Abstract
Human O6–alkylguanine-DNA alkyltransferase (AGT), which directly reverses endogenous alkylation at the O6-position of guanine, confers resistance to alkylation chemotherapies and is therefore an active anticancer drug target. Crystal structures of active human AGT and its biologically and therapeutically relevant methylated and benzylated product complexes reveal an unexpected zinc-stabilized helical bridge joining a two-domain α/β structure. An asparagine hinge couples the active site motif to a helix–turn–helix (HTH) motif implicated in DNA binding. The reactive cysteine environment, its position within a groove adjacent to the alkyl-binding cavity and mutational analyses characterize DNA-damage recognition and inhibitor specificity, support a structure-based dealkylation mechanism and suggest a molecular basis for destabilization of the alkylated protein. These results support damaged nucleotide flipping facilitated by an arginine finger within the HTH motif to stabilize the extrahelical O6–alkylguanine without the protein conformational change originally proposed from the empty Ada structure. Cysteine alkylation sterically shifts the HTH recognition helix to evidently mechanistically couple release of repaired DNA to an opening of the protein fold to promote the biological turnover of the alkylated protein.
Keywords: O6-alkylguanine alkyltransferase/DNA repair/protein crystallography
Introduction
O6–alkylguanine lesions in DNA, which result from endogenous sources such as S-adenosylmethionine (Rydberg and Lindahl, 1982) and environmental toxins (Beranek, 1990), are carcinogenic and mutagenic, causing GC to AT transition mutations (Kyrtopoulos et al., 1997). These cytotoxic lesions are the basis of anticancer chemo– therapies that employ DNA alkylating agents to methylate (dacarbazine, procarbazine and temozolomide) or chloroethylate (bis-chloroethylnitrosourea, clomesone and fotemustine) the guanine O6-position. Human O6–alkylguanine alkyltransferase (AGT), also called O6–methylguanine methyltransferase, repairs these lesions in both free guanine bases and modified oligodeoxynucleotides by transferring the O6–alkyl group to an active site cysteine (Cys145) in an irreversible and stoichiometric reaction (Pegg et al., 1995).
Increased AGT levels parallel acquired alkylation resistance in human tumors (Brent et al., 1985; Mattern et al., 1998), and transfection of AGT confers resistance to alkylation-sensitive cells (Kaina et al., 1991), demonstrating the role of AGT in therapeutic resistance. Therefore, AGT inhibitors are potential adjuvants for these anti– neoplastic therapies, and the stoichiometric nature of AGT activity is particularly amenable to depletion by covalent reaction with any O6–alkylguanine substrate. One such inhibitor, O6-benzylguanine (O6–BG), inactivates human AGT in vitro (Pegg et al., 1993) and in vivo (Moschel et al., 1992) and enhances the efficacy of chloroethylating antitumor drugs in clinical trials (Friedman et al., 1998; Spiro et al., 1999).
Despite the therapeutic potential of AGT inhibition, there are significant therapeutic limitations of the current inhibitor, O6–BG. First, O6–BG has low affinity for AGT compared with O6–alkylguanine residues in duplex DNA (Pegg et al., 1998). Secondly, O6–BG has a low bioavailability, poor water solubility and rapid plasma clearance (Dolan et al., 1994). Extensive analog screening of O6–alkylguanine compounds has as yet failed to identify notably superior inhibitors (Moschel et al., 1992; Chae et al., 1994, 1995). A detailed structural characterization of human AGT and the basis for recognition of O6–BG should assist the structure-based design of not only improved AGT inhibitors, but also AGT mutants resistant to O6–BG and subsequent inhibitors. Such mutants may ameliorate the toxicity of cancer chemotherapies via gene therapy (Encell et al., 1999).
From the C-terminal domain structure of Ada (Ada-C), an Escherichia coli AGT homolog, alkyl transfer was proposed to require significant protein conformational changes (Moore et al., 1994). The utility of the existing unmodified Ada-C structure for human AGT inhibitor design is also limited by its distinct specificity. O6–BG, one of the best inhibitors of human AGT, does not react with Ada (Pegg et al., 1993). The structure of an inhibited AGT complex should therefore provide both an experimental test of the proposed conformational change and have increased utility for inhibitor design. We report here three high-resolution AGT crystal structures: (i) unreacted, active AGT; (ii) AGT methylated by O6–methylguanine; and (iii) AGT benzylated by O6–BG. These three structures, coupled with analysis of mutations at Arg128 and Cys145, support a unified molecular basis for the recognition of extrahelical O6–alkylguanine nucleotides, inhibitor specificity, mechanism of action and post–alkylation fate of AGT.
Results and discussion
Overall structure of human AGT
Using a single gold derivative for multi-wavelength anomalous diffraction (MAD) phasing, we first determined the crystal structure of native AGT to 2.0 Å resolution (Table I). To elucidate the basis for guanine recog– nition by AGT, we soaked crystals of AGT with O6–methylguanine and O6–BG, which caused rapid loss of diffraction and physical crystal deterioration, evidently by an alkylation-induced protein conformational change or destabilization. Diffraction-quality crystals of alkylated AGT were therefore trapped by flash freezing after exposure to O6–BG for 2 min or O6–methylguanine for 1 h. Structures of methylated and benzylated AGT were then solved by molecular replacement with the native structure, and refined to 2.0 and 2.5 Å, respectively. No electron density was seen for the repaired guanine base. Occupancies calculated from the diffraction data were 72% for the methyl adduct and 84% for the benzyl adduct.
Table I. Crystallographic data collection and refinement statistics.
| AGT | Methyl-AGT | Benzyl-AGT | Au-AGTa | ||||
|---|---|---|---|---|---|---|---|
| X-ray diffraction data | |||||||
| Wavelength (Å) | 1.08 | 1.08 | 1.08 | 1.0781 | 1.0400 | 1.0397 | 1.0162 |
| Unit cellb (Å) | |||||||
| a, b | 71.28 | 71.40 | 71.60 | 71.75 | 71.75 | 71.75 | 71.75 |
| c | 73.61 | 76.33 | 76.04 | 75.08 | 75.08 | 75.08 | 75.08 |
| Resolution (Å) | 2.00 | 2.00 | 2.50 | 2.30 | 2.00 | 2.14 | 2.05 |
| Observations | 47 948 | 28 548 | 16 165 | 57 959 | 103 385 | 84 949 | 98 569 |
| Unique reflections | 14 538 | 15 222 | 7991 | 10 030 | 15 112 | 12 357 | 14 165 |
| I/σ | 17.1 | 13.5 | 8.9 | 22.1 | 25.6 | 24.3 | 24.8 |
| Completeness (%) | 97.0 | 97.2 | 98.9 | 95.7 | 96.7 | 96.5 | 97.1 |
| Final shell | 96.7 | 98.4 | 98.7 | 85.3 | 90.3 | 87.3 | 93.8 |
| Rsymc | 0.061 | 0.051 | 0.106 | 0.073 | 0.055 | 0.064 | 0.058 |
| Final shell | 0.367 | 0.255 | 0.444 | 0.310 | 0.204 | 0.332 | 0.293 |
| Mosaicity (°) | 0.96 | 0.52 | 0.73 | 0.74 | 0.54 | 0.59 | 0.56 |
| Model refinement | |||||||
| Resolution (Å) | 20.0–2.0 | 20.0–2.0 | 20.0–2.5 | ||||
| Rcrystd | 0.197 | 0.199 | 0.194 | ||||
| Rfreee | 0.218 | 0.229 | 0.230 | ||||
| Residues | 5–35, 45–181 | 5–35, 46–176 | 5–35, 45–179 | ||||
| Protein atoms | 1275 | 1235 | 1270 | ||||
| Solvent atoms | 153 | 182 | 86 | ||||
| R.m.s.d. bonds (Å) | 0.007 | 0.007 | 0.009 | ||||
| R.m.s.d. angles (°) | 1.40 | 1.35 | 1.37 |
aInitial figure of merit = 0.69, density modified = 0.82 (20.0–2.5 Å).
bSpace group = P3121.
cRsym = Σhkl ∣ I(hkl) – < I(hkl) > ∣ / Σ < I(hkl) >
dRcryst = Σhkl ∣ Fobs(hkl) – < Fcalc(hkl) > ∣ / Σhkl < Fobs(hkl) >
eRfree = Rcryst, calculated using the 10% of reflections against which the model was not refined.
