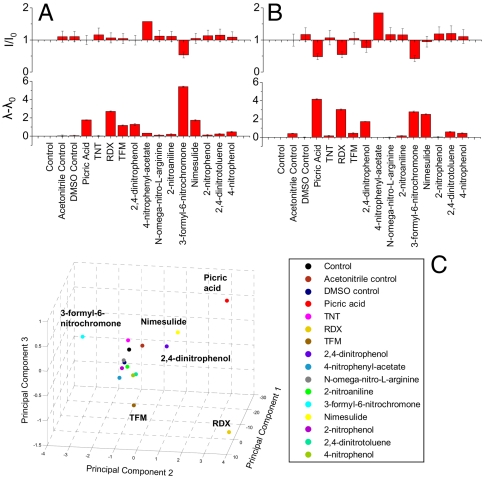
Exposing bombolitin-II–SWNT to a diverse set of nitro group compounds (Fig. 2 A and B) finds that 6 of 13 such analytes (Table S2) exhibit significant wavelength shifts with little concomitant attenuation, a relatively rare effect that suggests a significant change in the nanotube’s dielectric environment (15). The spectral changes differ among analytes, and different (n,m) nanotube species exhibit individualized detection signatures, where the intensity and wavelength changes vary across SWNT species. This variation is illustrated here for the (7,5) and (11,3) species, which possess different diameters (0.829 vs. 1.014 nm), chiral angles (24.5° vs. 11.74°), and optical bandgaps (1.211 vs. 1.036 eV).
Fig. 2.
Nitro compound detection and fingerprinting by bombolitin-II–SWNT. (A) Fluorescence intensity and wavelength response of bombolitin-II-solubilized nanotubes interrogated by 13 nitro group compounds. Responses of the (7,5) nanotube differ from (B) responses of the (11,3) nanotube to the same compounds. Intensity response (Upper) is normalized with respect to control intensity (I/I0). Wavelength shifting response (Lower) is presented as the peak wavelength in nanometers subtracted from that of the control (λ-λ0). (C) Principal components analysis plot using PL intensity and wavelength response inputs from eight (n, m) species of bombolitin-II-solubilized SWNT to the 13 nitro group compounds.
The analytes generate differentiable fingerprints via distinct spectral signatures and unique responses of several SWNT (n,m) species in a manner analogous to what we have shown for genotoxins (9). Principal components analysis, performed on the detection data and collected from eight different SWNT species, confirms unique signatures of the six analytes, denoted by their segregation into separate regions of the plot (Fig. 2C), allowing identification of the analytes by their responses. The analysis was conducted by compiling all eight nanotubes’ intensity change and wavelength shifting data for each analyte. The first three principal component scores, which account for a total of 99.5% of the total data variance, are shown. Loadings for the first three principal components are listed by input variable (Table S3).
All detected analytes, including explosive compounds RDX and picric acid, as well as pesticides 2,4-dinitrophenol and TFM, contain ring structures and nitro groups (Fig. S2). However, few other recognizable structural components or patterns are present in the responding set. Though bombolitin II is a relatively short peptide, it is difficult to predict binding events of such species, which accounts for the need for high-throughput selection methods such as phage display (18).
We find that SWNT clearly report changes in bombolitin conformation. The bombolitin family of oligopeptides contains five species with 17-residue sequences derived from bumblebee venom. Their conformations are helical but largely disordered in aqueous solution; in the presence of bilipid membranes, they take on a more ordered, alpha helical structure according to CD studies (19, 20). At high concentrations, above 2.5 mM, and in the presence of SDS, the peptides form aggregates (21). Bombolitin-II–SWNT, perturbed by SDS at concentrations much lower than the critical micelle concentration, causes profound changes to SWNT PL intensity and wavelength (Fig. S3 and Discussion of SDS Experiments).
