Abstract
The role of the intranuclear movement of chromatin in gene expression is not well-understood. Herpes simplex virus forms replication compartments (RCs) in infected cell nuclei as sites of viral DNA replication and late gene transcription. These structures develop from small compartments that grow in size, move, and coalesce. Quantitative analysis of RC trajectories, derived from 4D images, shows that most RCs move by directed motion. Directed movement is impaired in the presence of actin and myosin inhibitors as well as a transcription inhibitor. In addition, RCs coalesce at and reorganize nuclear speckles. Lastly, distinct effects of actin and myosin inhibitors on viral gene expression suggest that RC movement is not required for transcription, but rather, movement results in the bridging of transcriptionally active RCs with nuclear speckles to form structures that enhance export of viral late mRNAs.
Keywords: gene movement, nuclear export
Movement of chromatin and its interactions and location in the nucleus are believed to be important for regulation of gene expression (1). A number of studies have shown an association between the movement of chromosomal domains and changes in transcriptional activity. In yeast, activation of genes is associated with targeting of the gene to the nuclear periphery (2, 3), and this is believed to be because of the linkage of actively transcribed genes to the nuclear pore complex for RNA export (4). Lymphoid cell genes move to positions near centromeric heterochromatin coincident with transcriptional silencing (5). Transcriptional activation of the β-globin locus involves movement of the gene away from centromeric heterochromatin (6). During B-cell development, the IgH and Igκ loci are localized at the nuclear periphery in hematopoietic progenitor cells, but they move internally coincident with transcriptional activation in pro-B cells (7). Similarly, the β-globin gene relocalizes to the nuclear interior as it is activated during murine erythroid differentiation (8). Actively transcribed genes are also thought to be brought into close contact with nuclear speckles (9, 10), sites often regarded as hubs of RNA metabolism. Although these results link gene movement with changes in transcriptional activity, it is unclear whether intranuclear movement of chromatin is the cause or result of changes in transcriptional activity (1). Therefore, it is important to define systems in which the function of chromatin movement can be experimentally assessed.
Nuclear forms of actin and myosin have been implicated in mediating long-range directed movement (11–13), but the mechanism by which this putative nucleoskeletal system operates is unknown. In addition to their possible roles in mediating intranuclear movement, there is increasing evidence for roles of nuclear actin and myosin in cellular transcription, chromatin remodeling, and mRNA export (reviewed in ref. 14). How and whether their roles in movement are connected to regulation of gene activity are unclear.
Viruses are simple genetic entities and have often provided precise probes of cellular molecular mechanisms. Viral chromosomes of herpes simplex virus (HSV) are replicated and undergo late transcription in intranuclear structures called replication compartments (RCs) (15), which are known to move and coalesce in nuclei of infected cells during the course of infection (16, 17). The function and mechanism of RC movement are unknown. However, lepidopteran nucleopolyhedroviruses, large double-stranded DNA viruses that also replicate inside the host nucleus, have been shown to manipulate nuclear G- and F-actin for viral gene expression and progeny production (reviewed in ref. 18). Pseudorabies virus and HSV have also been shown to induce F-actin formation in nuclei of certain cells, and viral capsids seem to be associated with these structures (19, 20). Other studies have shown that HSV capsids undergo directed movement inside the nucleus (21), which is inhibited by the actin polymerization inhibitor latrunculin A (lat A) and the putative myosin inhibitor 2,3-butanedione monoxime (BDM) (22, 23). Based on the increasing evidence for the role of intranuclear movement in gene regulation and its involvement during viral infection, we were interested in the possible roles for movement in HSV-1 gene expression.
HSV-1 transcription, DNA synthesis, and capsid assembly all occur in the nucleus and are temporally regulated by a cascade of immediate-early (IE), early (E), and late (L) gene expression. As mentioned above, these processes are also spatially regulated in the form of intranuclear compartmentalization (24). RCs, which were first identified by the presence of infected cell protein (ICP) 8 (an HSV single-stranded DNA binding protein) (15), develop from small structures that grow in size, move, and coalesce, ultimately filling the entire nucleus and marginalizing host chromatin to the nuclear periphery (17, 25–27). RCs have also been shown to be transcriptionally active by the presence of a viral transactivator protein ICP4 (28, 29) and RNA pol II (30) that colocalize with ICP8. To better understand the mechanism and function of intranuclear movement of RCs, we carried out 4D imaging of cells infected with a recombinant HSV-1 strain expressing ICP8 fused to GFP to visualize RCs. Our results showed that the majority of RCs move by directed motion and require nuclear actin, myosin, and ongoing transcription. RC movement resulted in coalescence at nuclear speckles. Interestingly, inhibition of directed movement by lat A led to decreased accumulation of viral gene transcripts, whereas inhibition by BDM resulted in a defect in export of a late viral mRNA. Combined, our data suggest that HSV RC movement results in bridging transcriptional domains within RCs to RNA processing structures in the nucleus to enhance export of a subset of late viral mRNAs.
Results
RCs Move by an Active Process.
To better understand the process of RC movement in the context of the 3D nucleus, we carried out 4D (3D space and time) confocal imaging of cells infected with a recombinant HSV-1 strain expressing ICP8 fused to GFP (8GFP virus) (17). Cells were imaged from 4 to 7 h postinfection (hpi). ICP8-GFP localized to bright foci clustered within RCs. Smaller and dimmer individual foci, most likely representing viral prereplicative sites (15), were also present along the nuclear periphery and in the nucleoplasmic space between RCs (Fig. 1). These smaller individual foci exhibited highly constrained movement, whereas whole RCs displayed long-range movement, often to another RC (Fig. 1 and Movie S1). When the movement of RCs was tracked relative to the position of the nucleus in the 4D datasets, these structures moved at an average speed of 0.24 μm/min (up to 0.35 μm/min; SD = 0.056 μm/min). RCs traveled distances of up to 5.65 μm during the 3-h imaging period (as calculated by the shortest distance between the first and last frame for the longest track). The observed distances traveled by RCs and the average speed of movement are comparable with published reports of long-distance movement of chromosomal loci (11).
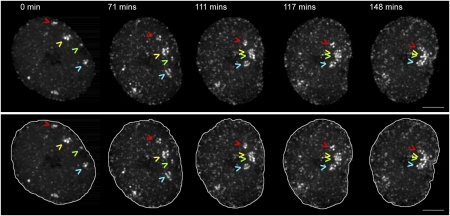
Fig. 1.
RCs move and coalesce inside the infected cell nucleus. A cell infected with ICP8-GFP virus was imaged in 4D starting at 4 hpi. The time series is displayed as a maximum intensity projection. Arrowheads point to four RCs that coalesce during the course of the movie. Lower panel shows the same time series with nuclear outlines. (Scale bar: 5 μm.)