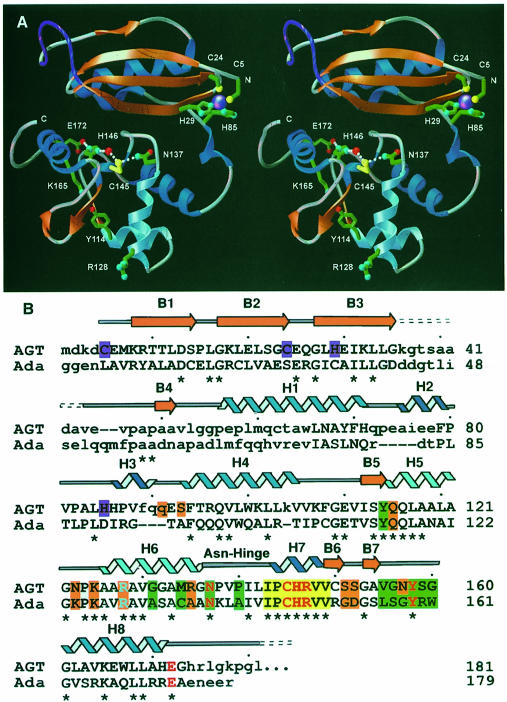
The structures of human AGT exhibit a two-domain α/β fold with overall dimensions of ∼20 × 35 × 40 Å (Figure 1A). The N-terminal domain α/β roll (residues 1–85) is comprised of a four-stranded anti-parallel β–sheet, a single α–helix and a 310–helix. AGT residues 36–44, part of a solvent-exposed and disordered loop following β3, correspond to the first Ada α–helix, a major difference between the human and bacterial structures (Figure 1B). Other differences include the tilting of AGT helix H1 by ∼30°, and the presence of two 310–helices (H2 and H3) in the AGT structure. The remaining secondary structure elements are surprisingly conserved between AGT and Ada despite low primary amino acid sequence homology (14% identity, 32% similarity). Helix H3 spans the cleft between the N- and C-terminal domains.
Fig. 1. Human AGT secondary structure, two-domain fold, zinc site, and location of structurally and catalytically critical residues. (A) Overall fold, displaying the active site cysteine (yellow) and its surrounding hydrogen-bond network, zinc (purple) ligands and Arg128 ‘arginine finger’. A central interdomain cleft separates the N-terminal α/β roll (α–helices in royal blue, β-strands in orange) from the C-terminal domain, which contains the active site and HTH DNA-binding motif (sky blue). For continuity, the location of the internal disordered loop (purple) has been interpolated. (B) Primary sequence, secondary structure and residue function of human AGT aligned with the E.coli Ada-C protein. Identity between the two sequences is shown with an asterisk, while corresponding residues that lie within 4.5 Å in the overlaid structures are given in upper case letters. The conserved active site motif (yellow boxes) is flanked by residues defining the O6–alkylguanine-binding channel (green boxes), anticipated DNA-binding residues (orange boxes), and HTH motif, with its associated Arg128 (sky blue). Colors distinguish residues participating in the active site hydrogen-bond network (red), zinc ligands (purple), α–helices (royal blue) and 310–helices (navy blue).
The C-terminal domain (residues 86–207), composed of a short two-stranded parallel β–sheet and five helices, contains the conserved active site cysteine motif (IPCHRV), the O6–alkylguanine-binding channel and the helix–turn–helix (HTH) DNA-binding motif. The second helix of the HTH motif (H6), termed the ‘recognition’ helix due to its interaction with the major groove of DNA in transcription factors (Wintjens and Rooman, 1996), precedes a 310–helix (H7) that bears the conserved IPCHRV active site motif (Figure 1A). The nucleophilic cysteine lies near the bottom of a solvent-accessible groove ∼7 Å wide, 9 Å deep and 14 Å long. Coupling helices H6 and H7 are two overlapping tight turns stabilized by Asn137, which has hydrogen bonds to the main chain of Val139 and Ile143 and the thiol of Cys145. This ‘Asn-hinge’ contacts β3 of the N-terminal domain through the inter– domain cleft and contributes >40% of the buried surface area between the two domains. Hydrophobic contacts that stabilize the domain interface between the Asn-hinge and the N-terminal domain include Val139, Ile141, Leu142, plus Ile143, which is a site of natural polymorphism in AGT (Deng et al., 1999).
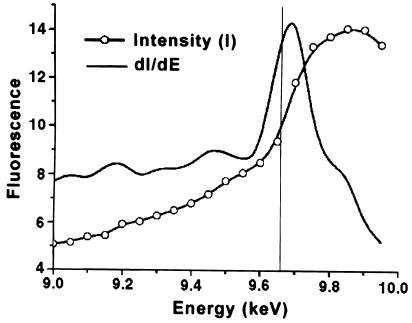
An unexpected Zn(II) ion bridges three strands of the β–sheet and the coil immediately preceding the domain-spanning helix H3 (Figure 1A). This metal atom lies on the active site face, but ∼20 Å from the reactive cysteine. The tetrahedral Cys5, Cys24, His29, His85 coordination (Figure 1A) and the X-ray fluorescence scan (Figure 2) are characteristic of a zinc atom. As zinc was not added during expression and purification, the protein evidently scavenged enough of the ion to contain zinc at 86.8 ± 1.2% occupancy, as determined by spectrophotometric assay (see Materials and methods). The role of this zinc atom is likely to be structural, serving to stabilize the interface between the N- and C-terminal domains by tethering the domain-spanning helix H3 to three strands of the N-terminal β–sheet. Notably, the four zinc ligands are conserved in higher eukaryotes and Leu84, adjacent to the ligand His85, is a site of genetic polymorphism (Deng et al., 1999).
Fig. 2. Identification of a zinc-binding site in human AGT. X-ray fluorescence of non-zinc-supplemented crystals of AGT was measured as a function of incident radiation. The position of the inflection point in the intensity (I) measurement and the maximum in the first derivative curve (dI/dE) correspond well to that expected for the K absorption edge for zinc (vertical line).
During review of this manuscript, the crystal structure of an N-terminally His-tagged, C-terminally truncated AGT construct was reported (Wibley et al., 2000), providing an experimentally independent structure for comparison to the three structures reported here. The secondary structure of the truncated construct agrees with our structure of native human AGT, with a root mean square deviation (r.m.s.d.) of 0.95 Å for all atoms and 0.48 Å for main chain atoms. Surprisingly, while they were obtained from distinct crystallization conditions, the two crystal forms have the same space group and comparable unit cell dimensions. Additionally, the structures were refined to nearly identical resolution and Rfree values, and both possess a disordered loop following β3. The striking difference is that the model of the His-tagged, truncated construct lacks a zinc atom, possibly due to its purification over metal affinity columns. Structurally, this apo-metal ion site results in local distortions in the N-terminal domain, including a slight opening of the zinc ligands (Cys5, Cys24, His29, His85) and surrounding residues (Glu6, His86). In addition, Pro56, adjacent to the zinc site, isomerizes from the cis conformation in our three AGT structures to the trans conformation in the zinc-free structure, which slightly opens the interdomain cleft of the apo protein. Loss of zinc is also presumably responsible for the increased disorder in the N-terminus of the truncated construct, in which Cys5, one of the zinc ligands in our structure, is not modeled. Comparison of the zinc-bound and apo structures suggests that the zinc site stabilizes the domain interaction and fold. Loss of zinc stabilization may therefore account for the ∼2-fold reduction in the apparent second-order rate constants observed for His-tagged relative to native recombinant AGT (Goodtzova et al., 1998).
