Abstract
The mammalian gastrointestinal tract harbors thousands of bacterial species that include symbionts as well as potential pathogens. The immune responses that limit access of these bacteria to underlying tissue remain poorly defined. Here we show that γδ intraepithelial lymphocytes (γδ IEL) of the small intestine produce innate antimicrobial factors in response to resident bacterial “pathobionts” that penetrate the intestinal epithelium. γδ IEL activation was dependent on epithelial cell-intrinsic MyD88, suggesting that epithelial cells supply microbe-dependent cues to γδ IEL. Finally, γδ T cells protect against invasion of intestinal tissues by resident bacteria specifically during the first few hours after bacterial encounter, indicating that γδ IEL occupy a unique temporal niche among intestinal immune defenses. Thus, γδ IEL detect the presence of invading bacteria through cross-talk with neighboring epithelial cells and are an essential component of the hierarchy of immune defenses that maintain homeostasis with the intestinal microbiota.
Keywords: antibacterial defense, mucosal immunity, microbiota
All mammals engage in a mutually beneficial symbiosis with a diverse microbial community that can change dynamically through acquisition of organisms from the environment (1, 2). Despite the mutually beneficial nature of the mammalian host–bacterial relationship, resident microbiota can invade intestinal tissues. This occurs during acquisition of new bacterial species from the environment, before the development of species-specific adaptive immune responses (3, 4). Tissue invasion can also occur in the presence of resident “pathobionts,” which have an intrinsic capacity to enter host cells (5). Thus, a key challenge for the mucosal immune system is to immediately detect barrier breach and rapidly limit invasion of bacteria into host tissue.
Large numbers of γδ T-cell receptor-bearing intraepithelial lymphocytes (γδ IEL) inhabit the body's epithelial barriers. γδ IEL intercalate between epithelial cells, and are thus poised to provide a first line of defense against environmental challenges. In the colon, γδ IEL function predominantly in the response to tissue injury, stimulating repair of damaged epithelia (6) and limiting bacterial penetration of injured tissue (7). However, γδ IEL are abundant even in undamaged intestinal tissues, suggesting that they perform homeostatic functions that extend beyond the response to tissue injury.
Here we show that γδ IEL are a critical component of the mucosal immune response against resident intestinal bacteria. We demonstrate that γδ IEL of the small intestine express innate antibacterial effectors, including the antibacterial lectin RegIIIγ, in response to a resident bacterial pathobiont that enters intestinal epithelial cells. The γδ IEL antibacterial response depends on bacterial stimulation of epithelial cell-intrinsic MyD88 signaling, indicating that epithelial cells supply microbe-dependent cues to γδ IEL. Finally, γδ T cells protect against tissue invasion by resident and pathogenic bacteria specifically during the first hours after bacterial encounter, suggesting that γδ IEL occupy a unique temporal niche among intestinal immune defenses. Together, our findings reveal that γδ IEL are one of the unique adaptations of the intestinal immune system that maintain homeostasis with a diverse resident microbiota.
Results
Intestinal Microbiota Direct an Antimicrobial Response in Small-Intestinal γδ IEL.
To characterize small-intestinal γδ IEL responses to the microbiota, we purified γδ IEL from germfree and conventionally raised mice. Similar numbers of small-intestinal γδ IEL were recovered from both groups of mice (Table S1), reflecting the fact that γδ IEL residence in the epithelium is independent of the microbiota (8). We established that our γδ IEL were free of contaminating epithelial cells by showing that the TCRδ+ cells expressed CD103, which marks lymphocytes but not epithelial cells (Fig. S1A).
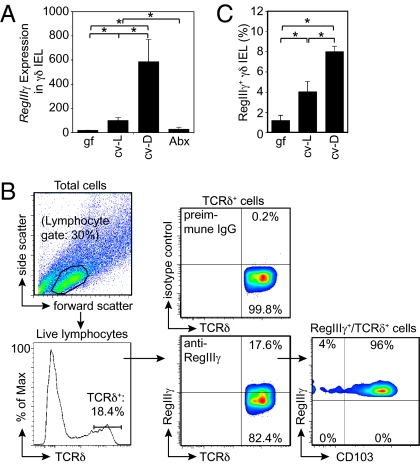
We next used microarrays to compare the gene expression profiles of these two γδ IEL populations. Of particular interest was the finding that intestinal microbiota elicited expression of a number of known or putative antimicrobial factors in small-intestinal γδ IEL (Fig. S1B). This response encompassed members of the regenerating islet-derived (Reg) protein family of C-type lectins, including RegIIIγ, which kills Gram-positive bacteria (9), and the related lectin RegIIIβ. In contrast to the colonic γδ IEL response to injury, we did not observe elevated expression of the proinflammatory cytokines KC, IL-1β, MIP-2α, or CXCL-9 (7). Elevated RegIIIγ and RegIIIβ expression was confirmed by quantitative PCR (Q-PCR) analysis of isolated γδ IEL (Fig. 1A and Fig. S2). To confirm that the observed RegIIIγ expression was in γδ IEL and did not result from epithelial cell contamination, we analyzed the RegIIIγ+/TCRδ+ population by flow cytometry. Virtually all RegIIIγ+/TCRδ+ cells expressed CD103, marking them as lymphocytes and not contaminating epithelial cells (Fig. 1B). These results indicate that γδ IEL respond to the microbiota not only in the context of injury but also under homeostatic conditions.
