Abstract
Circadian (24-hr) rhythms influence virtually every aspect of mammalian physiology. The main rhythm generation center is located in the suprachiasmatic nucleus (SCN) of the hypothalamus, and work over the past several years has revealed that rhythmic gene transcription and post-translational processes are central to clock timing. In addition, rhythmic translation control has also been implicated in clock timing; however the precise cell signaling pathways that drive this process are not well known. Here we report that a key translation activation cascade, the mammalian target of rapamycin (mTOR) pathway, is under control of the circadian clock in the SCN. Using phosphorylated S6 ribosomal protein (pS6) as a marker of mTOR activity, we show that the mTOR cascade exhibits maximal activity during the subjective day, and minimal activity during the late subjective night. Importantly, expression of S6 was not altered as a function of circadian time. Rhythmic S6 phosphorylation was detected throughout the dorsoventral axis of the SCN, thus suggesting that rhythmic mTOR activity was not restricted to a subset of SCN neurons. Rather, rhythmic pS6 expression appeared to parallel the expression pattern of the clock gene period1 (per1). Using a transgenic per1 reporter gene mouse strain, we found a statistically significant cellular level correlation between pS6 and per1 gene expression over the circadian cycle. Further, photic stimulation triggered a coordinate upregulation of per1 and mTOR activation in a subset of SCN cells. Interestingly, this cellular level correlation between mTOR activity and per1 expression appears to be specific, since a similar expression profile for pS6 and per2 or c-FOS was not detected. Finally, we show that mTOR activity is downstream of the ERK/MAPK signal transduction pathway. Together these data reveal that mTOR pathway activity is under the control of the SCN clock, and suggests that mTOR signaling may contribute to distinct aspects of the molecular clock timing process.
Keywords: circadian, SCN, S6, phosphorylation, per1, mTOR, ERK
Introduction
An inherent biological timing mechanism imparts 24 hr (i.e. circadian) rhythms on a vast array of physiological processes (Reppert and Weaver, 2002). For mammals, the dominant rhythm is generated by the suprachiasmatic nuclei (SCN), a relatively small brain region (~ 20,000 neurons/nucleus) located within the ventral hypothalamus (Ukai and Ueda, 2010). The rhythm-generating capacity of the SCN arises from a network of autonomous SCN oscillator neurons, which, through synaptic and paracrine-driven processes, forms a coherent and stable oscillator network (Welsh et al., 2010).
At the molecular level, the cellular clock is driven by several interlocking transcriptional/translational feedback loops (Ko and Takahashi, 2006). The major negative feedback loop is centered on the rhythmic regulation of period (per) and cryptochrome (cry) genes. Briefly, the transcription factors CLOCK and BMAL1 form heterodimers and activate transcription of the per and cry genes by binding to their E-box enhancers. As the levels of PER proteins increase, they form complexes with CRY, translocate into cell nucleus and associate with CLOCK–BMAL1 heterodimers which in turn leads to transcriptional repression of per and cry genes. Clock gene transcriptional inhibition is relieved via the posttranslational-component of the feedback loop. Along these lines, phosphorylation via casein kinase I (CKI) plays a key role in targeting in the PER/CRY complex for ubiquitin-mediated degradation, thereby allowing a new cycle of clock gene expression to occur (Gallego and Virshup, 2007).
Interestingly, a relatively new piece of this rhythm-generation puzzle has begun to emerge: translation regulation. In support of this concept, several studies have shown that microRNAs, which function as potent negative regulators of mRNA translation, influence clock timing (Cheng et al., 2007; Shi et al., 2009; Na et al., 2009; Nagel et al., 2009). Likewise, Nocturnin, a rhythmically expressed mRNA deadenylase, regulates circadian mRNA degradation (Douris and Green, 2009). Further, the rhythmically-expressed RNA-binding protein LARK (McNeil et al., 1998) regulates per mRNA translation and clock function (Kojima et al., 2007). Of particular relevance here, recent work in Drosophila has indicated a role for target of rapamycin (TOR) in clock physiology (Zheng and Sehgal, 2010).