Alkyl-binding pocket and substrate selectivity
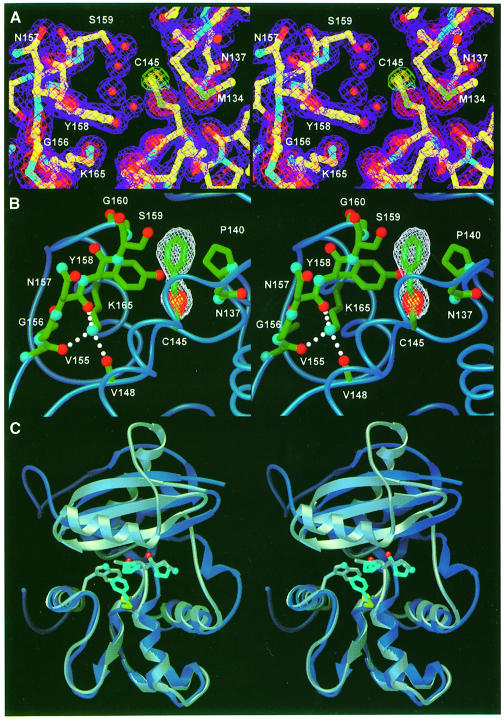
Structures of methylated and benzylated AGT, obtained by the reaction of AGT with O6–methylguanine and O6–BG, definitively establish the alkyl-binding pocket and suggest the structural basis for O6–alkylguanine recognition (Figure 3A and B). The benzyl group, covalently bound to Cys145, stacks between Pro140 and the Cβ of Ser159, and has an edge-on hydrophobic interaction with the Tyr158 side chain. AGT reacts with a variety of O6–BG analogs, preferring para- to meta-substituted derivatives, while ortho-substituted derivatives are markedly poorer substrates (Chae et al., 1994; Mineura et al., 1995). The structural basis for this substrate selectivity is clear from the benzylated product complex. The side chain of Tyr158 and helix H6 flank either side of the benzyl ring at ∼3.5 Å to preclude substituents at the ortho positions. The Gly160 Cα packs against one meta position at a distance of 4.2 Å, resulting in the tolerance of AGT toward a single meta-substituent, but the decrease in rate observed with two substituents. The para-carbon position lies exposed near the interdomain cleft, explaining the variety of bulky substituents that are permitted at this position.
Fig. 3. The methylated and benzylated AGT adducts identify the alkyl-binding pocket and suggest the bases for species-dependent substrate specificity. (A) Methylated AGT active site and electron density from a σA-weighted 2Fo – Fc map (2σ, purple; 4σ, red) and omit electron density excluding the methyl adduct (2σ, green; 3σ, gold). Four ordered water molecules adjacent to the active site cysteine have replaced the repaired guanine. (B) Omit electron density for benzylated AGT (2σ, white; 6σ, red) is shown for maps calculated excluding the benzyl adduct. The benzyl group stacks between the ring of Pro140 and the Cβ of Ser159, and is flanked by the side chains of Tyr158 and Asn137 (oxygens, red; nitrogens, blue; carbons, green). (C) The structural basis for differential affinity of O6–BG between human AGT (royal blue) and E.coli Ada-C (gray). The AGT alkyl-binding pocket, shown by the benzylated cysteine (yellow and sky blue), is partially filled by Trp161 in Ada-C. Additionally, alteration of Pro138 and Pro140 (sky blue) of AGT to Lys and Ala (gray), respectively, results in a narrowing of the alkyl-binding pocket.
Mutations affecting O6–BG sensitivity
Structural analysis of the active site channel structure provides a coherent understanding of numerous mutations that alter O6–alkylguanine recognition and repair (Figure 3B and C). Mutation of Pro140, which forms the floor of the alkyl-binding pocket, to lysine is the most O6–BG-resistant mutant yet identified, with an ED50 104-fold greater than that of wild-type protein (Xu-Welliver et al., 1998). Pro140Ala, the least resistant mutation at that site, still exhibits a 40-fold increase in ED50. Mutation of Pro140 disrupts the stacking between the benzyl group and the proline ring, thereby decreasing the affinity of the protein for O6–BG. In particular, extended and/or charged residues, such as lysine should effectively preclude binding of the benzyl group. Similarly, replacement of Tyr158, which defines one wall of the alkyl-binding pocket (Figure 3B), with histidine creates an unfavorable interaction between the charged imidazole ring and the closely packed benzyl group, raising the ED50 by 6.2 × 103 (Xu-Welliver et al., 1999b). Likewise, the introduced charge of Gly160His and Gly160Arg mutants would repel the benzyl group (Figure 3B), explaining the O6–BG resistance conferred by these mutations (Xu-Welliver et al., 1999a). The Gly160Trp mutation, adjacent to the alkyl-binding pocket, increases the sensitivity of AGT toward O6–BG (Edara et al., 1996), presumably by introducing a favorable hydrophobic packing interaction with the benzyl moiety.
Besides these side chain interactions with the O6–alkyl group, structure-based analysis of mutational data suggests that substitutions at Gly156 and Lys165 confer resistance to O6–BG through backbone distortions. Gly156 flanks the alkyl-binding pocket, between β5 and α7 (Figure 3B). All 11 tested mutations of Gly156 increase resistance to O6–BG (Xu-Welliver and Pegg, 2000). The allowed torsion angles of non-glycine residues require backbone conformational changes that prabably distort the adjacent loop (residues 158–160), which defines one wall of the benzyl-binding pocket. Similarly, all 19 mutations generated at Lys165 significantly increase resistance to O6–BG (Xu-Welliver et al., 2000) since the lysine side chain participates in a hydrogen bond with the carbonyls of Val148, Val155 and Asn157, stabilizing this active site loop (Figure 3B).
Differential inhibition of AGT and Ada by O6–BG
The active site channel of human AGT reveals the structural basis for differences in inhibitor specificity between the human and bacterial proteins. While the ED50 of O6–BG with AGT is 0.2 μM (Moschel et al., 1992), the E.coli Ada-C protein does not react with this compound (Pegg et al., 1993; Elder et al., 1994). The overlay of Ada-C and AGT indicates that Trp161 (corresponding to Gly160 in AGT) partially fills the benzyl-binding pocket (Figure 3C). Thus, Ada-C sterically excludes O6–BG from the binding pocket, while still repairing the smaller O6–methylguanine. Additionally, relaxation of the helix H6 C-terminus to α–helix in Ada due to the absence of prolines 138 and 140 (Lys139 and Ala141 in Ada-C) partially closes the alkyl-binding pocket, while loss of Pro140 removes hydrophobic stacking interactions. Therefore, the Ala141Pro/Trp161Ala double mutant increases the inhibition of Ada by O6–BG (Crone et al., 1995).
Guanine-binding pocket
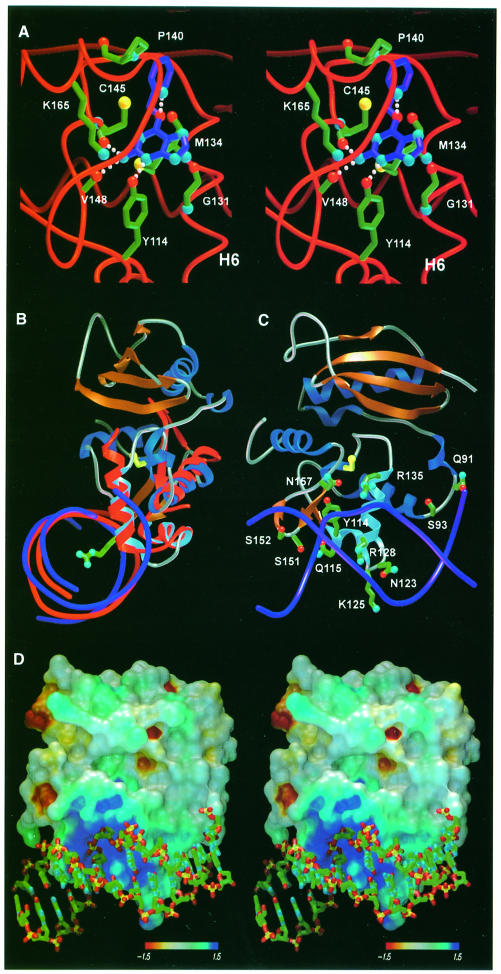
The position of alkylated Cys145 and conserved active site residues identifies the guanine-binding pocket and features that probably govern guanine recognition. Flanked on either side by the Tyr158 side chain and Asn137 backbone, the S-benzylcysteine residue is not free to rotate around the bond between the sulfur and the benzylic carbon (Figure 3B). Even the smaller methyl adduct occupies this same position, and is similarly unable to undergo bond rotation to reduce the close contact between the methyl group and the carbonyl of Met134, implying that this is the functionally relevant alkyl-binding orientation (Figure 3A). Superposition of an energy-minimized structure of O6–BG on the benzyl adduct, followed by translation of the substrate away from Cys145 to a minimum van der Waals distance, brings the O6-position of guanine within hydrogen-bond distance of the Ser159 backbone nitrogen (Figure 4A). The guanine base is then centered in the 8 Å groove defined by the backbone of Asn157/Tyr158 and helix H6, and packed between the main chain of Asn157, Tyr158, Gly131 and the side chain of Met134. This arrangement brings the guanine N3 within hydrogen-bond distance of the Tyr114 hydroxyl and positions the exocyclic amino group for hydrogen bonding to the carbonyls of Cys145 and Val148 (Figure 4A). The resulting positions of the O6, N2 and N3 guanine atoms correspond to well-ordered active site water molecules.