Fig. 1.
Intestinal microbiota direct RegIIIγ expression in small-intestinal γδ IEL. (A) RegIIIγ mRNA was quantified by Q-PCR of isolated small-intestinal γδ IEL from germfree (gf), conventionally raised (cv-L), conventionalized (cv-D), or antibiotic-treated (Abx) mice (n = 5 mice per group; representative of two experiments). (B) γδ IEL express RegIIIγ protein. Flow cytometry was performed on total small-intestinal IEL populations from cv-L mice. Intracellular staining was carried out with anti-RegIIIγ antibodies (9), and the gating strategy is shown. TCRδ+/RegIIIγ+ cells were analyzed to verify expression of the lymphocyte marker CD103. (C) Percentages of RegIIIγ+ γδ IEL were determined in gf, cv-L, and cv-D mice using the strategy shown in B. n = 5 mice per group; data are from two independent experiments. Error bars represent ±SEM. *P = 0.05.
We next asked whether microbial induction of the γδ IEL antibacterial response was reversible. Antibiotic treatment of conventional mice resulted in γδ IEL RegIIIγ transcript levels that were similar to those of germfree mice, demonstrating reversible expression upon microbiota depletion. Conversely, γδ IEL RegIIIγ expression was restored by “conventionalizing” germfree mice with a microbiota harvested from conventionally raised mice (Fig. 1 A and C). Conventionalization of germfree mice results in bacterial penetration of the epithelial barrier during the first 14 d after bacterial exposure, before the development of specific IgA (3). We noted higher RegIIIγ expression in conventionalized mice compared with mice that were conventional from birth (Fig. 1 A and C). Total numbers of small-intestinal bacteria did not differ between the two groups, arguing that this difference did not arise from higher bacterial numbers in the conventionalized mice. This suggests that enhanced γδ IEL antimicrobial responses in conventionalized compared with conventionally raised mice might be linked to this transient bacterial penetration of host tissue. We have addressed this possibility below.
RegIIIγ Expression Is Induced in γδ IEL by a Select Subset of the Microbiota.
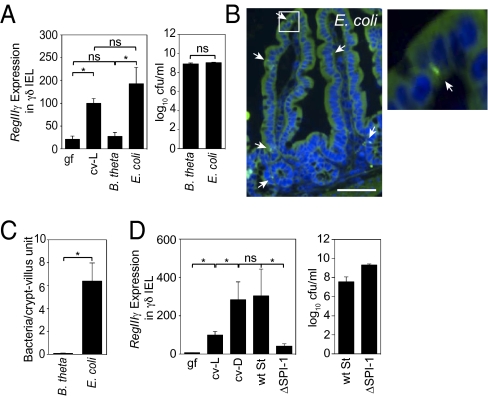
We next investigated which microbiota components elicit RegIIIγ expression in γδ IEL. Colonization with the Gram-negative symbiont Bacteroides thetaiotaomicron, a prominent member of the human and mouse intestinal microbiota, was not sufficient to trigger γδ IEL expression of RegIIIγ (Fig. 2A). This suggested that γδ IEL respond to a distinct subset of microbiota species rather than simply to the presence of bacteria. Th17 cells also respond to specific microbiota components, requiring segmented filamentous bacteria (SFB) for their differentiation (10). We therefore asked whether SFB could also activate γδ IEL. SFB are present in C57BL/6 mice from Taconic Farms but not in genetically identical mice from Jackson Laboratories (10). We verified increased SFB abundance in Taconic mice (Fig. S3A), but found no significant difference in RegIIIγ transcript abundance between γδ IEL isolated from Taconic and Jackson C57BL/6 mice (Fig. S3B). We therefore conclude that RegIIIγ expression by γδ IEL does not require SFB.
Fig. 2.