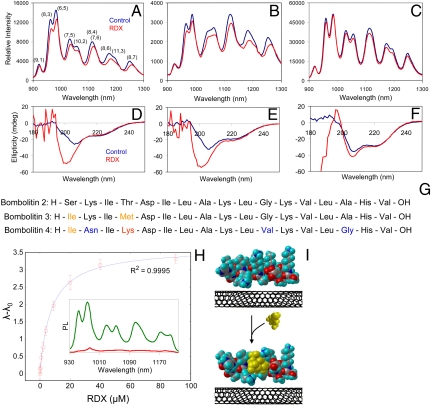
The bombolitin–SWNT response is highly dependent on peptide sequence. Aqueous suspensions of peptide-stabilized SWNT were synthesized with three of four members of the bombolitin family. The bombolitin I variant failed to solubilize nanotubes, whereas bombolitins II, III, and IV made highly NIR photoluminescent, stable solutions via probe-tip sonication in the presence of SWNT (Fig. 3 A–C). The NIR spectra of the solubilized nanotubes show variations in relative initial emission intensities and wavelengths of the PL maxima of the (n,m) SWNT species present in the sample. Upon introduction of RDX, the PL peaks of bombolitins II and III undergo distinct red shifts of up to 13 meV, with the magnitude of shift dependent on the (n,m) species (Fig. 3 A and B). The SWNT solubilized by bombolitin IV, however, exhibits no wavelength shifting behavior (Fig. 3C).
Fig. 3.
Peptide–SWNT chaperone sensor for RDX. Photoluminescence spectra of peptide-suspended nanotubes before (blue) and after (red) addition of RDX. PL spectra of nanotubes suspended by (A) bombolitin II, (B) bombolitin III, (C) bombolitin IV. (D–F) CD spectra of the same peptides in the absence of nanotubes before (blue) and after (red) addition of RDX. (G) Sequences of the three bombolitin peptides highlighting differences between the variants. (H) Center wavelength of the (11,3) nanotube peak of bombolitin-II-solubilized SWNT plotted versus RDX concentration (red circles). The data are fit to a first-order Langmuir adsorption isotherm (blue curve). (Inset) PL spectra of bombolitin-II-solubilized SWNT before (green) and after (red) dialysis using a 20,000 Da molecular weight cutoff membrane. (I) Energy-minimized structures of bombolitin II in water and in the presence of both water and RDX; a conformational change with RDX is computed.
A change in the secondary structure of only RDX-responsive peptide variants is evident. CD measurements conducted on three bombolitin variants, in the absence of nanotubes, show similar spectra for bombolitins II and III, but the bombolitin IV spectrum differs, more closely resembling the classical α-helical spectrum (Fig. 3 D–F). Upon introduction of 9 μM RDX (much higher concentrations blocked UV signal), the bombolitin II and III spectra exhibit a distinct blue-shift of their 206-nm peak and significant increase in negative ellipticity. The bombolitin IV spectrum, upon the same treatment with RDX, exhibits negligible changes. These data suggest RDX binding occurs to bombolitin II and III peptides, inducing a conformational change upon binding. The binding is sequence specific; bombolitin IV shows little conformational change upon exposure to RDX. The bombolitin IV sequence differs in only three residues that are conserved between bombolitin II and III variants: Asn2, Val11, and Gly15, which are responsible for the differences in peptide secondary structure modulation and sensor response (Fig. 3G).
The wavelength shifting of bombolitin-II-solubilized SWNT exhibits concentration dependence, which fits well to a first-order Langmuir adsorption isotherm (Fig. 3H), implying that the transition is reversible. The peptide is found to exhibit low affinity to SWNT because it can be dialyzed away, causing aggregation of the nanotubes, which is evident from PL quenching (Fig. 3H, Inset). This behavior suggests that the nanotube reports the conformation of the largely free peptide, which individually suspends the nanotubes as would a surfactant (22). The nanotube, although solubilized by the peptide, acts as a chaperone sensor, which indirectly detects the binding event by transducing changes to the secondary structure of the bombolitin variant. In addition to the usage described herein, it may allow researchers to detect misfolded proteins, among other applications. We propose that, in an aqueous solution, the lowest energy conformation results in the formation of a “binding pocket” within the bombolitin II where RDX noncovalently interacts. We computed energy-minimized structures of bombolitin II interacting with RDX in a water medium, resulting in the images presented in Fig. 3I and confirming an RDX-modulated peptide secondary structure change. The binding was calculated to yield a net free energy change of -6.35 kcal/mol, signifying an energetically favorable interaction.