Fig. 4. Structurally inferred guanine- and DNA-binding mode of AGT. (A) Guanine inserts into the active site channel along the recognition helix, stacking the aromatic base against Met134 and Gly131. Guanine-specific hydrogen bonds occur between the Watson–Crick base-pairing atoms of guanine and protein main chain atoms. The benzyl lesion is positioned adjacent to the active site cysteine, and oriented for optimal SN2 displace– ment. (B) Overlay of AGT and the HTH-containing DNA-binding region of the catabolite gene activator protein bound to DNA (both red), and proposed analogous binding of DNA (purple) by AGT. Arg128, extending from the HTH motif (sky blue), penetrates the base stack and is ideally positioned to facilitate flipping of target O6–alkylguanine nucleotides. (C) Potential DNA-contacting residues of AGT (same color scheme as for Figure 1A), rotated by 90° about the vertical axis relative to (B). The HTH motif (sky blue), lying within the major groove, provides several potential hydrophilic and electrostatic DNA contacts. The adjacent ‘wing’, corresponding to the turn between B6 and B7, contacts the minor groove through Ser151 and Ser152. (D) The electrostatic surface of AGT, oriented as (C) and colored by Coulombic electrostatic potential (kT/e–), displays a positively charged region centered around Arg128 complementary to the DNA phosphate backbone (oxygens, red; phosphates, yellow). Arg128, the ‘arginine finger’, facilitates damage repair by entering the base stack and displacing the extrahelical O6–alkylguanine nucleotide.
These structurally inferred interactions are consistent with activity studies of O6–BG analogs. Notably, removal of the exocyclic amino group, which probably participates in two hydrogen bonds, results in a 400-fold decrease in the inactivation of AGT by free O6–BG (Moschel et al., 1992), but has no effect on the binding of modified oligonucleotides (Spratt et al., 1999), where protein–DNA contacts compensate for the loss of these two hydrogen bonds. Furthermore, substituents at solvent-exposed C8 (Chae et al., 1995) and N9 (Chae et al., 1994) positions are tolerated. Finally, removal of the N3–Tyr114 hydrogen bond by mutation of the highly conserved Tyr114 results in a 100-fold decrease in reactivity with O6–BG (Goodtzova et al., 1998).
Structurally implicated DNA-binding mode
To elucidate the manner in which AGT binds DNA, we used the DALI structural comparison algorithm (Holm and Sander, 1993) to search for DNA-binding proteins with homology to AGT. Of the numerous HTH-containing proteins we identified, the E.coli catabolite gene activator protein (CAP; Protein Data Bank ID 2CGP) had the highest structural homology to AGT, with its three DNA-binding helices having a main chain r.m.s.d. of 0.93 Å from the 32 AGT residues of helices H4, H5 and H6 (Figure 4B). From our AGT structures and the CAP–DNA complex crystal structure (Schultz et al., 1991), a specific DNA-binding mode for AGT was inferred (Figure 4).
The implicated DNA-binding surface of AGT includes the HTH motif (H5, H6), the preceding helix (H4) and the B5–B6 β-turn (Figure 4B and C). The recognition helix, H6, is inserted into the DNA major groove, and the N-terminal residues of helices H4 and H5 interact with the phosphate backbone, as seen for CAP and structures of HTH-containing transcription factors (Wintjens and Rooman, 1996). Additionally, the AGT B5–B6 β-turn, analogous to the ‘wing’ of winged-HTH DNA-binding motif (Brennan, 1993), is poised to interact with the minor groove through Ser151 and Ser152 side chains (Figure 4C).
This testable motif-based DNA-binding mode is consistent with other pertinent structural results. First, the negatively charged DNA phosphodiester backbone matches the complementary positively charged surface of AGT centered at Arg128 (Figure 4B and D). Secondly, this surface shows a significantly higher evolutionary conservation than the remaining protein, indicating its importance in the biological function of AGT (Figures 1B and 4C). Finally, helix H6, lying within the major groove, presents residues lacking side chain hydrogen-bond capability (Ala127, Ala129, Gly131, Gly132) for sequence-independent DNA repair.
The overlay of AGT and CAP places Arg128, at the N-terminus of the recognition helix H6, within the DNA base stack (Figure 4B), suggesting that AGT employs an ‘arginine finger’ to extrude target O6–alkylguanine nucleotides from duplex DNA. O6–alkyl lesions occupy the major groove, and can induce a wobble base pair, further displacing the lesion into the major groove (Kalnik et al., 1989). Arg128, which enters the base stack from the major groove in our model, could therefore promote the extrusion of O6–alkylguanines and stabilize the gap left by the resultant extrahelical nucleotide. The ability of crystallized AGT to react with O6–alkylguanines without the conformationl change that was originally proposed (Moore et al., 1994) suggests that the target base must be extrahelical to reach the recessed AGT active site. DNA co-crystal structures of 3–methyladenine-DNA glycosylase (Lau et al., 1998) and uracil-DNA glycosylase (UDG) (Slupphaug et al., 1996) show similar penetration of DNA by single amino acid side chains to promote nucleotide flipping.
To test the role of conserved Arg128 in DNA binding, we made and characterized Arg128 mutations (Table II). Substitution of the arginine with lysine, which is long enough to penetrate the base stack but has a diminished hydrogen-bond capacity, results in a modest loss in activity with methylated duplex DNA. Arg128Leu, which may stack in the hydrophobic interior as seen for UDG (Parikh et al., 1998), but has no hydrogen-bond capability, shows a 5-fold reduction in dealkylation activity. Substitutions with smaller amino acids result in a dramatic loss of activity, with the Arg128Gly mutation showing <0.02% of wild-type activity. Notably, these mutations have no significant effect on the reaction of AGT with free O6–BG and have a minimal effect on DNA binding (Table II), demonstrating that they do not slow DNA association or the alkyl-transfer step of the reaction. Moreover, the relative retention of activity in the Arg128Leu mutations argues against general phosphate recognition by this residue. This mutational trend for Arg128 qualitatively mirrors that seen in the nucleotide-flipping enzyme UDG, where mutation of the analogous leucine finger to alanine decreases its DNA-binding ability (Parikh et al., 1998), while the corresponding arginine mutation increases DNA binding (Slupphaug et al., 1996).