RegIIIγ expression is induced in γδ IEL by a select subset of the microbiota. (A) γδ IEL expression of RegIIIγ is triggered by a resident E. coli strain but not by B. thetaiotaomicron (B. theta). Germfree wild-type mice were gavaged with 108 cfu of B. theta or an E. coli strain isolated from the microbiota of specified pathogen-free (SPF) mice (n = 4–5 mice; representative of two experiments). After 48 h, γδ IEL were isolated and RegIIIγ mRNA was quantified by Q-PCR. Colonizing bacteria were quantified by dilution plating. ns, not significant. (B) E. coli enters small-intestinal epithelial cells. Bacteria were detected by FISH using a probe against the 16S rRNA gene (green). Arrows indicate examples of tissue-associated bacteria. Cell nuclei were visualized with DAPI (blue). As previously reported, there is nonspecific diffuse autofluorescence of intestinal tissues when visualized with FITC filters (24). (Scale bar, 50 μm.) Boxed area is enlarged at right. (C) Tissue-associated bacteria were enumerated in 200 well-oriented crypt-villus units from 5 mice per group. (D) Wild-type (wt) S. typhimurium or the invasion-deficient mutant ΔSPI-1 (108 cfu) were introduced orally into germfree C57BL/6 mice. After 48 h, γδ IEL were isolated and RegIIIγ mRNA was quantified by Q-PCR. Colonization levels were determined by dilution plating. n = 4–5 mice per group. Error bars represent ±SEM. *P < 0.05.
We next searched for a microbiota species resident in our conventional mice with the capacity to activate the γδ IEL antimicrobial response. We cultured total small-intestinal bacteria and used 16S rRNA gene sequencing to identify a predominant facultative aerobe as an Escherichia coli strain (Fig. S4). We further characterized the localization of this E. coli isolate in small-intestinal tissue by fluorescence in situ hybridization (FISH) analysis of monocolonized mice, using a fluorescent probe directed against the bacterial 16S rRNA gene. We detected bacteria in the intestinal lumen, associated with epithelial cells, and in the lamina propria (Fig. 2B), but did not detect bacteria in spleen and blood. Enumeration of tissue-associated bacteria revealed ∼6 bacterial cells per crypt-villus unit (Fig. 2C). As some bacteria appeared to be located within epithelial cells, we further analyzed the tissues by confocal microscopy. Z stacks reconstructed from confocal images verified that bacteria were located within epithelial cells (Fig. S5A and Movie S1). The intraepithelial localization correlated with the ability of the E. coli strain to enter the mouse intestinal epithelial cell line MODE-K (Fig. S5B). In contrast, B. thetaiotaomicron was confined to the intestinal lumen and was not detected in tissue (Fig. 2C and Fig. S5 C and D).
To test whether the native E. coli strain was sufficient to trigger γδ IEL expression of RegIIIγ, we monocolonized germfree mice and quantified RegIIIγ mRNA in γδ IEL 48 h later. In contrast to B. thetaiotaomicron-monoassociated mice, γδ IEL from E. coli-monoassociated mice exhibited a sevenfold increase in RegIIIγ expression relative to germfree mice (Fig. 2A). We therefore conclude that this resident E. coli strain is one member of the indigenous microbiota that can induce γδ IEL to express RegIIIγ, and can in part account for the ability of the steady-state microbiota to activate γδ IEL.
The capacity of the native E. coli strain to cross the mucosal barrier was a distinctive characteristic relative to B. thetaiotaomicron. The correlation of bacterial invasion with γδ IEL activation suggested that mucosal invasion might be required to trigger the γδ IEL antimicrobial response. Because the molecular basis for the ability of this resident E. coli strain to invade epithelia is currently unknown, we addressed this question using Salmonella typhimurium, an enteric pathogen whose capacity to penetrate the epithelial barrier has a well-defined genetic basis. S. typhimurium invades intestinal epithelia as a first step in barrier translocation and systemic dissemination. This process requires genes contained within the SPI-1 pathogenicity island, and a mutant strain lacking SPI-1 (ΔSPI-1) thus cannot invade the epithelial barrier (11). Monocolonization of germfree mice with wild-type S. typhimurium elicited an ∼44-fold increase in the abundance of γδ IEL RegIIIγ transcripts (Fig. 2D). Wild-type and ΔSPI-1 strains each colonized the small intestine to equivalent levels, but wild-type S. typhimurium induced approximately eightfold higher γδ IEL RegIIIγ mRNA expression relative to the mutant (Fig. 2D). The RegIIIγ expression levels correlated with bacterial entry into the intestinal epithelium (Fig. S6 A and B). These data argue that bacterial penetration of the mucosal surface is required to trigger RegIIIγ production by γδ IEL.
Epithelial Cell MyD88 Is Required for RegIIIγ Expression in Small-Intestinal γδ IEL.