Mammalian target of rapamycin (mTOR) is a serine/threonine protein kinase that is inhibited by the antifungal metabolite rapamycin (Hay and Sonenberg, 2004). mTOR plays fundamental roles in regulating cell growth, metabolism and mRNA translation. It executes its function via two distinct multi-protein complexes: the rapamycin-sensitive mTOR Complex 1 (mTORC1), which contains Raptor, and the rapamycin-insensitive mTORC2, which contains Rictor (Wullschleger et al., 2006). mTOR functioning within mTORC1 regulates translational control by direct phosphorylation of two distinct translation effectors: S6 kinase1 (S6K1, including two isoforms, p70 S6K and p85 S6K) and eukaryotic initiation factor 4E-binding protein 1 (4E-BP1).
Our recent work has identified a light-regulated mTORC1 signal cascade in the SCN that influences clock entrainment (Cao et al., 2008, 2010). These studies also reported a relatively high level of basal mTOR activity in the SCN, thus raising the prospect that mTOR activity could be influenced by the core clock timing process. Here, we provide data revealing a robust circadian rhythm of mTOR activity in the SCN. Further, we found a cellular-level correlation between circadian Per1 gene expression and mTOR activity in the SCN. These data connect mTOR signaling to circadian cellular level clock timing, and as such, raise the prospect to mTOR contributes to clock physiology
Materials and Methods
Photic entrainment and tissue processing
Initially, adult (8~10-week-old) wild-type C57BL/6, mper1-Venus transgenic or mper2-DsRed transgenic mice (Cheng et al., 2009) were entrained to a 12h/12h light/dark (L/D) cycle for at least 2 weeks and then transferred to total darkness for two consecutive 24 h cycles. After dark-adaptation, animals were sacrificed under dim red light (Kodak series 2 filter <5 lux at cage level; Eastman Kodak, Rochester, NY) over the circadian cycle. Circadian times (CTs) were calculated based on Zeitgeber time (ZT) and the period length (tau value) of C57BL/6 mice (approximately 23 h 45 min) (Schwartz and Zimmerman, 1990) and mper transgenic mice (approximately 23 h 54 min) (Cheng et al., 2009) under free running conditions, with CT 0 denoting subjective daytime and CT 12 denoting the beginning of the subjective nighttime. For the light flash experiment, after dark-adaptation mper1-Venus animals received a single light exposure (100 lux, 15 min) at CT 15 and then were sacrificed 4 hr later at CT19. Control mice (no light exposure) were also sacrificed at CT19. Animals were killed via cervical dislocation, and brains were removed under red light. Brains were then placed in chilled, oxygenated physiological saline, cut into 1.5 mm coronal slices with a vibrotome (OTS 2000; Electron Microscopy Sciences, Fort Washington, PA), fixed in 4 % paraformaldehyde for 6 hr at room temperature and then transferred into 30% sucrose (w/v, with 2 mM sodium azide and 3 mM NaF) overnight at 4 °C. All procedures were in accordance with Ohio State University animal welfare guidelines and approved by the Institutional Animal Care and Use Committee.
Brain Infusion
Cannulation and infusion procedures were conducted as previously described (Cao et al., 2010). Briefly, adult (8~10-week-old) wildtype C57BL/6 mice were anesthetized and placed in a stereotaxic apparatus (Cartesian Research). The coordinates (posterior, 0.34 mm from bregma; lateral, 0.90 mm from the midline; and dorsoventral, −2.15 mm from bregma) were used to place the tip of a 24-gauge guide cannula into the lateral ventricle. After surgery animals were housed individually and allowed to recover for at least 2 weeks under a standard 12 h/12h L/D cycle. To disrupt the MAPK cascade, 2 µl of 1,4-diamino-2,3-dicyano-1,4-bis o-aminophenylmercapto butadiene (U0126, 10 mM; Calbiochem, La Jolla, CA) was infused 30 min before sacrifice. To inhibit the activity of mTOR, 2 µl of rapamycin (100 µM, Cell Signaling Technology, Beverly, MA) was infused 30 min before sacrifice. To inhibit the activity of AKT, 2 µl of Akti-1/2 (20 mM, Chemdea, Ridgewood, NJ) was infused 30 min before sacrifice. Control animals were infused with an equivalent volume of vehicle (DMSO).