Table II. DNA binding and repair by R128 mutants.
| Protein | Binding to O6–methyl-containing oligonucleotide |
Rate of repair |
||
|---|---|---|---|---|
| Double stranded KD (μM) | Single stranded KD (μM) | O6–BG k2 (M–1 min–1 × 10–2) | Methylated DNA k2 (M–1 min–1 × 10–6) | |
| Wild type | NMa | NMa | 6.1 | 38 |
| C145A | 0.13 | 0.69 | inactive | inactive |
| R128K | NMa | NMa | 7.3 | 24 |
| R128L | 2.6 | 6.1 | 7.1 | 10 |
| R128E | 3.4 | 11 | 7.8 | 0.6 |
| R128D | 4.2 | 22 | 5.1 | 0.03 |
| R128A | 3.1 | 28 | 8.4 | 0.02 |
| R128G | ND | ND | 6.1 | <0.006 |
ND, not determined.
aRate of repair prohibits the use of a gel shift assay. The inactive C145A mutant is included as a surrogate of these values. There was no difference in the binding of wild type and C145A to oligo– nucleotides that do not contain O6–methylguanine (KD = 0.6 μM for double-stranded DNA, 1.0 μM for single-stranded DNA).
Structural implications for alkyl transfer and protein destabilization
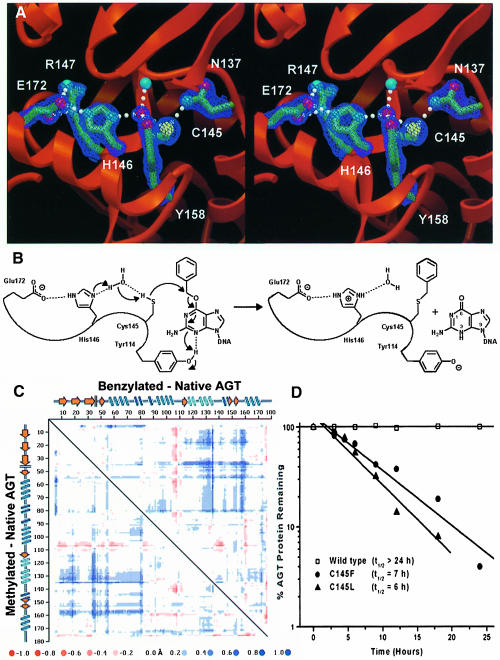
The Cys145 region displays an extensive hydrogen-bond network that is critical for protein stability (Crone et al., 1996), and poised to drive the dealkylation reaction. A well-ordered water molecule exhibits four short, nearly tetrahedral, hydrogen bonds to the side chains of Cys145 and Tyr158, the Nδ of His146 and the backbone nitrogen atom of Ile141 (Figure 5A). His146 stacks in a hydrophobic slot formed by the conserved side chains of Pro144, Arg147 and Leu168. In this hydrophobic environment, His146 is likely to be neutral, accepting a hydrogen bond from Wat1 and donating one to the negatively charged Glu172 carboxylate, which pairs with Arg147 in a salt bridge. We propose that proton abstraction occurs through this hydrogen-bond network to assist the nucleophilic attack by Cys145 (Figure 5B). The Tyr114 side chain may act as a general acid to protonate guanine at N3, consistent with the decrease in reaction rate that occurs when N3 is substituted with carbon (Spratt et al., 1999), or with mutations at Tyr114 (Goodtzova et al., 1998).
Fig. 5. AGT active site structure, dealkylation mechanism and stability of mutants mimicking alkylated AGT. (A) Active site hydrogen bond network, required for both stability and activity. Electron density from a σA-weighted 2Fo – Fc map contoured at 2σ is shown. (B) A proposed reaction mechanism for AGT. His146, acting as a water-mediated general base, deprotonates Cys145 to facilitate attack at the O6–alkyl carbon, with concomitant protonation of N3 by Tyr114. (C) Distance difference matrix plot of benzylated (upper right) and methylated (lower left) versus native AGT showing 0.5–1.5 Å shifts of helix H6 (residues 125–136) and the guanine-binding loop (residues 153–160) away from the N-terminal domain. This opening of the tertiary structure, accommodating the alkyl adducts, distorts the DNA-binding surface. (D) Instability of Cys145 mutants. Wild-type and C145F and C145L mutants were expressed in E.coli lacking endogenous AGT. Following arrest of protein synthesis, the presence of AGT was measured as a function of time by immunoblotting of cell lysates with anti-AGT antibodies. C145F and C145L mutants, which mimic alkylated C145, demonstrate in vivo instability.
Following alkylation of Cys145, AGT shows an increased Stokes radius (Hora et al., 1983) and increased susceptibility to proteolysis (Kanugula et al., 1998). Additionally, the inactivated protein is rapidly degraded in HT29 cells and cell-free extracts (Pegg et al., 1991b), probably due to its increased ubiquitylation (Srivenugopal et al., 1996). These results suggest a conformational change in AGT following DNA repair that may drive its proteolytic degradation. Comparison of the AGT structures before and after alkylation (Figure 5C) implies that the disruption of an essential hydrogen-bond network and a sterically driven helix displacement result in this conformational change, probably promoting release of repaired DNA and in vivo protein degradation.
The methyl and benzyl adducts of the two product complexes lie in close van der Waals contact (3.2 and 3.0 Å, respectively) with the carbonyl oxygen of Met134 within helix H6 of the Arg128-containing HTH motif (Figure 3A and B). In both the alkylated structures, movement of H6 away from the active site relative to the native structure has partially relieved the strain from this close contact. An unbiased difference distance matrix plot of the two alkylated proteins compared with the native structure reveals 0.5–1.5 Å Cα shifts of the recognition helix H6 away from the N-terminal domain of the protein in response to alkylation (Figure 5C). This expansion appears to be the maximum H6 movement allowed in the intact crystal and probably causes the rapid crystal decay resulting from exposure to inhibitors. Importantly, the helix H6 movement distorts the DNA-binding region and proposed DNA major groove interactions, providing a molecular mechanism to facilitate the dissociation of repaired DNA.
Alkylation of Cys145 and the resultant recognition helix displacement may help explain the in vivo destabilization of AGT. Sterically driven movement of the HTH helix H6, which is joined to the Asn-hinge by Asn137, and direct steric collision of Asn137 with the alkyl adduct probably disrupt the three hydrogen bonds of the Asn137 side chain and open the Asn-hinge (Figure 5C). Importantly, the Asn137 side chain is crucial for protein stability, as the Asn137Ala mutation results in dramatic protein destabilization (Crone et al., 1996). As the hydrophobic packing between the Asn-hinge and the N-terminal domain constitutes >40% of the domain interface, disrupting this contact should open the tertiary structure of AGT, providing a mechanism for protein destabilization. To partially test this hypothesis, we made and characterized Cys145Leu and Cys145Phe mutations, which mimic alkylated AGT, and found that they have dramatically shorter in vivo lifetimes than the native protein (Figure 5D). Therefore, the Asn-hinge, which joins the DNA-recognition helix to the Cys145-containing helix, may provide the structural basis to couple the dealkylation of DNA to both the release of repaired DNA and turnover of inactivated AGT.
Materials and methods
Expression of AGT and synthesis of O6–BG
Recombinant human AGT was expressed at high levels in E.coli using the pXC35 hAGT plasmid (Kanugula et al., 1998) and purified to homogeneity using Polymin P precipitation, ammonium sulfate fractionation and FPLC on a Mono S column as described previously (Pegg et al., 1993). O6–BG was prepared by treating 2-amino-6-chloropurine (1.2 mmol) with 2.5 equivalents of sodium benzyloxide in 8 ml of toluene at 90°C for 15 h. The reaction mixture was neutralized with glacial acetic acid and the product isolated following precipitation with diethyl ether. Crystallization of the precipitated solid from ethanol/water afforded O6–BG, confirmed by mass spectrometry and 1H NMR, in a 35% yield (Dolan et al., 1990).