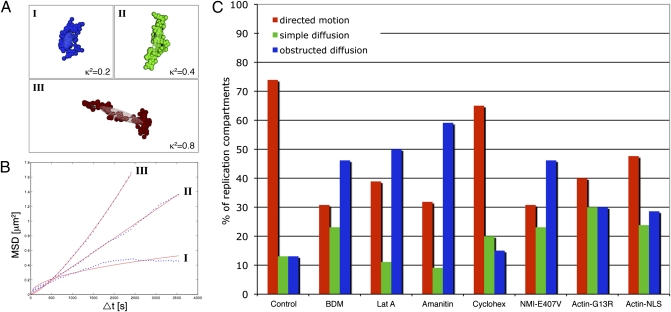
To quantify RC movement more rigorously, we developed a motion classification approach based on both the anomalous diffusion coefficient (α) (31) and 3D relative shape anisotropy (32, 33) (κ2; definitions in SI Results and SI Experimental Procedures). To obtain an estimate for α of each trajectory, we computed the mean square displacement (MSD) as a function of the time interval, Δt. To improve the accuracy of the estimates (31), we restricted the MSD calculations to Δt values less than N/3, where N is the total number of available positions in the trajectory. The motion properties of immobile RCs and those exhibiting either constrained or diffusive motion were already manifest over short-time intervals, and therefore, the computed α-value correlated well with these types of motion. However, because the motion properties of RCs displaying directed motion were evident only at a larger temporal scale, the computed value for α often did not characterize this type of motion accurately. Because the positions representing a directed motion trajectory exhibit a highly anisotropic scatter, a more effective way to detect this type of motion was to characterize the shape of the trajectory by its 3D relative shape anisotropy κ2 (32) (Fig. 2A shows sample RC trajectories, their ellipsoid of gyration, and κ2 values, which are defined as a function of the squared lengths of the semiaxes of the ellipsoid). Our hierarchical approach for determining the motion type of the RCs, therefore, first used κ2 to distinguish trajectories exhibiting directed motion from those displaying random motion. Trajectories with isotropic shapes were then additionally classified into confined diffusion, obstructed diffusion, or simple diffusion based on their α-values and the classification scheme by Bacher et al. (34) (Fig. 2B shows sample MSD curves that correspond to trajectories shown in Fig. 2A). Based on this hierarchical analysis approach, 74% of RCs appeared to undergo directed motion, 13% displayed simple diffusion, and 13% displayed obstructed diffusion (control; n = 5 nuclei, 23 RCs analyzed) (Fig. 2C). None of the RCs displayed confined diffusion. These results showed that the majority of the RCs moved by an active process.
Fig. 2.
Directed motion requires nuclear actin, myosin I, and transcription. Movement of RCs was analyzed in 4D and classified into directed motion, simple diffusion, or obstructed diffusion using a hierarchical approach based on both the 3D relative shape anisotropy (κ2) and anomalous diffusion coefficient (α). (A) Sample 3D trajectories and their ellipsoids of gyration are shown along with their κ2 values, which are defined as a function of the squared length of the semiaxes of the ellipsoid. Trajectories I and II represent nondirected motion, and trajectory III represents directed motion. (B) Corresponding MSD curves (blue) and fitted curves (red) are shown. Based on their α-values, trajectories I and II (A) are further classified as obstructed diffusion and simple diffusion, respectively. (C) Distribution of types of RC motion in control cells, cells treated with BDM or lat A at 3 hpi, cells treated with either α-amanitin or cycloheximide at 4 hpi, and cells transfected with plasmids encoding dominant negative nuclear myosin I (NMI-E407V) or actin (Actin-G13R) or with a plasmid encoding WT actin (Actin-NLS). Trajectories were classified as directed motion (red), simple diffusion (green), or obstructed diffusion (blue). The distributions of motion types are displayed as a percentage of the total number of RCs in each sample.
Active Movement of RCs Depends on Nuclear Actin, Myosin I, and Transcription.
Active movement of transgenes has been reported to involve nuclear actin and nuclear myosins such as nuclear myosin I (NMI) (11–13). To determine if nuclear myosin was involved in the active transport of RCs, we treated cells with BDM at 3 hpi and imaged the cells from 4 to 7 hpi. Because BDM also affects cytoplasmic myosin function, we treated cells with BDM starting at 3 hpi to allow unobstructed viral entry and minimize potential effects on cytoplasmic trafficking of incoming virions, which could, in turn, affect RC growth and dynamics. An average of 4 nuclei and 22 RCs were analyzed per treatment to determine the distribution of motion types as described in the previous section. BDM-treated cells showed a significant decrease in the fraction of RCs undergoing directed motion and an increase in RCs undergoing obstructed diffusion (P = 0.011) (Fig. 2C and Table S1). To determine the role of nuclear actin in RC movement, we also treated cells with lat A at 3 hpi. Treatment with this drug also led to a significant decrease in the fraction of RCs undergoing directed motion and an increase in those undergoing obstructed diffusion (P = 0.006) (Fig. 2C and Table S1). It should be noted that viral entry and cytoplasmic trafficking of HSV-1 in Vero cells, the cell type used in our experiments, occur independently of actin (35), consistent with the effects of lat A affecting nuclear but not cytoplasmic viral functions.
To determine whether NMI was specifically involved in the directed movement, we transfected cells with a plasmid expressing a dominant negative form of NMI (NMI-E407V). NMI-E407V is a mutant form of NMI that has impaired actin binding and motor activity, and it has been shown to reduce chromosomal loci movement (11). Expression of NMI-E407V led to a decreased number of RCs undergoing directed motion (P = 0.001) (Fig. 2C and Table S1). To confirm the effects of lat A, we also transfected cells with a construct encoding a dominant negative form of nuclear actin (actin-G13R). Actin-G13R contains a nuclear localization signal (NLS) and is a nonpolymerizable mutant form of actin, which has been shown to inhibit long-range movement of chromosomal loci (11). Expression of actin-G13R also led to a decrease in the fraction of RCs undergoing directed motion (P = 0.013) (Fig. 2C and Table S1). However, the effect was not caused entirely by the dominant negative mutation, because cells transfected with a plasmid encoding a YFP-tagged, nuclear-targeted WT actin (actin-NLS) also showed a decrease in the fraction of RCs undergoing directed motion (P = 0.038) (Fig. 2C and Table S1). All cells (including controls) were cotransfected with a plasmid encoding mCherry-tagged lamin A (mCh-LA) to correct for nuclear rotation and translation during RC tracking analysis. Therefore, the observed inhibitory effect was most likely not caused by nonspecific effects of overexpressing a nuclear protein.
Previous studies had shown that transcription is involved in long-range movement of chromosomal loci (12, 36). Therefore, we treated cells with the RNA pol II inhibitor, α-amanitin, at 4 hpi. The α-amanitin treatment led to a significant decrease in directed movement (P = 0.002) (Fig. 2C and Table S1). In contrast, treatment of cells with a protein synthesis inhibitor, cycloheximide, at 4 hpi did not lead to a significant change in the distribution of movements (P = 0.355) (Fig. 2C and Table S1). These results argued that directed RC movement involves nuclear actin, myosin, and ongoing transcription.
RCs Coalesce at Nuclear Speckles.