Different (n,m) nanotube species respond distinctly to RDX-induced conformational changes of the peptide. Certain SWNT species exhibit higher sensitivity to RDX interrogation, yet others exhibit a greater maximum shifting response (Fig. S4A). The dissociation constant, Kd, of the RDX response of the (11,3) nanotube species, calculated as the inverse of the Langmuir equilibrium constant, is 7.7 μM, which is on the order of the Kd of a TNT-binding antibody fragment interaction. Quantum dot-mediated detection of TNT using such a system resulted in a reported Kd of 15.7 μM (23). The maximum wavelength shift (λ-λ0 max) displays a different dependence on SWNT species (Fig. S4B). The λ-λ0 max maximizes when both the SWNT species diameter and chiral angle are large. Such a relationship follows if the bombolitin II, in its RDX-bound conformation, exhibited poorer stabilization of the larger, more chiral nanotubes as compared to the smaller SWNT with lower chiral angles. Absorption spectroscopy of bombolitin-II–SWNT-RDX interactions confirms variations in solvatochromic behavior among SWNT (n,m) species (Fig. S4C).
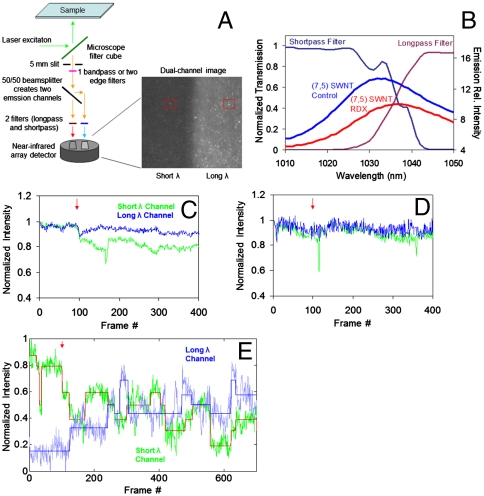
To image NIR PL spectral shifts of individual nanotubes, a detection scheme was developed (Fig. 4A). A split-channel microscope was constructed to divide an image into two channels that project adjacent spectral regions. The light from one isolated nanotube emission band is split into two beams that are filtered to permit only half of the peak’s emission to appear in each channel. One channel on a single NIR detector array projects the long-wavelength half of the emission band, and the other displays the short-wavelength half (Fig. 4B). Two images of the spectrally isolated (7,5) SWNT are collected transiently; concomitant intensity changes of the same nanotube in both channels are observed.
Fig. 4.
Single-molecule detection of RDX via split-channel microscope. (A) NIR split-channel microscope schematic. The microscope image shows the same location of the microscope field on both the short-λ and long-λ channels (red boxes), but the spectral region is adjacent in wavelength space. (B) Absorption curves of edgepass filters used in the dual-channel microscope measurements, plotted with the (7,5) SWNT PL curves before (blue) and after (red) introduction of 90 uM RDX. (C) Normalized intensity of short-λ (green) and long-λ (blue) channels of 100 averaged nanotube time traces upon addition of 9 μM RDX to surface-adsorbed bombolitin-II-bound SWNT. (D) Averaged normalized time traces of 100 nanotubes without introduction of RDX. (E) Time trace of the intensity of a single nanotube’s PL fit by an iterative error minimization. Addition of 9 μM RDX occurred at time 100 s (red arrow).