Crystallization and data collection
Crystals of human AGT were grown by hanging drop vapor diffusion at 25°C with 18 mg/ml purified protein against 1.5 M sucrose. A single isomorphous gold derivative was obtained by soaking crystals in mother liquor containing 1 mM Au(CN)2 for 24 h. Methylated and benzylated AGT products were obtained by soaking native crystals in mother liquor containing 2 mM O6–BG or commercially available O6–methylguanine for 2 min and 1 h, respectively. Longer soaking times resulted in loss of diffraction followed by crystal decay within 24 h. All crystals belonged to the space group P3121, with one molecule per asymmetric unit. Data for native, methylated and benzylated AGT crystals were collected at –180°C with a MAR research imaging plate from crystals flash-frozen in mother liquor at the Stanford Synchrotron Radiation Laboratory beamline 7-1. Au(CN)2 derivative data were collected at the peak absorption, inflection point, and high and low energy remote wavelengths, as determined by X-ray fluorescence, on a 2X2 Area Detector Systems Corporation CCD detector at the Advanced Light Source beamline 5.0.2. Data processing and reduction were performed with DENZO and SCALEPACK (Otwinowski and Minor, 1997).
Structure determination and analysis
Initial phases were calculated by MAD phasing with the SOLVE program package (Terwilliger and Berendzen, 1999) and phase improved by density modification with DM (Zhang and Main, 1990; Cowtan and Main, 1993). Model building and subsequent electron density map fitting were undertaken with XFIT (McRee, 1992). Refinements were carried out using Crystallography and NMR system (CNS) (Brunger et al., 1998), with overall anisotropic and bulk solvent correction applied to the data. All structures were inspected manually with simulated annealed omit and σA-weighted 2Fo – Fc and Fo – Fc electron density maps, and progress confirmed by the steady decrease in both Rcryst and Rfree. The highly flexible, glycine- and serine-rich C-terminal tail of AGT was not visible in the electron density for these three structures. However, truncation mutants have shown that these residues are not required for dealkylating or DNA-binding activities (Hazra et al., 1997).
For comparative analysis, structures of methylated AGT, benzylated AGT and E.coli Ada-C were superimposed using the CCP4 program suite (Collaborative Computational Project Number 4, 1994). A structure-based primary sequence alignment of AGT and Ada-C was constructed from the global alignment algorithm of SEQUOIA (http://www.scripps.edu/~bruns/sequoia.html). The surface electrostatic potential of AGT was calculated with protein and solvent dielectric constants of 2.0 and 78.0, respectively, using the University of Houston Brownian Dynamics Program (Davis et al., 1991). The catabolite gene activator protein was identified as one HTH-containing protein structurally similar to AGT by using the DALI distance matrix alignment algorithm (Holm and Sander, 1993).
Spectrophotometric zinc assay
The zinc occupancy of hydrolyzed AGT was quantitated by complexation with the zinc-binding chromophore 4-(2-pyridylazo)resorcinol (PAR) (Hunt et al., 1985; Casadevall and Sarkar, 1998). In brief, AGT was hydrolyzed for 17 h in 6 M HCl, and neutralized with NaOH. PAR was added to hydrolyzed protein (1–16 μM) to a final concentration of 100 μM, and the absorption measured at 500 nm. Using the absorption from zinc standards (1–16 μM), linear over this concentration range, the zinc occupancy was determined as 86.8 ± 1.2%. Control hydrolysis reactions without protein showed no absorption at 500 nm.
Mutant protein preparation
The C145A, R128K, R128L, R128E, R128D, R128A and R128G mutant proteins were prepared after expression as His-tagged proteins in E.coli strain JM 109 using the pQE30 vector as described (Edara et al., 1996). Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the cultures at a cell density equivalent to A600 of 0.5 and the cells were harvested 4 h later. The pellet from a 1–2 l culture was disrupted using a French Press at 4°C. After centrifugation at 17 000 g for 45 min at 4°C to remove cell debris, the supernatant was applied to a column of Talon IMAC resin (Clonetech) equilibrated with 20 mM Tris–HCl pH 8.0, 250 mM NaCl and the column washed with this buffer containing 10 mM imidazole. The AGT protein was then eluted using 200 mM imidazole and the fractions found to contain AGT by SDS–PAGE were pooled and dialyzed immediately against 50 mM Tris–HCl pH 7.6, 250 mM NaCl, 5 mM dithiothreitol (DTT) and 0.1 mM EDTA. The purity was >90% as judged by SDS–PAGE.
Kinetic measurements of AGT activity
The second-order rate constant for the repair of methylated DNA was determined by measuring the rate of transfer of methyl groups from a [methyl-3H]DNA substrate to the AGT protein (Goodtzova et al., 1998). The concentrations of AGT protein used were determined in preliminary experiments as those giving readily measurable rates under the assay conditions. The assay mixture contained AGT protein (in the range from 5 × 10–10 to 1.2 × 10–6 M depending on the mutation), 3.6 × 10–10 M O6-[3H]methylguanine in 3H–methylated DNA substrate and 50 μg of unlabeled calf thymus DNA in a buffer containing 50 mM Tris–HCl pH 7.6, 5 mM DTT and 0.5 mM EDTA. Since the reaction was found to be second order, the rate constant was determined by the following equation:
k(t) = 1/C0a – C0b ln(C0b(C0a – Ct)/C0a(C0b – Ct))
where C0a is the initial concentration of the AGT protein, C0b is the initial concentration of the methylated DNA substrate and Ct is the concentration of methylated AGT formed at a given time t.
The rate of reaction with O6–BG was measured by following the formation of [8-3H]guanine from O6-[8-3H]BG over a 20 min incubation in an assay mixture containing 0.5–3.0 μM AGT or mutant proteins, 1–4 μM O6-[8-3H]BG, 50 mM Tris–HCl pH 7.5, 0.1 mM EDTA and 5 mM DTT in a volume of 0.25 ml and the second-order rate constant calculated as described (Pegg et al., 1993).
Binding of AGT and mutants to DNA
The binding of AGT to double- and single-stranded oligodeoxynucleo– tides was measured by electrophoretic mobility shift assays with vary– ing amounts of protein using the 16mer 5′-AACAGCCATAT (O6-meG)GCCC alone and annealed to 5′-GGGCCATATGGCTGTT. The oligodeoxynucleotides were 5′-end labeled using [γ-32P]ATP and T4 polynucleotide kinase, and then annealed as indicated to the complementary oligodeoxynucleotide. Increasing concentrations of AGT protein were mixed with 10 nM 32P-labeled DNA probe in a 20 μl reaction mixture containing 20 mM Tris–HCl pH 7.6, 5 mM DTT, 0.1 mM EDTA and 3% glycerol. The reaction mixtures were incubated for 10 min on ice and loaded onto pre-run, pre-equilibrated (4°C) 6% (75:1 acrylamide:bis) polyacrylamide gels and electrophoresed in 10 mM Tris–acetate, 0.25 mM EDTA pH 7.6 buffer for 100 min at 125 V. The gels were vacuum dried and the amount of radioactivity in the unbound and bound DNA probe was quantified by PhosphorImager. Under conditions of excess protein, the dissociation constant (KD) was defined as the protein concentration required for half-maximal binding (Goodtzova et al., 1998).
Stability of C145 mutations
Wild-type, C145F and C145L AGT mutant proteins were produced using expression vectors derived from pINIII-A3(lppP-5) (Crone and Pegg, 1993) and expressed in E.coli strain GWR109, which lacks endogenous AGT. The half-life of the AGT proteins was determined by growing cells at 37°C in LB broth supplemented with 50 μg/ml ampicillin, 50 μg/ml kanamycin and 0.2 mM IPTG until the A600 reached 0.3 and then treating them with 50 μg/ml chloramphenicol to inhibit protein synthesis. Aliquots of 1–3 ml were taken through a 24 h period. Cell pellets were resuspended in 0.3 ml of 50 mM Tris–HCl pH 7.5, 0.1 mM EDTA, 5 mM DTT and extracts prepared by sonication on ice. After removal of cell debris by sedimentation at 12 000 g in a microcentrifuge for 15 min at 4°C, the supernatant was saved and total protein content measured. The AGT protein content was determined by Western blotting as described previously (Crone et al., 1996) with antiserum MAP-1, which was raised to peptide KRTTLDSPLGKLE corresponding to residues 8–20 of the human AGT (Pegg et al., 1991a). Results were expressed as the percentage of the AGT protein present at the time of chloramphenicol addition.