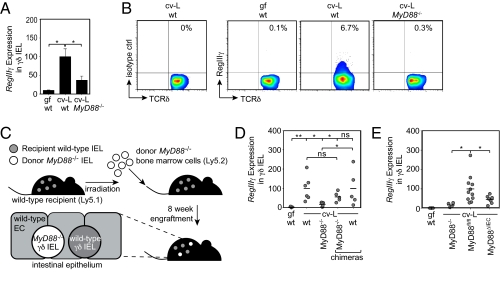
We next examined the host factors that regulate bacterial induction of the γδ IEL antimicrobial response. We have previously shown that small-intestinal Paneth cells, specialized epithelial cells, detect intestinal bacteria through Toll-like receptors (TLRs) that require the signaling adaptor protein MyD88. Paneth cell TLR engagement elicits production of antimicrobial proteins, including RegIIIγ, that promote homeostasis with the microbiota (12). We therefore tested whether MyD88 governs RegIIIγ expression in small-intestinal γδ IEL. Similar numbers of γδ IEL were recovered from wild-type and MyD88−/− mice (Table S1), in agreement with published data (13) and with the fact that germfree mice harbor normal numbers of γδ IEL. However, γδ IEL from MyD88−/− mice showed decreased expression of RegIIIγ relative to wild-type mice (Fig. 3A), and flow cytometry confirmed fewer RegIIIγ+ γδ IEL from MyD88−/− mice (Fig. 3B). The related antibacterial lectin RegIIIβ showed a similar dependence on MyD88 (Fig. S2). Thus, expression of RegIIIγ and RegIIIβ in small-intestinal γδ IEL requires MyD88.
Fig. 3.
Epithelial cell MyD88 is required for RegIIIγ expression in small-intestinal γδ IEL. RegIIIγ was quantified by (A) Q-PCR of sorted small-intestinal γδ IEL and (B) flow cytometry of total IEL, performed as in Fig. 1. Gated γδ IEL populations are shown, and percentages of gated populations are given. Four to five mice were pooled per group; results are representative of three independent experiments. (C) Schematic of bone marrow chimera experiment. Ly5.1 wild-type mice were irradiated and reconstituted with bone marrow cells from Ly5.2 MyD88−/− mice. The chimeric intestines retained residual recipient cells while also supporting engraftment of transplanted cells (40% donor/60% recipient γδ IEL). EC, epithelial cells. (D) Eight weeks after reconstitution, Ly5.1 and Ly5.2 γδ IEL were isolated by FACS and analyzed individually for RegIIIγ mRNA by Q-PCR. Each point represents an individual mouse, and the conventional experimental groups were cohoused for 5 d before sacrifice to ensure a shared microbiota. (E) Q-PCR for RegIIIγ expression in sorted small-intestinal γδ IEL. MyD88ΔIEC, mice with an epithelial cell-specific MyD88 deletion; MyD88fl/fl, littermates harboring two floxed MyD88 alleles. Littermates were cohoused to ensure a shared microbiota. Each point represents an individual mouse. Error bars represent ±SEM. *P < 0.05; **P < 0.01; ns, not significant.
We next addressed the cellular origin of the MyD88 signaling that triggers γδ IEL production of RegIIIγ. As a first step, we generated bone marrow chimeric mice in which MyD88 was predominantly expressed in nonhematopoietic lineages such as epithelial cells (Fig. 3C). Although we were also able to generate the reverse chimera, we were unable to recover viable γδ IEL from these mice, suggesting that nonhematopoietic MyD88 might be important for γδ IEL viability after irradiation. As previously reported, chimerism was lower in the intestine than in the circulation (40% donor γδ IEL in intestine compared with ∼90% donor lymphocytes in the circulation) (14). Therefore, the chimeric mice had a mixed population of recipient wild-type γδ IEL and donor MyD88−/− γδ IEL that could be tracked based on the congenic markers Ly5.1 and Ly5.2 (Fig. 3C). Adoptive transfer of MyD88−/− bone marrow into lethally irradiated wild-type mice restored expression of RegIIIγ in donor MyD88−/− γδ IEL to wild-type levels (Fig. 3D). Thus, RegIIIγ expression does not depend on γδ IEL MyD88, establishing that bacterial activation of γδ IEL is cell-nonautonomous.