This microscope detects minute SWNT spectral shifts which signify RDX binding to bombolitin II. Nanotubes are immobilized on a glass surface and imaged in the presence of 8 μM of the peptide in Tris buffer. The emission of each PL spot in the detector is binned in a 2 × 2 pixel area and measured in each channel over the course of a movie captured at one frame per second. An aliquot of RDX is introduced to the peptide solution above the immobilized nanotubes during the course of the movie, resulting in a final concentration of 9 μM. The aggregate behavior of the emission of the nanotubes in each channel shows expected anticorrelated behavior. Upon averaging all data from 100 traces, the normalized data show that the short-wavelength channel emission decreases concomitantly with the increase of the long-λ channel after the addition of RDX (Fig. 4C), whereas the 100 control traces show no changes (Fig. 4D). The simultaneous anticorrelated behavior of the split-channel nanotube emission after RDX addition demonstrates the effect of solvatochromic shifting of individual, surface-adsorbed SWNT by RDX. An example trace of a single nanotube observed with the split-channel microscope upon introduction of RDX exhibits stepwise correlated and anticorrelated behavior (Fig. 4E and Fig S5A). The data were fit by an error-minimizing algorithm which distinguishes true steps from gradual intensity changes (24). The result suggests that solvatochromic shifts occur as discrete events on single carbon nanotubes and can be employed for molecular detection of analytes.
Polymer-Mediated Analyte Selectivity.
The polymer encapsulating a carbon nanotube significantly modulates analyte response and selectivity. Nanotubes solubilized by poly(vinyl alcohol) (PVA–SWNT), and screened against the aforementioned 42 analyte library, undergo intensity attenuation without shifting upon exposure to certain redox-active compounds (ascorbic acid, NADH, dopamine, L-thyroxine, melatonin, and seratonin) (Fig. S6A). We have previously hypothesized that such reducing agents quench excitons in nanotubes via nonradiative Auger recombination (25). Introduction of the same analytes to nanotubes encapsulated by the ss(AT)15 oligonucleotide (ss(AT)15–SWNT) (Fig. S6B) results in a different profile, with riboflavin, alpha tocopherol, and TNT exhibiting a quenching response and concomitantly preventing others. The disparities in PL responses of ss(AT)15–SWNT relative to those of PVA–SWNT are likely caused by a combination of steric effects, due to the tight, nucleobase-mediated wrapping of short oligonucleotides (26–28) and p doping of the nanotube by DNA, which has been documented (29).
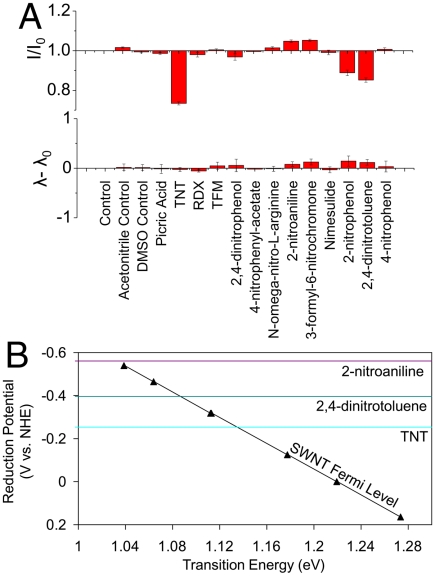
We find that encapsulation of SWNT by ss(AT)15 allows selective molecular recognition of TNT among nitroaromatics. Upon probing the complex with 13 nitro group compounds (Fig. 5A), the PL exhibits quenching in response to TNT and attenuates to a lesser degree by 2,4-dinitrotoluene and 2-nitrophenol, to the exclusion of the other analytes. Relative responses are predicted by literature values (30–32) of three analytes’ reduction potentials plotted relative to SWNT Fermi levels of several (n,m) SWNT species (from ref. 10) in Fig. 5B. The SWNT Fermi levels have not been adjusted according to polymer type. The PL-attenuating analytes TNT and 2,4-dinitrotoluene are positioned to withdraw electron density from several SWNT species, whereas 2-nitroaniline is not; the PL data show agreement.
Fig. 5.
Screening of analyte responses against polymer-encapsulated SWNT. (A) Photoluminescence intensity and wavelength responses of the (7,5) SWNT species, encapsulated by ss(AT)15, to nitro group compounds. (B) The reduction potentials of three nitroaryl compounds compared to the SWNT Fermi level. The reduction potentials of TNT and 2,4-dinitrotoluene suggest they will oxidize several SWNT species, whereas 2-nitroanaline will not.