Acknowledgments
Acknowledgements
We thank S.S.Parikh, M.E.Stroupe and A.J.Das for reading of the manuscript, C.D.Putnam and D.S.Shin for helpful discussion, and R.C.Moschel for the providing O6-[8-3H]BG. We also acknowledge Dr Dale Boger and the staffs of the Advanced Light Source and the Stanford Synchrotron Radiation Laboratory for use of facilities. This work was supported by the National Institutes of Health (GM46312 to J.A.T; CA-18137, CA-71676 to A.E.P), the Skaggs Institute for Chemical Biology, a Graduate Research Fellowship from the National Science Foundation (D.S.D.) and a Special Fellowship from the Leukemia & Lymphoma Society (C.D.M.).
References
- Beranek D.T. (1990) Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat. Res., 231, 11–30. [DOI] [PubMed] [Google Scholar]
- Brennan R.G. (1993) The winged–helix DNA-binding motif: another helix–turn–helix takeoff. Cell, 74, 773–776. [DOI] [PubMed] [Google Scholar]
- Brent T.P., Houghton,P.J. and Houghton,J.A. (1985) O6-Alkylguanine-DNA alkyltransferase activity correlates with the therapeutic response of human rhabdomyosarcoma xenografts to 1-(2-chloroethyl)-3-(trans-4–methylcyclohexyl)-1-nitrosourea. Proc. Natl Acad. Sci. USA, 82, 2985–2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger A.T., et al. (1998)Crystallography and NMR system (CNS): A new software system for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Casadevall M. and Sarkar, B. (1998) Effect of redox conditions on the DNA-binding efficiency of the retinoic acid receptor zinc-finger. J. Inorg. Biochem., 71, 147–152. [DOI] [PubMed] [Google Scholar]
- Chae M.-Y., McDougall, M.G., Dolan, M.E., Swenn, K., Pegg, A.E. and Moschel, R.C. (1994) Substituted O6-benzylguanine derivatives and their inactivation of human O6–alkylguanine alkyltransferase. J. Med. Chem., 37, 342–347. [DOI] [PubMed] [Google Scholar]
- Chae M.-Y., Swenn, K., Kanugula, S., Dolan, M.E., Pegg, A.E. and Moschel, R.C. (1995) 8-Substituted O6-benzylguanine, substituted 6(4)-(benzyloxy)pyrimidine and related derivatives as inactivators of human O6–alkylguanine-DNA alkyltransferase. J. Med. Chem., 38, 359–365. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4 (1994) The CCP4 Suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763.. [DOI] [PubMed] [Google Scholar]
- Cowtan K.D. and Main, P. (1993) Improvement of macromolecular electron-density maps by the simultaneous application of real and reciprocal space constraints. Acta Crystallogr. D, 49, 148–157. [DOI] [PubMed] [Google Scholar]
- Crone T.M. and Pegg, A.E. (1993) A single amino acid change in human O6–alkylguanine-DNA alkyltransferase decreasing sensitivity to inactivation by O6-benzylguanine. Cancer Res., 53, 4750–4753. [PubMed] [Google Scholar]
- Crone T.M., Kanugula, S. and Pegg, A.E. (1995) Mutations in the Ada O6–alkylguanine-DNA alkyltransferase conferring sensitivity to inactivation by O6-benzylguanine and 2,4-diamino-6-benzyloxy-5-nitrosopyrimidine. Carcinogenesis, 16, 1687–1692. [DOI] [PubMed] [Google Scholar]
- Crone T.M., Goodtzova, K. and Pegg, A.E. (1996) Amino acid residues affecting the activity and stability of human O6–alkylguanine-DNA alkyltransferase. Mutat. Res., 363, 15–25. [DOI] [PubMed] [Google Scholar]
- Davis M.E., Madura, J.D., Luty, B.A., McCammon, J.A. (1991) Electrostatics and diffusion of molecules in solution: simulations with the University of Houston Brownian Dynamics Program. Comput. Phys. Commun., 62, 187–197. [Google Scholar]
- Deng C., Xie, D., Capasso, H., Zhao, Y., Wang, L.-D., Hong, J.-Y. (1999) Genetic polymorphism of human O6–alkylguanine-DNA alkyltransferase: identification of a missense variation in the active site region. Pharmacogenetics, 9, 81–87. [DOI] [PubMed] [Google Scholar]
- Dolan M.E., Moschel, R.C. and Pegg, A.E. (1990) Depletion of mammalian O6–alkylguanine-DNA alkyltransferase activity by O6-benzylguanine provides a means to evaluate the role of this protein in protection against carcinogenic and therapeutic alkylating agents. Proc. Natl Acad. Sci. USA, 87, 5368–5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan M.E., Chae, M.-Y., Pegg, A.E., Mullen, J.H., Friedman, H.S. and Moschel, R.C. (1994) Metabolism of O6-benzylguanine, an inactivator of O6–alkylguanine-DNA alkyltransferase. Cancer Res., 54, 5123–5130. [PubMed] [Google Scholar]
- Edara S., Kanugula, S., Goodtzova, K. and Pegg, A.E. (1996) Resistance of the human O6–alkylguanine-DNA alkyltransferase containing arginine at codon 160 to inactivation by O6-benzylguanine. Cancer Res., 56, 5571–5575. [PubMed] [Google Scholar]
- Elder R.H., Margison, G.P. and Rafferty, J.A. (1994) Differential inactivation of mammalian and Escherichia coli O6–alkylguanine-DNA alkyltransferases by O6-benzylguanine. Biochem. J., 298, 231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encell L.P., Landis, D.M. and Loeb, L.A. (1999) Improving enzymes for cancer gene therapy. Nature Biotechnol., 17, 143–147. [DOI] [PubMed] [Google Scholar]
- Friedman H.S., et al. (1998)Phase I trial of O6-benzylguanine for patients undergoing surgery for malignant glioma. J. Clin. Oncol., 16, 3570–3575. [DOI] [PubMed] [Google Scholar]
- Goodtzova K., Kanugula, S., Edara, S. and Pegg, A.E. (1998) Investigation of the role of tyrosine-114 in the activity of human O6–alkylguanine-DNA alkyltransferase. Biochemistry, 37, 12489–12495. [DOI] [PubMed] [Google Scholar]
- Hazra T.K., Roy, R., Biswas, T., Grabowski, D.T., Pegg, A.E. and Mitra, S. (1997) Specific recognition of O6–methylguanine in DNA by active site mutants of human O6–methylguanine-DNA methyltransferase. Biochemistry, 36, 5769–5776. [DOI] [PubMed] [Google Scholar]
- Holm L. and Sander, C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. [DOI] [PubMed] [Google Scholar]
- Hora J.F., Eastman,A. and Bresnick,E. (1983) O6-Methylguanine methyltransferase in rat liver. Biochemistry, 22, 3759–3763. [DOI] [PubMed] [Google Scholar]
- Hunt J.B., Neece, S.H. and Ginsburg, A. (1985) The use of 4-(2-pyridylazo)resorcinol in studies of zinc release from Escherichia coli aspartate transcarbamoylase. Anal. Biochem., 146, 150–157. [DOI] [PubMed] [Google Scholar]
- Kaina B., Fritz, G., Mitra, S. and Coquerelle, T. (1991) Transfection and expression of human O6–methylguanine-DNA methyltransferase (MGMT) cDNA in Chinese hamster cells: the role of MGMT in protection against the genotoxic effects of alkylating agents. Carcinogenesis, 12, 1857–1867. [DOI] [PubMed] [Google Scholar]
- Kalnik M.W., Li,B.F., Swann,P.F. and Patel,D.J. (1989) O6-Ethylguanine carcinogenic lesions in DNA: an NMR study of O6etG.C pairing in dodecanucleotide duplexes. Biochemistry, 28, 6182–6192. [DOI] [PubMed] [Google Scholar]