Immunohistochemistry
Coronal brain slices (1.5 mm) containing the SCN were cut into thin sections (40 µm) using a freezing microtome and placed in PBS containing 2 mM sodium azide and 3 mM NaF, pH 7.4. For immunohistochemical staining, sections were first treated with 0.3 % H2O2 and 20 % methanol in PBS for 10 min to deactivate endogenous peroxidases and to permeabilize the tissue. The tissue was then blocked for 1 h in 10% goat serum/PBS and incubated in rabbit anti-phospho-S6 ribosomal protein (pS6, Ser-240/244) (1:1000; Cell Signaling Technology), mouse anti-S6 ribosomal protein (1:500; Cell Signaling Technology) or rabbit anti-phosphorylated ERK (pERK, Thr-202, Tyr-204) (1:2000; Cell Signaling Technology) antibody overnight at 4°C. Next, tissue was incubated for 1.5 h in biotinylated secondary antibody (1:200; Vector Laboratories, Burlingame, CA) at room temperature and then placed in an avidin/biotin/HRP complex for 1 h (Vector Laboratories). Sections were washed in PBS (three times, 10 min per wash) between each labeling step. The signal was visualized using nickel-intensified DAB substrate (Vector Laboratories) and sections were mounted on slides with Permount media (Fisher Scientific, Houston, TX).
For immunofluorescence labeling, tissue was permeabilized with PBST (PBS with 1 % Triton X-100) for 30 min, blocked as described above and then incubated (overnight, 4° C) in 5 % goat serum/PBS with a combination of two of the following antibodies: mouse anti-pERK (Thr-202, Tyr-204) (1:300; Cell Signaling Technology), rabbit polyclonal anti-pS6 (Ser-240/244) (1:300; Cell Signaling Technology), chicken anti-GFP (1:2,000; Abcam, Cambridge, MA), rabbit polyclonal anti-DsRed (1:200; Clontech), goat polyclonal anti-c-Fos (1:500; Calbiochem) or guinea pig anti-vasopressin (1:500; Abcam). The following day, sections were incubated (3 h, room temperature) in Alexa Fluor-conjugated secondary antibodies (1:500; Molecular Probes, Eugene OR) directed against the IgG domains of the primary antibodies.
Of note, double labeling DsRed or c-Fos with pS6 employed an Alexa Fluor-488-conjugated rabbit polyclonal anti-pS6 (Ser-240/244) (1:200; Cell Signaling Technology). Labeling (3h at room temperature) with the Fluor-conjugated pS6 antibody was performed after all of the DsRed and c-Fos labeling steps were completed. Brain sections were washed in PBS (three times, 10 min per wash) between each labeling step. For some experiments, sections were incubated (10 min) in PBS containing the DNA stain DRAQ5 (1:3000; Biostatus Limited, Leicestershire, UK) before the final PBS wash. Sections were mounted on slides with Cytoseal 60 (Richard-Allan Scientific, Kalamazoo, MI).
Bright-field photomicrographs were captured using a 16 bit digital camera (Micromax YHS 1300; Princeton Instruments, Trenton, NJ) mounted on an inverted Leica microscope (DM IRB; Nussloch, Germany); images were acquired with Metamoph software (Molecular Devices, Sunnyvale CA). Fluorescence images were captured using a Zeiss 510 Meta confocal microscope (Oberkochen, Germany). All confocal parameters (pinhole, contrast, brightness, etc.) were held constant for all data sets from the same experiment.