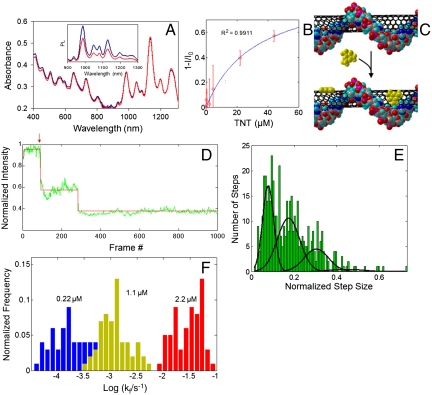
Absorption spectroscopy of ss(AT)15–SWNT interacting with TNT does not exhibit attenuation of the SWNT bands (Fig. 6A), as compared to a significant drop in PL emission on exposure to the same conditions (Fig. 6A, Inset), denoting a lack of spectral bleaching. We thus infer the sensing mechanism to be excitonic PL quenching of SWNT (13) (mechanism 2 from Fig. 1) and conclude that doping of the adsorbed oligonucleotide modulates the exciton quenching. The response fits a first-order Langmuir adsorption isotherm (Fig. 6B), suggesting reversible behavior and confirming the picture of direct binding of TNT to the SWNT surface without interacting with the encapsulating DNA (Fig. 6C).
Fig. 6.
Single-molecule, reversible excitonic quenching of ss(AT)15–SWNT by TNT. (A) Absorption spectra of ss(AT)15–SWNT before (blue) and after (red) introduction of 44 μM TNT. (Inset) PL spectra under same conditions. (B) Langmuir adsorption isotherm of TNT binding to the ss(AT)15-encapsulated (8,7) SWNT species. (C) Depiction of direct TNT binding to the SWNT surface of the ss(AT)15–SWNT complex. (D) Single-molecule TNT detection by ss(AT)15–SWNT complexes bound to glass. The trace shows the PL of a single complex acquired under 658-nm excitation. TNT (220 nM) was added at frame 100 (red arrow); one frame was acquired every 500 ms. Intensity fluctuations were fit by an error-minimizing algorithm (red). (E) A histogram of fitted intensity fluctuations from multiple nanotube intensity traces upon exposure of SWNT to 1.1 μM TNT. The histogram, fit by a series of Gaussian curves, elucidates quantization into single, double, and triple steps. (F) Histogram of quenching rates of individual nanotubes exposed to three different concentrations of TNT.
Stochastic, single-molecule detection of TNT is achieved by real-time PL measurement of ss(AT)15–SWNT adhered to a glass surface. The time trace of the emission of one SWNT (Fig. 6D and Fig. S5B) exhibits discrete quenching and dequenching steps with quantized heights, signifying single-molecule adsorption and desorption events, and confirming sensor reversibility. Traces containing at least one step were fit by an error-minimizing, step-finding algorithm as used previously (24). A histogram of fitted step heights upon exposure to 1.1 μM TNT (Fig. 6E) illustrates quantization—three separate regions of probability density occur due to single, double, and triple exciton quenching/dequenching events occurring within the time resolution of the experiment (500 ms), denoted by clustering at integer-multiple step heights. The plot was fit by a series of Gaussian curves using an automated curve-fitting function. Such quantization of normalized step heights is observed by our group (8) and others (33) in the case of single-molecule fluorescence quenching of SWNT. However, the histogram herein combines steps from multiple traces of individual nanotubes; therefore, we expect a greater dispersity of step sizes due to the presence of different (n,m) species, diverse biomacromolecule wrapping conformations, and inhomogeneities in the excitation field.
Exposure of surface-adsorbed ss(AT)15–SWNT to three different TNT concentrations results in differentiable quenching rates. Histograms (Fig. 6F) illustrate the quenching rate (kf) of each PL-emitting nanotube during the course of a 1,000-s measurement upon introduction of TNT. (Data analysis is detailed in SI Methods.) The clustering of rates from each TNT perfusion experiment into different regions denotes an increase of stepwise quenching rate with TNT concentration. Control experiments found relatively few quenching steps of any rate compared to TNT exposure. Reverse quenching (dequenching) rates of SWNT emission (kr) show less dependence on TNT concentration, consistent with earlier studies (8) (Fig. S5C).