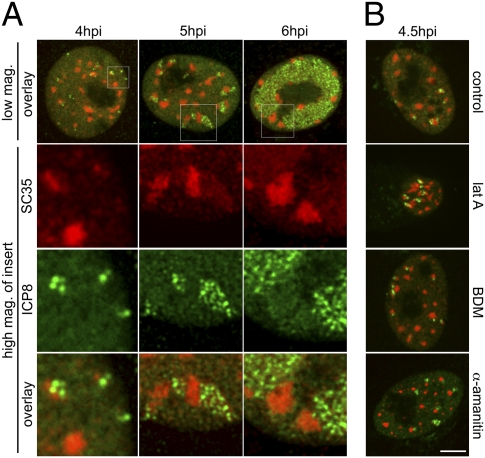
Previous studies have reported the clustering of coregulated genes at nuclear speckle bodies (9, 13, 37). To determine whether RCs were coalescing at speckles, we first carried out indirect immunofluorescence analysis of infected cells at various times postinfection using antibodies specific for SC35, one of the major components of speckles, and ICP8. In the resulting time series of images (Fig. 3A), RCs were frequently found adjacent to SC35-positive speckles. In some cases, speckles were associated with more than one RC, and some RCs were associated with more than one speckle (Fig. 3A, 5 hpi panel). Both RCs and speckles were reorganized so that by 6 hpi, many cells contained large RCs bordered by a few large speckles (Fig. 3A, 6 hpi panel). To determine if movement or transcription played a role in this association, we treated cells with either lat A, BDM, or control medium at 1 hpi or with α-amanitin at 4 hpi. When fixed and processed at 4.5 hpi, all cells showed RCs associated with speckles, including those treated with the various drugs (Fig. 3B). Speckle bodies appeared rounder and smaller in α-amanitin–treated cells, and lat A-treated cells exhibited nuclei that were smaller and more convoluted; however, the majority of RCs were still found adjacent to speckle bodies. These results showed that the initial association between RCs and speckles did not depend on actin or myosin.
Fig. 3.
RCs are associated with nuclear speckles. (A) Cells were processed for indirect immunofluorescence at various times postinfection to detect nuclear speckles and RCs using SC35 (red) and ICP8 (green) antibodies, respectively. High-magnification images of Insets are shown in rows 2–4. (B) Cells were treated with control medium, lat A, or BDM at 1 hpi or with α-amanitin at 4 hpi and processed for indirect immunofluorescence to detect SC35 (red) and ICP8 (green) at 4.5 hpi. (Scale bar: 5 μm.)
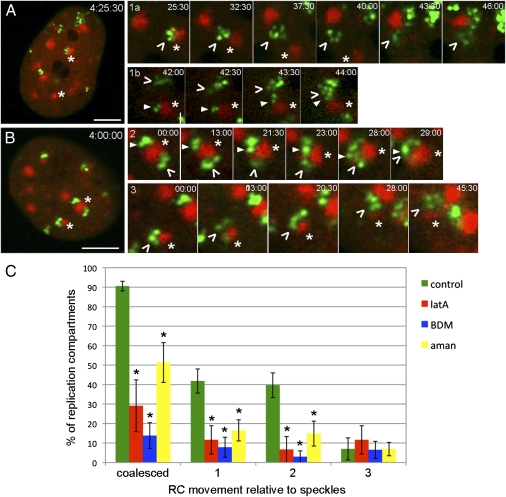
To examine potential interactions between speckles and RCs in live cells, we transfected cells with a plasmid encoding mCherry-tagged SC35 protein (mCh-SC35). At 24 h posttransfection, we infected the cells with 8GFP virus and started imaging them at 4 hpi (Fig. 4 A and B and Movie S2). mCh-SC35 localized to structures that resembled speckle bodies. At 4 hpi, the majority (78%) of RCs were adjacent to these SC35-positive speckles. Live-cell imaging revealed that ∼91% of RCs coalesced, and 100% of these coalescence events occurred in association with a speckle body. Three main types of RC movement patterns relative to speckles were observed: (i) independent movement of RCs (type 1) that resulted in contact either with a speckle (Fig. 4A, 1a) or with an RC that was associated with a speckle (Fig. 4A, 1b), (ii) movement of RC on the surface of a speckle (type 2) (Fig. 4B, 2), and (iii) movement of RCs with speckles (type 3) (Fig. 4B, 3). Speckles were also observed to merge during type 3 movement. Approximately 42% of RCs moved independently, ∼40% moved on the surface of a speckle, and ∼7% moved with a speckle (Fig. 4C). The number of RCs moving on the surface of a speckle may be underestimated, because such movement was only identifiable when there were at least two RCs on a common speckle where one could serve as a fiducial mark. To determine which of these three types of movement required actin, myosin, or transcription, we treated cells with control medium, lat A, or BDM at 3 hpi or with α-amanitin at 4 hpi (Movies S3, S4, and S5). An average of 7 nuclei and 44 RCs was analyzed per treatment. Lat A, BDM, and α-amanitin treatment all led to significant reductions in the number of coalescence events (Fig. 4C). In addition, all three treatments also led to significant reductions in both types 1 and 2 movements (Fig. 4C), indicating that actin, myosin, and transcription could be involved in mediating these movements. These drugs did not have a significant effect on type 3 movement. However, the frequency of this type of movement was also low in control cells compared with types 1 and 2.
Fig. 4.
RCs coalesce at nuclear speckles. (A and B) Cells were transfected with a plasmid encoding mCh-SC35 for 24 h, and then, they were infected with ICP8-GFP virus and imaged at 4 hpi. Four major types of RC movement (arrowheads) relative to speckles (asterisks) are shown in rows 1–3. Rows 1a and 1b are high-magnification time-series images of the cell shown in A, and they show RCs (green) that move alone either to a speckle (red; 1a) or another RC (1b). Rows 2 and 3 are high-magnification time-series images of the cell in B, and they represent RCs that move on speckles or with speckles, respectively. Time stamps in A and B show hour:minute:second postinfection. Time stamps in rows 1–3 show minute:second (at 4 hpi) (Movie S2). (Scale bar: 5 μm.) (C) mCh-SC35 transfected cells were treated with control medium, lat A, or BDM at 3 hpi or with α-amanitin at 4 hpi, and they were imaged at 4 hpi. Numbers of RC coalescence events and types 1–3 movement were determined manually. The data are presented as mean percentage ± SEM per nucleus (∼7 nuclei and ∼44 RCs were analyzed per treatment). Asterisks mark values that are statistically different from control medium-treated cells (P < 0.05) (Movies S2, S3, S4, and S5).
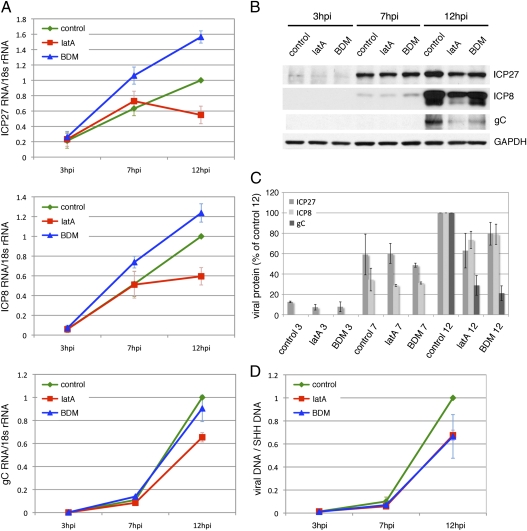
Effects of Directed RC Movement on Viral Gene Expression and DNA Replication.