Materials
Unless otherwise indicated, all reagents were obtained from Sigma.
Data analysis
All photomicrographic data sets were statistically analyzed using Adobe Photoshop software (Adobe Systems Incorporated, San Jose, CA). For the pS6 intensity analysis (figure 1), the SCN from 10X images were digitally outlined and the mean pixel values determined. Next, the pixel value within a digital oval (150×200 pixels) in the adjacent lateral hypothalamus determined and the ratio of the pS6 in the SCN to the lateral hypothalamus was determined. Data were averaged bilaterally, and from 3 central SCN sections per animal. For pERK intensity analysis (figure 5), SCN were digitally outlined and the mean pixel values (0–255 scale) determined. Next, a digital oval (150×200 pixels) was placed on the adjacent lateral hypothalamus and this mean value was subtracted from the adjacent SCN signal to provide a normalized SCN intensity value. As with the pS6 analysis, the data were averaged bilaterally, and from 3 central SCN sections per animal to generate a mean value. Mean values from different animals were pooled into treatment groups and analyzed by one-way ANOVA and compared via the SNK (Student-Newman-Keuls) post-hoc test. The values are presented as the mean ± standard error of mean (SEM). For the pS6 and Venus or DRAQ5 cellular correlation study, confocal images (40X magnification) of pS6 immunolabeling were collected and 20 Venus-positive SCN cells were randomly picked from each image and manually outlined. Next, a threshold filter was applied to eliminate non-specific background labeling, and densitometry values for red (for pS6), green (for Venus) and blue (for DRAQ5) channels of the outlined cells were determined. Pearson's correlation analysis was performed between pS6 and Venus data or DRAQ5 and Venus data. P < 0.05 was accepted as statistically significant. All statistical analysis was performed using SPSS software (SPSS Inc, Chicago, IL).
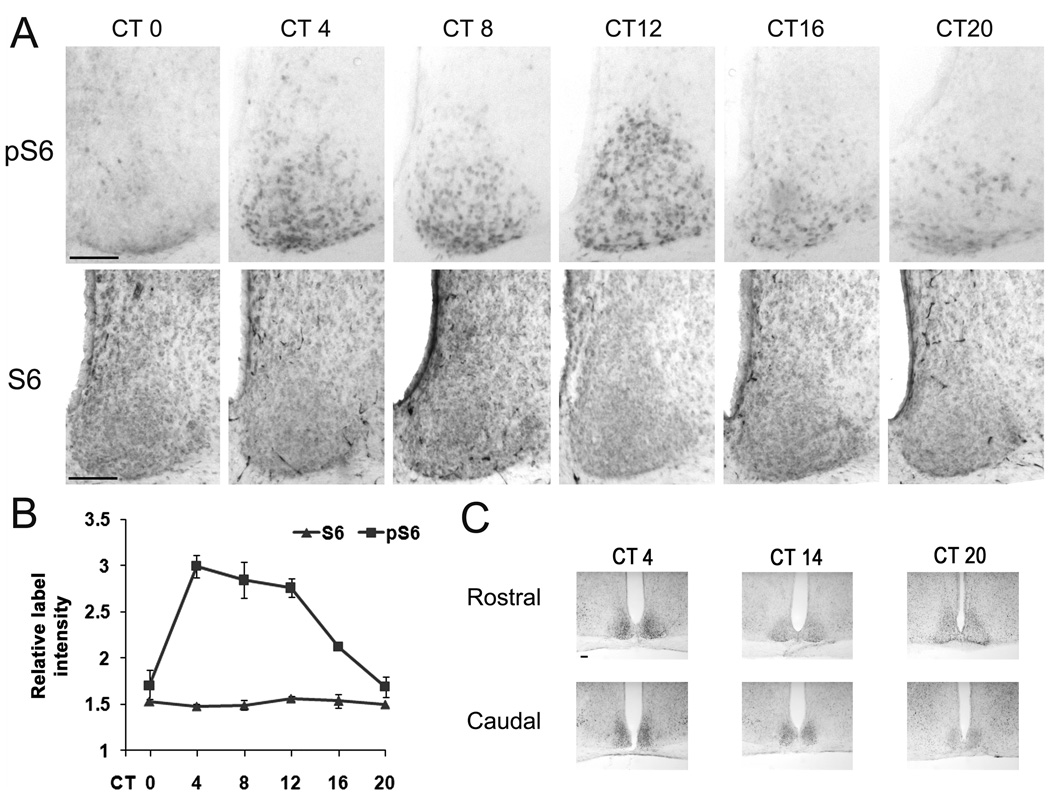
Figure 1. Circadian expression of phosphorylated S6 ribosomal protein (pS6) in the SCN.