Conclusions
This work has introduced the concept of a chaperone sensor, where an analyte is detected indirectly via the optical transduction of the secondary structure changes to a polypeptide in solution. Variants of the bombolitin class of amphipathic, bee-venom-derived peptides, not previously known for nitroaromatic recognition, undergo a unique sequence-dependent conformational change upon binding, resulting in a specific analyte response involving wavelength shifting of the SWNT emission. The induced wavelength shift permits both the fingerprinting of the analyte via analysis of the response of different SWNT species, as well as imaging of the solvatochromic shifting of single nanotubes. The imaging of single-nanotube shifts was conducted using a unique split-channel microscope to observe solvatochromic events by converting a wavelength shift into an anticorrelated intensity fluctuation that can be monitored spatially and in real time. In addition to the above mechanism, electronic and steric effects of an adsorbed biopolymer are shown to create a binding site for selective detection of a nitroaromatic analyte via excitonic quenching on the nanotube sidewall. In this case, the ss(AT)15 oligonucleotide encapsulation of SWNT results in a selective optical sensor for TNT with single-molecule resolution.
Methods
Preparation of SWNT Suspensions.
Nanotubes were suspended with DNA and peptides by sonication with PVA by dialysis (details provided in SI Methods). Bombolitin sequences used were bombolitin I, IKITTMLAKLGKVLAHV; bombolitin II, SKITDILAKLGKVLAHV; bombolitin III, IKIMDILAKLGKVLAHV; and bombolitin IV, INIKDILAKLVKVLGHV (AnaSpec).
Spectroscopy.
NIR PL spectra were obtained using 658- or 785-nm excitation and an Acton SP-150 spectrograph coupled to a Princeton Instruments OMA V InGaAs detector. Absorption measurements were obtained using a Shimadzu UV-3101PC UV-visible-NIR scanning spectrophotometer in a cuvette with a 1-cm path length. CD measurements were obtained using an Aviv Model 202 CD Spectrometer in a 1-mm path length strain-free cuvette.
High-Throughput Analyte Screening.
Analyte screening was conducted in a 96-well plate containing polymer-encapsulated nanotubes and interrogated by 42 analytes plus controls, or 13 nitro group compounds and controls (Tables S1 and S2), and measured via NIR PL spectroscopy with 785-nm excitation. Spectra were fit to a series of eight Lorentzian peaks to give analyte responses of each (n,m) SWNT species.
RDX Binding Experiments.
Bombolitin–SWNT solutions were interrogated with concentrations of up to 90 uM RDX dissolved in acetonitrile. NIR PL spectra were obtained 30 min to 1 h after mixing. Control spectra included the same acetonitrile concentrations.
Microscopy of Single SWNT.
Nanotubes were imaged on a glass-bottom Petri dish (MatTek) under 658-nm excitation using a Zeiss Axio Observer D1 microscope. Light was collected using a 256 × 320 pixel InGaAs array detector (PI Acton). Additional experimental details appear in SI Methods.
Supplementary Material
Acknowledgments.
The authors thank D. Pheasant for experimental assistance and P. Jena for aiding instrumentation design. M.S.S. acknowledges support from the National Science Foundation, an Arnold and Mabel Beckman Young Investigator Award, and the Institute for Soldier Nanotechnologies at Massachusetts Institute of Technology. The latter is funded by a grant from the Army Research Office. The Biophysical Instrumentation Facility for the Study of Complex Macromolecular Systems (National Science Foundation 0070319 and National Institutes of Health GM68762) is gratefully acknowledged.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1005512108/-/DCSupplemental.