γδ IEL make direct contact with neighboring epithelial cells, suggesting that epithelial cells might provide the MyD88 signals that activate γδ IEL. To test this idea, we generated mice with an epithelial cell-specific deletion of MyD88 (MyD88ΔIEC). We crossed mice carrying the epithelial cell-restricted Villin-Cre transgene (15) with mice harboring a loxP-flanked MyD88 allele (MyD88fl/fl) (16). This yielded progeny with an epithelial cell-specific MyD88 deletion (Fig. S7). RegIIIγ expression in γδ IEL from MyD88ΔIEC mice was decreased relative to MyD88fl/fl littermates (Fig. 3E). This indicates that RegIIIγ induction in γδ IEL requires epithelial cell MyD88, and suggests that epithelial cells supply γδ IEL with microbe-dependent cues that govern the expression of antimicrobial factors.
γδ T Cells Limit Bacterial Penetration at Early Time Points After Bacterial Exposure.
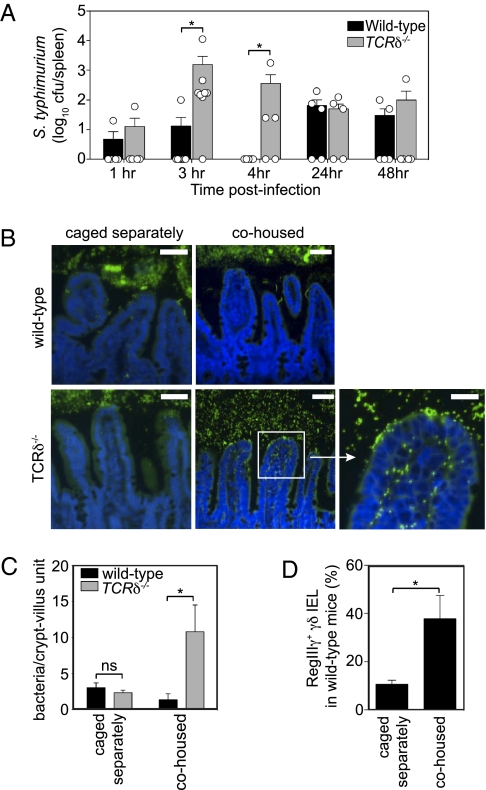
The finding that penetrant bacteria stimulate an antibacterial response in γδ IEL suggested that a key function of γδ IEL might be to limit bacterial invasion of mucosal tissues. To test this idea, we studied TCRδ−/− mice, which lack all γδ T cells including γδ IEL (17). We orally inoculated conventional wild-type and TCRδ−/− mice with 108 cfu of S. typhimurium and then quantified splenic bacteria at various times after inoculation. At 3 h there was a 100-fold increase in splenic S. typhimurium in TCRδ−/− compared with wild-type mice (Fig. 4A). This difference was limited to early time points, as numbers of S. typhimurium recovered from spleen at 24 and 48 h after infection were similar in TCRδ−/− and wild-type mice (Fig. 4A). The increased splenic dissemination at 3 h correlated with increased bacterial entry into epithelial cells in TCRδ−/− mice (Fig. S8A). The elevated bacterial loads in spleen were not due to increased luminal loads in TCRδ−/− mice (Fig. S8B) or to impaired bacterial killing in the spleens of TCRδ−/− mice (Fig. S8C). These results support the idea that γδ T cells, and most likely γδ IEL, limit S. typhimurium penetration and dissemination immediately following bacterial encounter in the intestinal mucosa.
Fig. 4.
γδ T cells limit bacterial penetration of the intestinal mucosa at early time points after bacterial challenge. (A) Wild-type S. typhimurium (108 cfu) were gavaged into conventional wild-type and TCRδ−/− mice, and splenic bacteria were quantified by dilution plating. Each point represents one mouse; data are from two independent experiments. (B) FISH analysis of conventional wild-type and TCRδ−/− mice. Wild-type and TCRδ−/− mice caged separately by genotype were either analyzed immediately upon removal from the SPF facility or were cohoused in the same cage for 4 h before sacrifice. Small-intestinal tissues were probed with a universal bacterial FISH probe (green) and counterstained with DAPI (blue). (Scale bars, 50 μm.) Results are representative of all mice from two independent cohousing experiments (10 mice per genotype). (C) Tissue-associated bacteria were enumerated by counting bacteria within 200 well-oriented crypt-villus units from 5 mice per genotype. (D) The percentage of γδ IEL expressing RegIIIγ was determined in wild-type mice that were caged separately from TCRδ−/− mice and compared with wild-type mice that were not cohoused. RegIIIγ expression was determined by intracellular staining. n = 3–4 mice per group; results are representative of two independent cohousing experiments. Error bars represent ±SEM. *P < 0.05; ns, not significant.