Entrained animals were dark-adapted for 2 d and killed under dim red light every 4 h over a 24 h cycle. A, Upper row: representative immunohistochemical labeling for pS6 in central SCN coronal sections across the circadian cycle. Of note relatively high levels of pS6 labeling were observed in the SCN during the mid- to late- subjective day time points (i.e., CT 4, 8 and 12). Lower row: representative images of total S6 labeling across the circadian cycle. B, Quantitation of normalized pS6 and S6 expressions in the central SCN over a 24h cycle. Please see the Methods section for a description of the quantitative analysis. Error bars denote standard error of mean. A minimum of four mice were used for each time point. C, Circadian variations in pS6 expression were also observed in the rostral and caudal SCN sections. Scale bars: 100 microns.
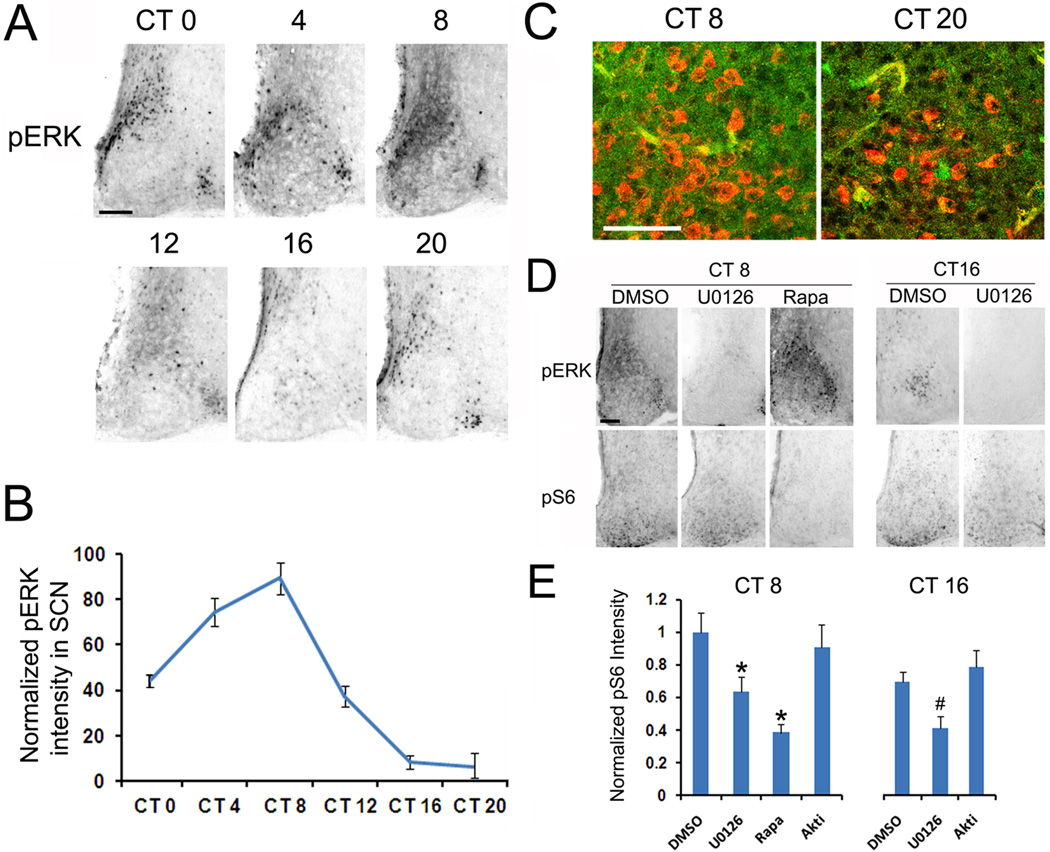
Figure 5. The MAPK cascade stimulates mTOR activation.