References
- 1.O’Connell MJ, et al. Band gap fluorescence from individual single-walled carbon nanotubes. Science. 2002;297:593–596. doi: 10.1126/science.1072631. [DOI] [PubMed] [Google Scholar]
- 2.Lefebvre J, Fraser JM, Homma Y, Finnie P. Photoluminescence from single-walled carbon nanotubes: A comparison between suspended and micelle-encapsulated nanotubes. Appl Phys A Mater Sci Process. 2004;78:1107–1110. [Google Scholar]
- 3.Barone PW, Baik S, Heller DA, Strano MS. Near-infrared optical sensors based on single-walled carbon nanotubes. Nat Mater. 2005;4:86–92. doi: 10.1038/nmat1276. [DOI] [PubMed] [Google Scholar]
- 4.Strano MS, et al. The role of surfactant adsorption during ultrasonication in the dispersion of single-walled carbon nanotubes. J Nanosci Nanotechnol. 2003;3:81–86. doi: 10.1166/jnn.2003.194. [DOI] [PubMed] [Google Scholar]
- 5.Kim JH, et al. The rational design of nitric oxide selectivity in single-walled carbon nanotube near-infrared fluorescence sensors for biological detection. Nat Chem. 2009;1:473–481. doi: 10.1038/nchem.332. [DOI] [PubMed] [Google Scholar]
- 6.Shan W, et al. Pressure dependence of optical transitions in semiconducting single-walled carbon nanotubes. Phys Status Solidi B. 2004;241:3367–3373. [Google Scholar]
- 7.Cognet L, et al. Stepwise quenching of exciton fluorescence in carbon nanotubes by single-molecule reactions. Science. 2007;316:1465–1468. doi: 10.1126/science.1141316. [DOI] [PubMed] [Google Scholar]
- 8.Jin H, Heller DA, Kim JH, Strano MS. Stochastic analysis of stepwise fluorescence quenching reactions on single-walled carbon nanotubes: Single molecule sensors. Nano Lett. 2008;8:4299–4304. doi: 10.1021/nl802010z. [DOI] [PubMed] [Google Scholar]
- 9.Heller DA, et al. Multimodal optical sensing and analyte specificity using single-walled carbon nanotubes. Nat Nanotechnol. 2009;4:114–120. doi: 10.1038/nnano.2008.369. [DOI] [PubMed] [Google Scholar]
- 10.O’Connell MJ, Eibergen EE, Doorn SK. Chiral selectivity in the charge-transfer bleaching of single-walled carbon-nanotube spectra. Nat Mater. 2005;4:412–418. doi: 10.1038/nmat1367. [DOI] [PubMed] [Google Scholar]
- 11.Tu XM, Pehrsson PE, Zhao W. Redox reaction of DNA-Encased HiPCO carbon nanotubes with hydrogen peroxide: A near infrared optical sensitivity and kinetics study. J Phys Chem C. 2007;111:17227–17231. [Google Scholar]
- 12.Strano MS, et al. Reversible, band-gap-selective protonation of single-walled carbon nanotubes in solution. J Phys Chem B. 2003;107:6979–6985. [Google Scholar]
- 13.Satishkumar BC, et al. Reversible fluorescence quenching in carbon nanotubes for biomolecular sensing. Nat Nanotechnol. 2007;2:560–564. doi: 10.1038/nnano.2007.261. [DOI] [PubMed] [Google Scholar]
- 14.Choi JH, Strano MS. Solvatochromism in single-walled carbon nanotubes. Appl Phys Lett. 2007;90:223114. [Google Scholar]
- 15.Heller DA, et al. Optical detection of DNA conformational polymorphism on single-walled carbon nanotubes. Science. 2006;311:508–511. doi: 10.1126/science.1120792. [DOI] [PubMed] [Google Scholar]
- 16.Jin H, et al. Divalent ion and thermally induced DNA conformational polymorphism on single-walled carbon nanotubes. Macromolecules. 2007;40:6731–6739. [Google Scholar]
- 17.Yoon H, et al. Periplasmic binding proteins as optical modulators of single-walled carbon nanotube fluorescence: Amplifying a nanoscale actuator. Angew Chem, Int Ed. 2011;50:1828–1831. doi: 10.1002/anie.201006167. [DOI] [PubMed] [Google Scholar]