To determine the possible role(s) for directed movement of RCs in viral gene expression, we infected cells with HSV-1 and treated them with lat A, BDM, or control medium at 3 hpi. We harvested cells at 3, 7, and 12 hpi and measured RNA and protein levels of three genes, ICP27, ICP8, and gC, representative of the IE, E, and L HSV genes, respectively. RNA levels were measured by RT quantitative real-time PCR (RT-qPCR), and protein levels were determined by SDS-PAGE and Western blot analysis. At 12 hpi, lat A-treated cells contained reduced levels of all three transcripts compared with control medium-treated cells (Fig. 5A). Furthermore, there was a corresponding decrease in levels of the three proteins, with the largest decrease in gC level at 12 hpi (71% decrease) (Fig. 5 B and C). In contrast, BDM treatment resulted in increased accumulation of both ICP27 and ICP8 mRNAs by 12 hpi compared with control medium (Fig. 5A). gC mRNA levels in BDM-treated cells were similar to control medium-treated cells (Fig. 5A). However, the effects of BDM on protein levels were similar to lat A-treated cells with modest reductions in ICP27 and ICP8 and a significant reduction in gC level (79% reduction) (Fig. 5 B and C). Therefore, lat A treatment reduced viral RNA levels generally, whereas BDM treatment specifically inhibited the protein levels of gC.
Fig. 5.
Effect of directed RC movement on viral gene expression and DNA replication. Infected cells were incubated with lat A, BDM, or control medium at 3 hpi and harvested at 3, 7, and 12 hpi for RNA, protein, and DNA analysis. ICP27, ICP8, and gC were chosen for analysis to represent the IE, E, and L gene classes, respectively. (A) RNA samples were analyzed by RT-qPCR. Viral RNA levels were normalized to 18S rRNA levels and further normalized to control 12 hpi values. Mean values ± SEM are shown. (B) Western blot analysis of ICP27, ICP8, and gC protein levels. (C) Quantification of bands in Western blots. Shorter exposure blots with values within the linear range of the film were used for quantitations. Values were normalized to GAPDH and then, to control 12 hpi values for each drug treatment. Mean values ± SEM are shown. (D) Viral DNA levels were analyzed by qPCR using primers specific for the ICP27 gene. Levels were normalized to cellular sonic hedgehog gene (SHH) levels and then, to control 12 hpi values. Mean values ± SEM are shown.
To determine if inhibition of directed RC movement affected viral DNA replication, we harvested cells treated with lat A, BDM, or control medium at 3, 7, and 12 hpi for DNA isolation. DNA samples were analyzed by real-time PCR using primers specific for the ICP27 gene to determine viral DNA levels. At 7 hpi, lat A- and BDM-treated cells contained ∼59% and ∼69% of viral DNA found in control cells, respectively (Fig. 5D). At 12 hpi, lat A-treated cells contained ∼68% of viral DNA found in control cells, and BDM-treated cells contained ∼67% (Fig. 5D). The reduced level of viral DNA in these cells may be a result of the observed decreased levels of IE and E proteins that regulate viral DNA replication and/or direct inhibition of replication by the drugs.
Effects of Directed RC Movement on Nuclear Export of gC mRNA.
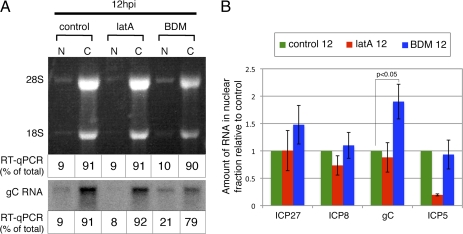
The contrasting effects of BDM on gC mRNA and protein levels led us to examine whether inhibition of RC movement by BDM had an effect on nuclear export of gC mRNA. We incubated cells with lat A, BDM, or control medium at 3 hpi as described above, harvested the cells at 12 hpi for isolation of nuclear and cytoplasmic RNA, and determined the amounts of RNA by Northern blotting or RT-qPCR to determine relative amounts of gC transcripts (Fig. 6A). Lat A treatment did not significantly affect the nuclear–cytoplasmic distribution of gC transcripts. In contrast, BDM treatment led to an approximately twofold increase in the percent of gC mRNA present in the nucleus compared with cells treated with control medium. BDM treatment did not affect the percentage of ICP27 and ICP8 transcripts in the nucleus (Fig. 6B). The percentage of ICP5 mRNA encoded by a leaky-late gene in the nucleus was also not affected by BDM (Fig. 6B). These results suggested a role for myosin-mediated RC movement in the nuclear export of a subset of viral late mRNAs (perhaps true late gene transcripts such as gC).
Fig. 6.
Role of directed RC movement in nuclear export of viral RNA. Cells were treated with control medium, lat A, or BDM at 3 hpi and harvested at 12 hpi for nuclear (N) and cytoplasmic (C) RNA. (A) Ethidium bromide-stained RNA is shown in Upper panel to confirm fractionation by the presence of 28S and 18S rRNA in the cytoplasm. Northern blot analysis for gC RNA is shown in Lower panel. RNA samples used in the Northern blot were analyzed by RT-qPCR, and the values are displayed on the bottom of each lane as a percentage of total RNA (N + C). (B) Fractionated RNA was analyzed by RT-qPCR for the presence of ICP27, ICP8, gC, and ICP5 RNA. Amounts were normalized to 18S rRNA. Lat A and BDM values were further normalized to control values, and mean ± SEM are shown (n = 3 experiments).
Discussion
Movement of RCs.
Our results show that approximately three-quarters of HSV RCs move in infected cell nuclei by directed motion. The distance (up to 5.65 μm) and average speed of RC movement (0.24 μm/min) are comparable with values reported for long-range movement of chromosomal loci (11). RC movement requires nuclear actin, myosin, and ongoing transcription. Furthermore, movement results in coalescence of RCs at speckles and reorganization of speckle bodies. Because speckles are considered to be hubs of transcription, RNA splicing, processing, and export (reviewed in refs. 37 and 38), the observed interactions between RCs and speckles further support the idea that RC movement is related to RNA metabolism events.
Association with Nuclear Speckles.
HSV infection has long been known to reorganize speckles into larger bodies (39), with ICP27 being required for this reorganization (40, 41). Reorganization of speckles may be the result of (i) inhibition of host RNA splicing events, which in turn, contribute to the inhibition of cellular gene expression (reviewed in ref. 42), (ii) an indirect effect of inhibiting cellular transcription, or (iii) an active process that is at least in part caused by the RC and speckle movement shown here. Unlike cellular pol II transcripts, the majority of HSV transcripts are intronless. Because splicing and RNA processing events are closely linked to nuclear export, in the absence of splicing, RCs may need to reorganize and associate with speckle bodies to recruit RNA processing and nuclear export factors to sites of viral true late gene transcription on progeny viral DNA molecules.