We next investigated whether γδ T cells also limit penetration of resident microbiota. We performed FISH analysis on wild-type and TCRδ−/− mice to visualize the locations of intestinal bacteria relative to tissue. We observed small numbers of invading bacteria in the intestines of wild-type mice (∼3–4 bacteria per crypt-villus unit), consistent with the stimulation of γδ IEL antibacterial responses by the microbiota in conventional mice (Fig. 4 B and C). However, we did not detect increased numbers of tissue-associated bacteria in TCRδ−/− mice (Fig. 4 B and C). Given that γδ T cells limit S. typhimurium dissemination at early time points after infection, we reasoned that perhaps these cells also function specifically at early time points following exposure to new resident bacteria. Germfree TCRδ−/− mice are not available, precluding analysis of the protective effects of γδ T cells against mucosal penetration by single resident bacterial species. However, cohousing of mice that have been previously caged separately introduces new resident bacterial species into the gastrointestinal tract and/or alters the abundances of species already present (3, 18, 19). We therefore used cohousing to test whether γδ T cells protect against penetration of resident bacteria during acquisition of new resident species from other mice and from the environment. Wild-type and TCRδ−/− mice that had been separately caged were cohoused for 4 h and their small intestines were examined by FISH. Numbers of intracellular bacteria increased in TCRδ−/− mice (∼11 bacteria per crypt-villus unit; Fig. 4 B and C). In contrast, numbers of penetrant bacteria in wild-type mice remained similar to the numbers observed before cohousing (Fig. 4 B and C). Further, γδ IEL isolated from wild-type mice after cohousing showed increased RegIIIγ protein relative to noncohoused mice (Fig. 4D). Measurements of serum fluorescence following FITC-dextran administration revealed that the bacterial penetration in the TCRδ−/− mice did not arise from increased nonspecific barrier permeability (Fig. S9). These results demonstrate that γδ IEL play an essential role in limiting mucosal penetration by intestinal bacteria during shifts in microbiota composition and/or acquisition of new organisms from the environment.
Discussion
γδ IEL represent a large proportion of intestinal T cells, yet their biological functions are poorly understood. Here we show that the biological functions of γδ IEL extend beyond contributions to epithelial wound repair and immunoregulation to include a general role in maintaining homeostasis with the intestinal microbiota. γδ IEL provide a rapid first line of mucosal defense that promotes homeostasis with the microbiota by detecting and limiting bacterial penetration of intestinal tissue. The mucosal protection afforded by γδ IEL is of critical importance during the first hours after bacterial exposure, suggesting that γδ IEL occupy a unique temporal niche among mucosal immune responses.
Our studies have yielded key mechanistic insights into how γδ IEL sense intestinal bacteria. First, our data argue that γδ IEL respond specifically to invasive microorganisms, whether they are resident pathobionts or exogenous overt pathogens. Second, we found that bacterial stimulation of γδ IEL was indirect, requiring activation of MyD88 signaling in neighboring epithelial cells. Interestingly, the MyD88 dependence of small-intestinal γδ IEL RegIIIγ expression contrasts with γδ IEL from chemically injured colon, which exhibit MyD88-independent RegIIIγ expression (7). Such differences suggest that γδ IEL function may differ in the two organs, or that γδ IEL may be regulated by distinct mechanisms in damaged and undamaged mucosal tissues. Analysis of skin γδ T cells suggests that γδ IEL–epithelial cell crosstalk could occur via recognition of epithelial cell stress antigens or bacterial antigens by γδ IEL T-cell receptors (20, 21). Other studies indicate that epithelial cell cytokines could stimulate γδ IEL antibacterial responses (22, 23). Further investigation will be required to determine which of these mechanisms induces γδ IEL antimicrobial responses to intestinal bacteria.
Epithelial cells produce antimicrobial proteins that defend the apical surfaces of epithelial cells against bacterial attachment and invasion (12, 24). Our findings suggest that by signaling to γδ IEL, epithelial cells could also direct the expression of antibacterial effectors that defend the intraepithelial and basolateral compartments against microbial invasion. An important unresolved issue is whether antibacterial factors, such as RegIIIγ and RegIIIβ, account for the protective functions of γδ IEL. These effectors would likely be secreted into the intraepithelial and basolateral spaces, thus suggesting a molecular mechanism by which γδ IEL could limit bacterial penetration into mucosal tissue. A second possibility is that γδ IEL cytolytic functions promote the removal of infected epithelial cells, thus preventing further bacterial invasion (25). Testing these models awaits the development of new tools for cell-specific genetic manipulations of γδ IEL in mice.
Mucosal immune responses that follow a bacterial challenge are complex, evolving sequentially over many hours and days and involving contributions from multiple cells and effectors. We show here that γδ IEL provide essential antibacterial protection of the mucosal surface specifically during the first hours following bacterial exposure. Like cohoused mice, humans are continually exposed to new intestinal bacteria through ingestion of food and water, contact with other humans, and exposure to organisms in the surrounding environment. Our findings suggest that γδ IEL could be critical mediators of homeostasis between humans and their microbiota, and thus represent attractive therapeutic targets for the management of intestinal inflammatory diseases.