- 18.Fairbrother WJ, et al. Novel peptides selected to bind vascular endothelial growth factor target the receptor-binding site. Biochemistry. 1998;37:17754–17764. doi: 10.1021/bi981931e. [DOI] [PubMed] [Google Scholar]
- 19.Schievano E, Mammi S, Monticelli L, Ciardellaj M, Peggion E. Conformational studies of a bombolitin III-derived peptide mimicking the four-helix bundle structural motif of proteins. J Am Chem Soc. 2003;125:15314–15323. doi: 10.1021/ja0300970. [DOI] [PubMed] [Google Scholar]
- 20.Battistutta R, Pastore A, Mammi S, Peggion E. Conformational properties of the amphipathic lytic polypeptide bombolitin II—a circular-dichroism NMR and computer-simulation study. Macromol Chem Phys. 1995;196:2827–2841. [Google Scholar]
- 21.Peggion E, Mammi S, Schievano E. Conformation and interactions of bioactive peptides from insect venoms: The bombolitins. Biopolymers. 1997;43:419–431. doi: 10.1002/(SICI)1097-0282(1997)43:6<419::AID-BIP4>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 22.Dieckmann GR, et al. Controlled assembly of carbon nanotubes by designed amphiphilic peptide helices. J Am Chem Soc. 2003;125:1770–1777. doi: 10.1021/ja029084x. [DOI] [PubMed] [Google Scholar]
- 23.Goldman ER, et al. A hybrid quantum dot-antibody fragment fluorescence resonance energy transfer-based TNT sensor. J Am Chem Soc. 2005;127:6744–6751. doi: 10.1021/ja043677l. [DOI] [PubMed] [Google Scholar]
- 24.Kerssemakers JWJ, et al. Assembly dynamics of microtubules at molecular resolution. Nature. 2006;442:709–712. doi: 10.1038/nature04928. [DOI] [PubMed] [Google Scholar]
- 25.Zhang J, et al. Single molecule detection of nitric oxide enabled by d(AT)15 DNA adsorbed to near infrared fluorescent single-walled carbon nanotubes. J Am Chem Soc. 2011;133:567–581. doi: 10.1021/ja1084942. [DOI] [PubMed] [Google Scholar]
- 26.Zheng M, et al. DNA-assisted dispersion and separation of carbon nanotubes. Nat Mater. 2003;2:338–342. doi: 10.1038/nmat877. [DOI] [PubMed] [Google Scholar]
- 27.Dukovic G, et al. Racemic single-walled carbon nanotubes exhibit circular dichroism when wrapped with DNA. J Am Chem Soc. 2006;128:9004–9005. doi: 10.1021/ja062095w. [DOI] [PubMed] [Google Scholar]
- 28.Strano MS, et al. Understanding the nature of the DNA-assisted separation of single-walled carbon nanotubes using fluorescence and Raman spectroscopy. Nano Lett. 2004;4:543–550. [Google Scholar]
- 29.Shoda M, Bandow S, Maruyama Y, Iijima S. Probing interaction between ssDNA and carbon nanotubes by Raman scattering and electron microscopy. J Phys Chem C. 2009;113:6033–6036. [Google Scholar]
- 30.Hofstetter TB, Heijman CG, Haderlein SB, Holliger C, Schwarzenbach RP. Complete reduction of TNT and other (poly)nitroaromatic compounds under iron reducing subsurface conditions. Environ Sci Technol. 1999;33:1479–1487. [Google Scholar]
- 31.Kumagai Y, Kikushima M, Nakai Y, Shimojo N, Kunimoto M. Neuronal nitric oxide synthase (nNOS) catalyzes one-electron reduction of 2,4,6-trinitrotoluene, resulting in decreased nitric oxide production and increased nNOS gene expression: Implication for oxidative stress. Free Radic Biol Med. 2004;37:350–357. doi: 10.1016/j.freeradbiomed.2004.04.023. [DOI] [PubMed] [Google Scholar]
- 32.Meaney MS, McGuffin VL. Investigation of common fluorophores for the detection of nitrated explosives by fluorescence quenching. Anal Chim Acta. 2008;610:57–67. doi: 10.1016/j.aca.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 33.Cognet L, et al. Stepwise quenching of exciton fluorescence in carbon nanotubes by single-molecule reactions. Science. 2007;316:1465–1468. doi: 10.1126/science.1141316. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.