Similar reorganization of nuclear speckles is known to occur in response to heat shock (reviewed in ref. 43) and inhibition of transcription by α-amanitin (44, 45). Recent studies showed that heat shock-induced Hsp70 transgenes associated with nuclear speckles in a dynamic fashion (46), and this association was promoter- and transcription-dependent (47). Despite the dependence on transcription, the level of accumulated nascent transcript did not seem to be critical for speckle association. Other studies involving the association of heat-shock genes with speckles showed that this association was independent of the presence of introns (48). However, the functional significance of the association remains unclear. Other studies have shown that, in cells where transcription was inhibited by α-amanitin or heat shock, RNA pol II and splicing factors accumulated in speckles (45). We speculate that these results reflect the role of speckles as recycling centers for transcription and splicing factors, especially during periods of transcriptional inactivation. Therefore, association of HSV-1 RCs with nuclear speckles may be a method to take advantage of the heightened concentration of these factors in speckles during infection when host transcription is shut down.
We found that the initial association of RCs with speckles did not require actin- and myosin-mediated movement, because treatment of cells with lat A or BDM at 1 hpi did not inhibit the association between individual RCs and speckle bodies by 4.5 hpi. This initial association could be formed either by diffusion or de novo formation of speckles adjacent to RCs. Such de novo formation of speckle bodies has been reported for cellular transgenes (46). Promyelocytic leukemia nuclear bodies (PML) have also been shown to form de novo and in association with viral genomes at early times postinfection (49). This association is part of an intrinsic cellular defense mechanim that represses viral transcription and is counteracted by the viral regulatory protein ICP0, which induces subsequent degradation of PML (50). It will be interesting to determine if these early viral genomes associated with nuclear domain 10 sites are also associated with speckle bodies, and if so, the spatial and temporal relationships among these three structures. In contrast to the initial actin- and myosin-independent association, coalescence of RCs at speckles requires actin- and myosin-mediated movement. These results suggest that it is not the mere association with speckles but rather movement of RCs postassociation with speckles and the act of coalescence on the surface of speckles that enhance gC mRNA export. Recent studies showed the presence of myosin Va (51) and proteins that regulate actin polymer dynamics, such as PIP2, profilin and protein 4.1, in nuclear speckles (reviewed in ref. 52). These observations present interesting possibilities regarding the recruitment of myosin and F-actin regulators from speckles to initiate RC movement.
Role for Directed RC Movement in Viral Late mRNA Export.
Surprisingly, lat A and BDM treatment, both of which inhibited movement, resulted in differing effects on viral gene expression, with lat A leading to a general reduction in the accumulation of viral transcripts and BDM leading to a defect in the nuclear export of gC mRNA. The effect of BDM on nuclear export may be specific for true late transcripts, because export of the leaky late ICP5 mRNA was not affected. This result shows that the inhibition of gC mRNA export is most likely a result of inhibiting RC movement and not a result of general export inhibition by BDM at late times postinfection. BDM treatment may also inhibit other posttranscriptional processing events, because the approximately twofold increase in nuclear accumulation of gC transcript may not be sufficient to explain the ∼79% reduction in gC protein levels at 12 hpi. Alternatively, because degradation of unprocessed mRNA precursors is known to occur in the nucleus (53), our results could be an underestimation of the defect in export. Interestingly, lat A treatment seemed to lead to decreased accumulation of nuclear ICP5 mRNA. This result raises the possibility that not all viral genes share a requirement for actin and/or myosin.
We hypothesize that the differing effects on gene expression of two drugs that both inhibit movement of RCs could be that the inhibition of movement by one is indirect and by the other is direct. Actin has been shown previously to be part of preinitiation complexes and required for transcription by RNA pol II (54). In our model, lat A treatment, which inhibited transcription, could inhibit movement indirectly. In contrast, BDM treatment, which inhibited nuclear export, may inhibit movement directly. We speculate that myosin is recruited specifically to RCs involved in transcription of true late genes, triggering directed movement to hubs of RNA processing and export (Fig. 7). It is surprising that BDM treatment did not lead to decreased levels of mRNA, because NMI has been shown previously to stimulate basal transcription by RNA pol II (55). This may indicate differences between cellular and viral gene transcription, or it may indicate that BDM treatment may affect a different or broader range of myosin isoforms. Alternatively, BDM has also been shown to affect the recruitment of proteins involved in actin polymerization such as the Arp 2/3 complex (23). It is possible that the inhibition of RC movement by BDM is related to its effects on actin polymer dynamics, which is distinct from lat A-mediated effects on actin, and that this property, in turn, is involved in posttranscriptional events.
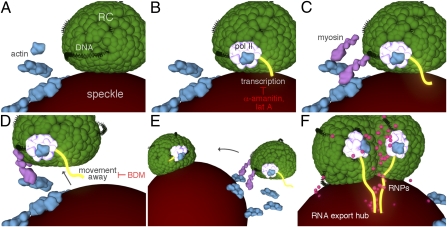
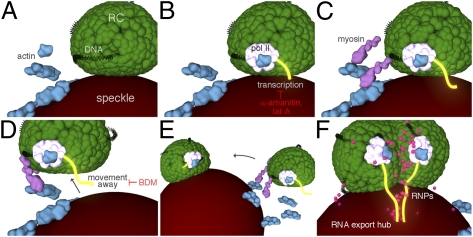
Fig. 7.
Actively transcribing RCs move to and coalesce at speckles to form RNA export/processing structures. (A and B) Progeny DNA (black) in a stationary RC undergoes actin- (blue) and RNA pol II (white) -mediated transcription (yellow) of true late genes such as gC. (C and D) Myosin (pink) is recruited to the transcribing RC and triggers movement of the RC away from the associated speckle. (E) The mobile RC moves to another RC-associated speckle and moves on the surface of the speckle to merge with the other RC. (F) Coalescence of RCs on the speckle surface leads to a reorganization of RCs, allowing for efficient incorporation of RNA processing factors and the processing of true late transcripts in the speckle, which ultimately create an RNA export/processing hub.
Regardless of the mechanism behind the effects of lat A and BDM on RC movement, the inhibition of movement by α-amanitin treatment combined with the differing effects on viral gene expression by lat A and BDM treatments argue that movement is not required for transcription but rather, that transcription is required for movement (Fig. 7). Our results are consistent with previous studies showing the involvement of transcription in mediating movement of genetic loci (12, 36, 47 and reviewed in ref. 1). Furthermore, our results showing that BDM treatment inhibits movement without reducing transcript levels raise the possibility that transcription alone does not induce movement but rather, that the recruitment of posttranscriptional processing factors, which are dependent on transcription, may be the trigger for movement. Determination of the exact mechanisms by which actin, myosin, transcription, and RNA processing events cause movement will require additional studies.