Materials and Methods
Animals.
Wild-type C57BL/6 mice, MyD88−/− mice, Villin-Cre mice, MyD88fl/fl mice, and TCRδ−/− mice were bred at the University of Texas Southwestern Medical Center. For certain experiments, C57BL/6 mice were purchased from Jackson Laboratories and Taconic Farms and used immediately. Germfree C57BL/6 mice were maintained in isolators as described (9). All protocols were approved by the Institutional Animal Care and Use Committees of the University of Texas Southwestern Medical Center.
Bacterial Strains.
B. thetaiotaomicron (VPI-5482) and S. enterica serovar typhimurium (SL1344) and its isogenic mutant ΔSPI-1 were cultured as described (12). An E. coli strain native to the mouse microbiota was isolated by plating total small-intestinal contents on LB agar and growing aerobically at 37 °C. The species was identified by amplifying the 16S rRNA gene using primers that anneal to conserved regions (forward: 5′-ACTCCTACGGGAGGCAGCAGT-3′; reverse: 5′-ATTACCGCGGCTGCTGGC-3′) (24). The amplicon sequence was compared with all known rRNA genes through BLAST, revealing 100% identity with rRNAs from multiple E. coli strains (Fig. S4).
γδ IEL Isolation.
Intestines were flushed with PBS, everted, and washed in cold PBS. Tissues were gently agitated for 30 min at 37 °C in 25 mL extraction buffer (1 mM EDTA, 1% BSA, 1 mM DTT, PBS). IEL were shaken off the intestinal lining by vigorous vortexing for 2 min, and then were filtered through a 100 μM cell strainer, a 40 μM strainer, and a glass wool column. Total IEL were stained with phycoerythrin-labeled anti-TCRδ (GL3; BD Pharmingen), and γδ IEL were purified on a Beckman Coulter MoFlo cell sorter. γδ IEL purity was assayed postsorting and was ≥98% (Fig. S1A).
Quantitative PCR.
Total RNA was isolated from purified γδ IEL using the Arcturus PicoPure RNA Kit and was used to generate template cDNA. mRNA levels were normalized to 18S rRNA abundance and are expressed relative to conventionally raised mice. Sequences of RegIIIγ and 18S rRNA primers are published (9).
Flow Cytometry.
For surface staining, IEL were suspended in PBS/0.5% BSA and stained for 20 min with PE-conjugated anti-TCRγδ (BD Pharmingen). For intracellular staining, cells were fixed using the BD Pharmingen Cytofix/Cytoperm Kit, and stained with preimmune IgG or anti-RegIIIγ (9), followed by anti-rabbit secondary antibody (Jackson Immunoresearch) and anti-CD103 (eBioscience).
Antibiotic Treatment.
Conventional C57BL/6 mice were given ampicillin (1 mg/mL), vancomycin (0.50 mg/mL), neomycin sulfate (1 mg/mL), and metronidazole (1 mg/mL) in drinking water for 4 wk. All antibiotics were from Sigma. Microbiota depletion was verified by aerobic and anaerobic culture of intestinal contents.
Bone Marrow Chimeras.
Wild-type C57BL/6 Ly5.1 mice were γ-irradiated at 9 Gy and reconstituted with 5 × 106 bone marrow cells from MyD88−/− Ly5.2 mice. Eight weeks posttransfer, γδ IEL of Ly5.1 (wild-type) and Ly5.2 (MyD88−/−) origin were isolated by flow cytometry. Chimeras were cohoused with wild-type and MyD88−/− controls before sacrifice to ensure that all experimental groups shared a microbiota.
FISH Analysis.
Small intestines were prepared by fixation in Carnoy's fixative (Ricca Chemical) and embedded in paraffin. Tissues were sectioned at 5 μm and hybridized to a bacterial 16S rRNA gene probe: [AminoC6+Alexa488]-GCTGCCTCCCGTAGGAGT-[AmC7∼Q+Alexa488]. Hybridizations were carried out as described (24) and visualized on a Zeiss AxioImager M1 microscope.
Statistics.
Statistical significance was determined by unpaired two-tailed Student's t test.
Supplementary Material
Acknowledgments
We thank A. Mobley, S. Murray, and C. Clements for their assistance with various aspects of this work. This work was supported by National Institutes of Health (NIH) Grant R01 DK070855 (to L.V.H.), the Crohn's and Colitis Foundation of America (L.V.H.), a Crohn's and Colitis Foundation of America fellowship award (to S.V.), a Burroughs Wellcome Foundation New Investigators in the Pathogenesis of Infectious Diseases award (to L.V.H.), and NIH Grants T32 AI005284 (to A.S.I.) and P01 AI078869 (to A.D.).