- Kanugula S., Goodtzova, K. and Pegg, A.E. (1998) Probing the conformational changes in human O6–alkylguanine-DNA alkyl– transferase protein in its alkylated and DNA-bound states by limited proteolysis. Biochem. J., 329, 545–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrtopoulos S.A., Anderson, L.M., Chhabra, S.K., Souliotis, V.L., Pletsa, V., Valavanis, C. and Georgiadis, P. (1997) DNA adducts and the mechanism of carcinogenesis and cytotoxicity of methylating agents of environmental and clinical significance. Cancer Detect. Pev., 21, 391–405. [PubMed] [Google Scholar]
- Lau A.Y., Scharer, O.D., Samson, L., Verdine, G.L. and Ellenberger, T. (1998) Crystal structure of a human alkylbase-DNA repair enzyme complexed to DNA: mechanisms for nucleotide flipping and base excision. Cell, 95, 249–258. [DOI] [PubMed] [Google Scholar]
- Mattern J., Eichhorn,U., Kaina,B. and Volm,M. (1998) O6-Methyl– guanine-DNA methyltransferase activity and sensitivity to cyclophosphamide and cisplatin in human lung tumor xenografts. Int. J. Cancer, 77, 919–922. [DOI] [PubMed] [Google Scholar]
- McRee D.E. (1992) XtalView: a visual protein crystallographic software system for X11/XView. J. Mol. Graph., 10, 44–47. [Google Scholar]
- Mineura K., Fukuchi, M., Kowada, M., Terashima, I. and Kohda, K. (1995) Differential inactivation of O6–methyguanine-DNA methyltransferase activity by O6-arylmethylguanines. Int. J. Cancer, 63, 148–151. [DOI] [PubMed] [Google Scholar]
- Moore M.H., Gulbis, J.M., Dodson, E.J., Demple, B. and Moody, P.C.E. (1994) Crystal structure of a suicidal DNA repair protein: the Ada O6–methylguanine-DNA methyltransferase from E.coli. EMBO J., 13, 1495–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moschel R.C., McDougall, M.G., Dolan, M.E., Stine, L. and Pegg, A.E. (1992) Structural features of substituted purine derivatives compatible with depletion of human O6–alkylguanine-DNA alkyltransferase. J. Med. Chem., 35, 4486–4491. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor, W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Parikh S.S., Mol, C.D., Slupphaug, G., Bharati, S., Krokan, H.E. and Tainer, J.A. (1998) Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. EMBO J., 17, 5214–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg A.E., Wiest, L., Mummert, C. and Dolan, M.E. (1991a) Production of antibodies to peptide sequences present in human O6–alkylguanine-DNA alkyltransferase and their use to detect this protein in cell extracts. Carcinogenesis, 12, 1671–1677. [DOI] [PubMed] [Google Scholar]
- Pegg A.E., Wiest, L., Mummert, C., Stine, L., Moschel, R.C. and Dolan, M.E. (1991b) Use of antibodies to human O6–alkylguanine-DNA alkyltransferase to study the content of this protein in cells treated with O6-benzylguanine or N–methyl-N′-nitro-N-nitrosoguanidine. Carcinogenesis, 12, 1679–1683. [DOI] [PubMed] [Google Scholar]
- Pegg A.E., Boosalis, M., Samson, L., Moschel, R.C., Byers, T.L., Swenn, K. and Dolan, M.E. (1993) Mechanism of inactivation of human O6–alkylguanine-DNA alkyltransferase by O6-benzylguanine. Biochemistry, 32, 11998–12006. [DOI] [PubMed] [Google Scholar]
- Pegg A.E., Dolan, M.E. and Moschel, R.C. (1995) Structure, function and inhibition of O6–alkylguanine-DNA alkyltransferase. Prog. Nucleic Acid Res. Mol. Biol., 51, 167–223. [DOI] [PubMed] [Google Scholar]
- Pegg A.E., Kanugula, S., Edara, S., Pauly, G.T., Moschel, R.C. and Goodtzova, K. (1998) Reaction of O6-benzylguanine-resistant mutants of human O6–alkylguanine-DNA alkyltransferase with O6-benzyl– guanine in oligodeoxyribonucleotides. J. Biol. Chem., 273, 10863–10867. [DOI] [PubMed] [Google Scholar]
- Rydberg B. and Lindahl, T. (1982) Nonenzymatic methylation of DNA by the intracellular methyl group donor S-adenosyl-l–methionine is a potentially mutagenic reaction. EMBO J., 1, 211–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz S.C., Shields, G.C. and Steitz, T.A. (1991) Crystal structure of a CAP–DNA complex: the DNA is bent by 90°. Science, 253, 1001–1007. [DOI] [PubMed] [Google Scholar]
- Slupphaug G., Mol, C.D., Kavli, B., Arvai, A.S., Krokan, H.E. and Tainer, J.A. (1996) A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature, 384, 87–92. [DOI] [PubMed] [Google Scholar]
- Spiro T.P., Gerson,S.L., Liu,L., Majka,S., Haaga,J., Hoppel,C.L., Ingalls,S.T., Pluda,J.M. and Willson,J.K. (1999) O6-Benzylguanine: a clinical trial establishing the biochemical modulatory dose in tumor tissue for alkyltransferase-directed DNA repair. Cancer Res., 59, 2402–2410. [PubMed] [Google Scholar]
- Spratt T.E., Wu, J.D., Levy, D.E., Kanugula, S. and Pegg, A.E. (1999) Reaction and binding of oligodeoxynucleotides containing analogues of O6–methylguanine with wild-type and mutant human O6–alkylguanine-DNA alkyltranferase. Biochemistry, 38, 6801–6806. [DOI] [PubMed] [Google Scholar]
- Srivenugopal K.S., Yuan, X.-H., Friedman, H.S., Ali-Osman, F. (1996) Ubiquitination-dependent proteolysis of O6–methylguanine-DNA methyltransferase in human and murine tumor cells following inactivation with O6-benzylguanine or 1,3-bis(2-chlorethyl)-1-nitrosourea. Biochemistry, 35, 1328–1334. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen, J. (1999) Automated structure solution for MIR and MAD. Acta Crystallogr. D, 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibley J.E.A., Pegg, A.E. and Moody, P.C.E. (2000) Crystal structure of the human O6–alkylguanine-DNA alkyltransferase. Nucleic Acids Res., 28, 393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintjens R. and Rooman, M. (1996) Structural classification of HTH DNA-binding domains and protein–DNA interaction modes. J. Mol. Biol., 262, 294–313. [DOI] [PubMed] [Google Scholar]
- Xu-Welliver M. and Pegg,A.E. (2000) Point mutations at multiple sites including highly conserved amino acids maintain activity but render O6–alkylguanine-DNA alkyltransferase insensitive to O6-benzyl– guanine. Biochem. J., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu-Welliver M., Kanuguloa, S. and Pegg, A.E. (1998) Isolation of human O6–alkylguanine-DNA alkyltransferase mutants highly resistant to inactivation by O6-benzylguanine. Cancer Res., 58, 1936–1945. [PubMed] [Google Scholar]
- Xu-Welliver M., Leitao, J., Kanugula, S., Meehan, W.J. and Pegg, A.E. (1999a) Role of codon 160 in the sensitivity of human O6–alkylguanine-DNA alkyltransferase to O6-benzylguanine. Biochem. Pharmacol., 58, 1279–1285. [DOI] [PubMed] [Google Scholar]
- Xu-Welliver M., Leitao, J., Kanugula, S. and Pegg, A.E. (1999b) Alteration of the conserved residue tyrosine-158 to histidine renders human O6–alkylguanine alkyltransferase insensitive to the inhibitor O6–alkylguanine. Cancer Res., 59, 1514–1519. [PubMed] [Google Scholar]
- Xu-Welliver M., Kanugula,S., Loktionova,N.A., Crone,T.M. and Pegg,A.E. (2000) Conserved residue lysine-165 is essential for the ability of O6–alkylguanine-DNA–alkyltransferase to react with O6-benzylguanine. Biochem. J., in press. [DOI] [PMC free article] [PubMed]
- Zhang K. and Main, P. (1990) Histogram matching as a new density modification technique for phase refinement and extension of protein molecules. Acta Crystallogr. A, 46, 41–46. [Google Scholar]