Finally, our live-cell imaging experiments show that the majority of RC movement events result in fusion of two or more RCs at speckle bodies. We hypothesize that the fusion of RCs could lead to a structural reorganization in both compartments, allowing for more efficient incorporation of RNA processing factors and/or the translocation of RNAs into the associated speckle body for processing, thus leading to the formation of a hub that is optimized for processing and export of a subset of late mRNAs (Fig. 7). It will be important to determine if RC movement affects export of other viral transcripts and if so, to determine whether they share similar RNA export or processing requirements.
The highly dynamic HSV RCs provide a useful tool for investigating the mechanism and function of intranuclear movement and interchromosomal interactions in living cells. Our results support the idea that the dynamic nature and positioning of genetic material within the complex nuclear architecture play an essential role in the expression of eukaryotic genes. The movement of actively transcribed genes to sites of RNA processing and export may be a general mechanism that eukaryotic cells and viruses use to optimize the efficiency of coupling of transcription to RNA processing and nuclear export.
Experimental Procedures
Cells and Viruses.
Vero cells, which were used for RNA, protein and DNA isolation, and immunofluorescence analysis, were grown and maintained in DMEM with 5% FBS, 5% bovine calf serum (BCS), and glutamine (DMEM+ 10% S). For live-cell imaging studies, an ICP8-complementing cell line, V529 (56), was used. V529 cells were grown and maintained in DMEM+10%S containing 400 μg/mL G418. The ICP8-GFP recombinant virus (8GFP) used for live-cell imaging experiments is described by Taylor et al. (17). The 8GFP virus was propagated on V529 cells. The HSV-1 WT strain KOS used in the rest of the experiments was propagated on Vero cells.
Infection.
For live-cell imaging experiments, V529 cells were infected with the 8GFP virus at multiplicity of infection of 10. Vero cells were infected with KOS at an multiplicity of infection of 10. At 1 h postincubation with either virus, unabsorbed virus was inactivated by treatment with acid wash (40 mM citric acid, 135 mM NaCl, 10 mM KCl, pH 3.0). The acid wash was then replaced with DMEM with 1% BCS, glutamine, streptomycin, and penicillin (DMEV). For live-cell imaging, DMEV was replaced with phenol-free L-15, 1% BCS, and glutamine at 4 hpi for the duration of the imaging period.
Transfections.
Cells were transfected with plasmids using the QIAGEN Effectene transfection reagent 24 h before infection. For tracking experiments, cells were transfected with either mCh-LA alone (for drug treatments) or mCh-LA and an actin/myosin plasmid (for dominant negative experiments). Details regarding the plasmids used can be found in SI Experimental Procedures.
Drugs.
Cells were treated with DMEV containing 0.03% DMSO (control medium), 1 mM lat A (Invitrogen), 20 mM 2,3-Butanedione monoxime (BDM; Sigma-Aldrich), 10 mM α-amanitin (Sigma-Aldrich), or 100 μg/mL cycloheximide (Sigma-Aldrich). For live-cell imaging experiments, drugs were included in the imaging medium.
Microscopy.
The imaging system consisted of a fully motorized, inverted microscope (Zeiss Axiovert 200M) with a piezo-driven stage (Applied Scientific Instrument) that was enclosed in an environmental chamber and coupled to a computer-controlled spherical aberration correction device (Intelligent Imaging Innovations.), a Yokogawa spinning disk confocal head (Perkin-Elmer), and a back-illuminated EM-CCD camera (Cascade; Roper Scientific). Solid-state 50-mW lasers (488 and 568 nm; Crystal Laser) controlled by an acousto-optical tunable filter were used for illumination. The image acquisition software used was SlideBook 4.2 (Intelligent Imaging Innovations). A Plan Apochromat 63× 1.4 N.A. lens (Carl Zeiss) was used. For 4D imaging, 3D stacks consisting of ∼30 optical z slices that were 0.36 μm apart were captured every 30 s to 1 min for periods of up to 3 h.
4D Tracking and Motion Analysis.
Methods used for RC tracking and motion analyses are described in SI Experimental Procedures.
Indirect Immunofluorescence.
WT HSV-1–infected Vero cells were fixed with 3.65% formaldehyde, permeabilized with 0.1% Triton-X 100, and incubated with antibodies for 30 min at 37 °C. Stained cells were mounted in ProLong Gold antifade reagent (Invitrogen). A list of antibodies that were used and their dilutions can be found in SI Experimental Procedures.
RC–Speckle Dynamics Analysis.
To determine the distribution of the three types of movement in control vs. drug-treated cells, ∼7 nuclei (∼44 RCs) were analyzed per treatment. The three types of movement were defined qualitatively, and the number of events was counted manually. The number of RCs undergoing each type of movement was divided by the total number of RCs in each cell and presented as a percentage value. Student t test was used to determine the statistical significance of the differences between control and drug-treated samples.
Western Blots.
Cells were lysed in SDS-PAGE loading buffer, and proteins were resolved by SDS-PAGE. Western blots were developed using Pierce ECL Western blotting substrate (Thermo Fisher Scientific). ImageJ software (http://rsb.info.nih.gov/ij/) was used to quantify the band intensities. Serial dilutions of the control 12 hpi protein sample were analyzed for each blot and each antibody to ensure linearity. Values were normalized to GAPDH and then, to 12 hpi values for each drug treatment. Experiments were conducted three times, and their values were averaged. A list of antibodies and the dilutions used can be found in SI Experimental Procedures.
Viral RNA Analysis by qPCR.
Total RNA was extracted using the TRI-Reagent solution (Ambion) and DNase treated using the DNA-free kit (Ambion). DNase-treated RNA (80 ng/sample) was then reverse-transcribed and quantified by real-time PCR (qPCR) as described by Cliffe et al. (57). Mock reverse-transcribed samples were included as negative controls. For nuclear–cytoplasmic fractionation, cells were lysed and fractionated according to the supplemental protocol accompanying the QIAGEN RNeasy kit. RNA was extracted using the QIAGEN RNeasy mini kit, DNase-treated as above, and quantified using the Power SYBR Green RNA to CT one-step kit and a Prism 7300 sequence detection system (Applied Biosystems). All reactions were carried out in duplicate, and relative copy numbers were determined by comparison with standard curves. Viral RNA levels were normalized to 18S rRNA levels and further normalized to control 12 hpi values. Experiments were conducted three times, and the values were averaged. The Student t test was used to determine the statistical significance of differences between nonnormalized control and drug-treated samples. Primer sequences can be found in SI Experimental Procedures.
Viral DNA Analysis by qPCR.
We used the QIAGEN DNeasy Blood and Tissue kit for DNA isolation. DNA amounts were analyzed using the Power SYBR Green PCR master mix and a Prism 7300 sequence detection system (Applied Biosystems). qPCR reactions were carried out in duplicate, and relative copy numbers were determined by comparison with standard curves. Viral DNA levels were normalized to cellular sonic hedgehog gene (SHH) levels and then, to control 12 hpi values. Experiments were conducted three times, and the values were averaged. Primer sequences can be found in SI Experimental Procedures.
Northern Blot Hybridization and Analysis.