Footnotes
The authors declare no conflict of interest.
Data deposition: Array data are deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE28437).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1019574108/-/DCSupplemental.
References
- 1.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shroff KE, Meslin K, Cebra JJ. Commensal enteric bacteria engender a self-limiting humoral mucosal immune response while permanently colonizing the gut. Infect Immun. 1995;63:3904–3913. doi: 10.1128/iai.63.10.3904-3913.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 5.Chow J, Mazmanian SK. A pathobiont of the microbiota balances host colonization and intestinal inflammation. Cell Host Microbe. 2010;7:265–276. doi: 10.1016/j.chom.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y, Chou K, Fuchs E, Havran WL, Boismenu R. Protection of the intestinal mucosa by intraepithelial γ δ T cells. Proc Natl Acad Sci USA. 2002;99:14338–14343. doi: 10.1073/pnas.212290499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ismail AS, Behrendt CL, Hooper LV. Reciprocal interactions between commensal bacteria and γ δ intraepithelial lymphocytes during mucosal injury. J Immunol. 2009;182:3047–3054. doi: 10.4049/jimmunol.0802705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bandeira A, et al. Localization of γ/δ T cells to the intestinal epithelium is independent of normal microbial colonization. J Exp Med. 1990;172:239–244. doi: 10.1084/jem.172.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eichelberg K, Galán JE. Differential regulation of Salmonella typhimurium type III secreted proteins by pathogenicity island 1 (SPI-1)-encoded transcriptional activators InvF and hilA. Infect Immun. 1999;67:4099–4105. doi: 10.1128/iai.67.8.4099-4105.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci USA. 2008;105:20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iiyama R, et al. Normal development of the gut-associated lymphoid tissue except Peyer's patch in MyD88-deficient mice. Scand J Immunol. 2003;58:620–627. doi: 10.1111/j.1365-3083.2003.01346.x. [DOI] [PubMed] [Google Scholar]
- 14.Hall JA, et al. Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity. 2008;29:637–649. doi: 10.1016/j.immuni.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Madison BB, et al. cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J Biol Chem. 2002;277:33275–33283. doi: 10.1074/jbc.M204935200. [DOI] [PubMed] [Google Scholar]
- 16.Hou B, Reizis B, DeFranco AL. Toll-like receptors activate innate and adaptive immunity by using dendritic cell-intrinsic and -extrinsic mechanisms. Immunity. 2008;29:272–282. doi: 10.1016/j.immuni.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Itohara S, et al. T cell receptor δ gene mutant mice: Independent generation of α β T cells and programmed rearrangements of γ δ TCR genes. Cell. 1993;72:337–348. doi: 10.1016/0092-8674(93)90112-4. [DOI] [PubMed] [Google Scholar]
- 18.Ivanov II, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fushuku S, Fukuda K. Inhomogeneity of fecal flora in separately reared laboratory mice, as detected by denaturing gradient gel electrophoresis (DGGE) Exp Anim. 2008;57:95–99. doi: 10.1538/expanim.57.95. [DOI] [PubMed] [Google Scholar]
- 20.Jameson JM, Cauvi G, Witherden DA, Havran WL. A keratinocyte-responsive γ δ TCR is necessary for dendritic epidermal T cell activation by damaged keratinocytes and maintenance in the epidermis. J Immunol. 2004;172:3573–3579. doi: 10.4049/jimmunol.172.6.3573. [DOI] [PubMed] [Google Scholar]
- 21.Jameson J, et al. A role for skin γδ T cells in wound repair. Science. 2002;296:747–749. doi: 10.1126/science.1069639. [DOI] [PubMed] [Google Scholar]
- 22.Laky K, et al. Enterocyte expression of interleukin 7 induces development of γδ T cells and Peyer's patches. J Exp Med. 2000;191:1569–1580. doi: 10.1084/jem.191.9.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu Q, et al. MyD88-dependent signaling for IL-15 production plays an important role in maintenance of CD8αα TCRαβ and TCRγδ intestinal intraepithelial lymphocytes. J Immunol. 2006;176:6180–6185. doi: 10.4049/jimmunol.176.10.6180. [DOI] [PubMed] [Google Scholar]
- 24.Salzman NH, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2010;11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lefrancois L, Goodman T. In vivo modulation of cytolytic activity and Thy-1 expression in TCR-γ δ+ intraepithelial lymphocytes. Science. 1989;243:1716–1718. doi: 10.1126/science.2564701. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.