The gC gene hybridization probe was prepared by cleavage of the pCI-gC plasmid (58). [32P]dCTP labeling was carried out as described by Fontaine-Rodriguez and Knipe (59). Fractionated cytoplasmic and nuclear RNA samples from infected cells (described above) were resolved with a 1.5% agarose gel prepared with NorthernMax-Glyc gel prep/running buffer (Ambion). Resolved samples were transferred to a nylon membrane using the NorthernMax transfer buffer by Ambion. Incubation with the probe was carried out in 5× saline-sodium phosphate-EDTA buffer (SSPE) (Boston BioProducts), 5× Denhardt's solution (Boston BioProducts), 50% formamide (Sigma-Aldrich), 0.5% SDS, and 0.1 mg/mL ssDNA (Invitrogen) for 48 h at 42 °C. Membranes were exposed to film for autoradiography.
Supplementary Material
Acknowledgments
We thank Dr. Robert D. Goldman and Dr. Takeshi Shimi at Northwestern University Medical School for providing the mCh-lamin A construct and Dr. Prasanth V. Kannanganattu at University of Illinois at Urbana–Champaign and Dr. David L. Spector at Cold Spring Harbor Laboratory for providing the YFP-SC35 plasmid. We also thank the New England Regional Center of Excellence for Biodefense and Emerging Infectious Diseases (National Institutes of Health Grant AI057159) imaging facility at the Immune Disease Institute for use of the spinning disk confocal microscope. Funding for this work was provided by National Institutes of Health Grant AI63106 (to D.M.K.) and R01GM080587 (to P.d.L.). Support for the project VIROQUANT (0313923) by German Federal Ministry of Education and Research (BMBF) (FORSYS) is also acknowledged.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
See Author Summary on page 8539.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1103411108/-/DCSupplemental.
References
- 1.Hübner MR, Spector DL. Chromatin dynamics. Annu Rev Biophys. 2010;39:471–489. doi: 10.1146/annurev.biophys.093008.131348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brickner JH, Walter P. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2004;2:e342. doi: 10.1371/journal.pbio.0020342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casolari JM, et al. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–439. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- 4.Blobel G. Gene gating: A hypothesis. Proc Natl Acad Sci USA. 1985;82:8527–8529. doi: 10.1073/pnas.82.24.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown KE, Baxter J, Graf D, Merkenschlager M, Fisher AG. Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol Cell. 1999;3:207–217. doi: 10.1016/s1097-2765(00)80311-1. [DOI] [PubMed] [Google Scholar]
- 6.Schübeler D, et al. Nuclear localization and histone acetylation: A pathway for chromatin opening and transcriptional activation of the human beta-globin locus. Genes Dev. 2000;14:940–950. [PMC free article] [PubMed] [Google Scholar]
- 7.Kosak ST, et al. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- 8.Ragoczy T, Bender MA, Telling A, Byron R, Groudine M. The locus control region is required for association of the murine beta-globin locus with engaged transcription factories during erythroid maturation. Genes Dev. 2006;20:1447–1457. doi: 10.1101/gad.1419506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown JM, et al. Association between active genes occurs at nuclear speckles and is modulated by chromatin environment. J Cell Biol. 2008;182:1083–1097. doi: 10.1083/jcb.200803174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spector DL, Lamond AI. Nuclear speckles. Cold Spring Harb Perspect Biol. 2011;3:a000646. doi: 10.1101/cshperspect.a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chuang CH, et al. Long-range directional movement of an interphase chromosome site. Curr Biol. 2006;16:825–831. doi: 10.1016/j.cub.2006.03.059. [DOI] [PubMed] [Google Scholar]
- 12.Dundr M, et al. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J Cell Biol. 2007;179:1095–1103. doi: 10.1083/jcb.200710058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Q, et al. Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc Natl Acad Sci USA. 2008;105:19199–19204. doi: 10.1073/pnas.0810634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Visa N, Percipalle P. Nuclear functions of actin. Cold Spring Harb Perspect Biol. 2010;2:a000620. doi: 10.1101/cshperspect.a000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quinlan MP, Chen LB, Knipe DM. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell. 1984;36:857–868. doi: 10.1016/0092-8674(84)90035-7. [DOI] [PubMed] [Google Scholar]
- 16.Sourvinos G, Everett RD. Visualization of parental HSV-1 genomes and replication compartments in association with ND10 in live infected cells. EMBO J. 2002;21:4989–4997. doi: 10.1093/emboj/cdf458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor TJ, McNamee EE, Day C, Knipe DM. Herpes simplex virus replication compartments can form by coalescence of smaller compartments. Virology. 2003;309:232–247. doi: 10.1016/s0042-6822(03)00107-7. [DOI] [PubMed] [Google Scholar]
- 18.Volkman LE. Baculovirus infectivity and the actin cytoskeleton. Curr Drug Targets. 2007;8:1075–1083. doi: 10.2174/138945007782151379. [DOI] [PubMed] [Google Scholar]
- 19.Feierbach B, Piccinotti S, Bisher M, Denk W, Enquist LW. Alpha-herpesvirus infection induces the formation of nuclear actin filaments. PLoS Pathog. 2006;2:e85. doi: 10.1371/journal.ppat.0020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ecob-Johnston MS, Whetsell WO., Jr Host-cell response to herpes virus infection in central and peripheral nervous tissue in vitro. J Gen Virol. 1979;44:747–757. doi: 10.1099/0022-1317-44-3-747. [DOI] [PubMed] [Google Scholar]
- 21.Forest T, Barnard S, Baines JD. Active intranuclear movement of herpesvirus capsids. Nat Cell Biol. 2005;7:429–431. doi: 10.1038/ncb1243. [DOI] [PubMed] [Google Scholar]
- 22.Ostap EM. 2,3-Butanedione monoxime (BDM) as a myosin inhibitor. J Muscle Res Cell Motil. 2002;23:305–308. doi: 10.1023/a:1022047102064. [DOI] [PubMed] [Google Scholar]
- 23.Yarrow JC, Lechler T, Li R, Mitchison TJ. Rapid de-localization of actin leading edge components with BDM treatment. BMC Cell Biol. 2003;4:5. doi: 10.1186/1471-2121-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roizman B, Knipe DM, Whitley RJ. Herpes simplex viruses. In: Knipe DM, Howley PM, editors. Fields Virology. 5th Ed. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 2501–2602. [Google Scholar]
- 25.de Bruyn Kops A, Knipe DM. Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell. 1988;55:857–868. doi: 10.1016/0092-8674(88)90141-9. [DOI] [PubMed] [Google Scholar]
- 26.Simpson-Holley M, Baines J, Roller R, Knipe DM. Herpes simplex virus 1 U(L)31 and U(L)34 gene products promote the late maturation of viral replication compartments to the nuclear periphery. J Virol. 2004;78:5591–5600. doi: 10.1128/JVI.78.11.5591-5600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monier K, Armas JC, Etteldorf S, Ghazal P, Sullivan KF. Annexation of the interchromosomal space during viral infection. Nat Cell Biol. 2000;2:661–665. doi: 10.1038/35023615. [DOI] [PubMed] [Google Scholar]
- 28.Knipe DM, Senechek D, Rice SA, Smith JL. Stages in the nuclear association of the herpes simplex virus transcriptional activator protein ICP4. J Virol. 1987;61:276–284. doi: 10.1128/jvi.61.2.276-284.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Bruyn Kops A, Uprichard SL, Chen M, Knipe DM. Comparison of the intranuclear distributions of herpes simplex virus proteins involved in various viral functions. Virology. 1998;252:162–178. doi: 10.1006/viro.1998.9450. [DOI] [PubMed] [Google Scholar]
- 30.Rice SA, Long MC, Lam V, Spencer CA. RNA polymerase II is aberrantly phosphorylated and localized to viral replication compartments following herpes simplex virus infection. J Virol. 1994;68:988–1001. doi: 10.1128/jvi.68.2.988-1001.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saxton MJ, Jacobson K. Single-particle tracking: Applications to membrane dynamics. Annu Rev Biophys Biomol Struct. 1997;26:373–399. doi: 10.1146/annurev.biophys.26.1.373. [DOI] [PubMed] [Google Scholar]
- 32.Theodorou DN, Suter UW. Shape of unpreturbed linear polymers: Polypropylene. Macromolecules. 1985;18:1206–1214. [Google Scholar]
- 33.Saxton MJ. Lateral diffusion in an archipelago. Single-particle diffusion. Biophys J. 1993;64:1766–1780. doi: 10.1016/S0006-3495(93)81548-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bacher CP, Reichenzeller M, Athale C, Herrmann H, Eils R. 4-D single particle tracking of synthetic and proteinaceous microspheres reveals preferential movement of nuclear particles along chromatin—poor tracks. BMC Cell Biol. 2004;5:45. doi: 10.1186/1471-2121-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sodeik B, Ebersold MW, Helenius A. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J Cell Biol. 1997;136:1007–1021. doi: 10.1083/jcb.136.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tumbar T, Belmont AS. Interphase movements of a DNA chromosome region modulated by VP16 transcriptional activator. Nat Cell Biol. 2001;3:134–139. doi: 10.1038/35055033. [DOI] [PubMed] [Google Scholar]
- 37.Hall LL, Smith KP, Byron M, Lawrence JB. Molecular anatomy of a speckle. Anat Rec A Discov Mol Cell Evol Biol. 2006;288:664–675. doi: 10.1002/ar.a.20336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhong XY, Wang P, Han J, Rosenfeld MG, Fu XD. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol Cell. 2009;35:1–10. doi: 10.1016/j.molcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin TE, Barghusen SC, Leser GP, Spear PG. Redistribution of nuclear ribonucleoprotein antigens during herpes simplex virus infection. J Cell Biol. 1987;105:2069–2082. doi: 10.1083/jcb.105.5.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phelan A, Carmo-Fonseca M, McLaughlan J, Lamond AI, Clements JB. A herpes simplex virus type 1 immediate-early gene product, IE63, regulates small nuclear ribonucleoprotein distribution. Proc Natl Acad Sci USA. 1993;90:9056–9060. doi: 10.1073/pnas.90.19.9056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sandri-Goldin RM, Hibbard MK, Hardwicke MA. The C-terminal repressor region of herpes simplex virus type 1 ICP27 is required for the redistribution of small nuclear ribonucleoprotein particles and splicing factor SC35; however, these alterations are not sufficient to inhibit host cell splicing. J Virol. 1995;69:6063–6076. doi: 10.1128/jvi.69.10.6063-6076.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sandri-Goldin RM. The many roles of the regulatory protein ICP27 during herpes simplex virus infection. Front Biosci. 2008;13:5241–5256. doi: 10.2741/3078. [DOI] [PubMed] [Google Scholar]
- 43.Lamond AI, Spector DL. Nuclear speckles: A model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- 44.Carmo-Fonseca M, Pepperkok R, Carvalho MT, Lamond AI. Transcription-dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J Cell Biol. 1992;117:1–14. doi: 10.1083/jcb.117.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeng C, Kim E, Warren SL, Berget SM. Dynamic relocation of transcription and splicing factors dependent upon transcriptional activity. EMBO J. 1997;16:1401–1412. doi: 10.1093/emboj/16.6.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu Y, Kireev I, Plutz M, Ashourian N, Belmont AS. Large-scale chromatin structure of inducible genes: Transcription on a condensed, linear template. J Cell Biol. 2009;185:87–100. doi: 10.1083/jcb.200809196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu Y, Plutz M, Belmont AS. Hsp70 gene association with nuclear speckles is Hsp70 promoter specific. J Cell Biol. 2010;191:711–719. doi: 10.1083/jcb.201004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jolly C, Vourc'h C, Robert-Nicoud M, Morimoto RI. Intron-independent association of splicing factors with active genes. J Cell Biol. 1999;145:1133–1143. doi: 10.1083/jcb.145.6.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Everett RD, Murray J. ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J Virol. 2005;79:5078–5089. doi: 10.1128/JVI.79.8.5078-5089.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Everett RD, et al. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J Virol. 1998;72:6581–6591. doi: 10.1128/jvi.72.8.6581-6591.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pranchevicius MC, et al. Myosin Va phosphorylated on Ser1650 is found in nuclear speckles and redistributes to nucleoli upon inhibition of transcription. Cell Motil Cytoskeleton. 2008;65:441–456. doi: 10.1002/cm.20269. [DOI] [PubMed] [Google Scholar]
- 52.Gieni RS, Hendzel MJ. Actin dynamics and functions in the interphase nucleus: Moving toward an understanding of nuclear polymeric actin. Biochem Cell Biol. 2009;87:283–306. doi: 10.1139/O08-133. [DOI] [PubMed] [Google Scholar]
- 53.Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 54.Hofmann WA, et al. Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II. Nat Cell Biol. 2004;6:1094–1101. doi: 10.1038/ncb1182. [DOI] [PubMed] [Google Scholar]
- 55.Hofmann WA, Johnson T, Klapczynski M, Fan JL, de Lanerolle P. From transcription to transport: Emerging roles for nuclear myosin I. Biochem Cell Biol. 2006;84:418–426. doi: 10.1139/o06-069. [DOI] [PubMed] [Google Scholar]
- 56.Da Costa XJ, Kramer MF, Zhu J, Brockman MA, Knipe DM. Construction, phenotypic analysis, and immunogenicity of a UL5/UL29 double deletion mutant of herpes simplex virus 2. J Virol. 2000;74:7963–7971. doi: 10.1128/jvi.74.17.7963-7971.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cliffe AR, Garber DA, Knipe DM. Transcription of the herpes simplex virus latency-associated transcript promotes the formation of facultative heterochromatin on lytic promoters. J Virol. 2009;83:8182–8190. doi: 10.1128/JVI.00712-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Godowski PJ, Knipe DM. Transcriptional control of herpesvirus gene expression: Gene functions required for positive and negative regulation. Proc Natl Acad Sci USA. 1986;83:256–260. doi: 10.1073/pnas.83.2.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fontaine-Rodriguez EC, Knipe DM. Herpes simplex virus ICP27 increases translation of a subset of viral late mRNAs. J Virol. 2008;82:3538–3545. doi: 10.1128/JVI.02395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]