Entrained animals were dark-adapted for 2d and killed under dim red light every 4 h over a 24 h cycle. A, Representative immunohistochemical labeling for pERK over the circadian cycle. Scale bar: 100 microns. B, Quantitation of circadian pERK expression in the SCN. Error bars denote the SEM: data were averaged from 4 animals per time point. Please see the Methods section for a description of the quantitation technique. C, Representative confocal images of the central SCN immunolabeled for pERK (green) and pS6 (red) at CT 8 and CT20. Scale bar: 50 microns. D, Representative immunohistochemical labeling of SCN tissue labeled for pERK and pS6 following infusion with U0126 (10 mM, 2 µl), rapamycin (100 µM, 2 µl) or DMSO (2 µl). Animals were infused 30 min before sacrifice at CT8 or CT16. Of note, infusion of U0126 repressed pERK expression and attenuated pS6 expression at both day and night time points. Rapamycin repressed pS6 expression but not pERK expression at CT8. E, Quantitative presentation of the effects of U0126, rapamycin and the Akt inhibitor Akti-1/2 (20 mM, 2 µl) on pS6 expression at CT 8 and CT 16 in the SCN. Of note, infusion of U0126 but not Akti-1/2 significantly inhibited circadian pS6 expression at both day and night time points. Error bars denote SEM. At least three animals were used for each condition. * p<0.05 vs DMSO group at CT8; # p<0.05 vs DMSO group at CT16.
RESULTS
Circadian mTOR activity in the SCN
To test whether mTOR activity is under control of the circadian clock, mice maintained on a standard 12 hr light/dark cycle were transferred to total darkness for 2 days, and then sacrificed at 4 hr intervals over a 24 hr period. SCN-containing tissue was then fixed and immunolabled for the expression of the Ser 240/244-phosphorylated form of S6 ribosomal protein (pS6). The rationale for monitoring the phosphorylation state of Ser 240/244 is based on work showing that these post-translational modifications are mediated specifically via mTOR-dependent S6K1 kinase activity. Importantly, data provided here (Fig. 5D and 5E: described below), along with our previous work (Cao et al., 2008) reveals that microinfusion of the mTOR inhibitor rapamycin into the SCN leads to a rapid and potent suppression of S6 phosphorylation. Hence, pS6 immunolabeling serves as an excellent marker of relative mTOR activity. Examination of the labeling pattern revealed a statistically-significant circadian oscillation in pS6 expression in the SCN (Fig. 1A and 1B). Specifically, relatively little pS6 was detected at the beginning of the subjective day (i.e., Circadian Time 0: CT 0); however, at the CT 4 time point, a marked increase in S6 phosphorylation was detected. Elevated labeling persisted throughout the subjective day, and then declined, reaching nadir during the late subjective night time points (CT 20-0). Starting at CT4, the expression of pS6 showed progressive spread from the ventral to the dorsal SCN, culminating in robust SCN-wide pS6 expression at CT12. Of note, total S6 levels were not altered as a function of circadian time, indicating that the change in pS6 expression reflected a circadian-regulated change in mTOR/S6K1 kinase activity (Fig. 1A and 1B). Together these data indicate that mTOR signaling is under control of the SCN circadian clock.
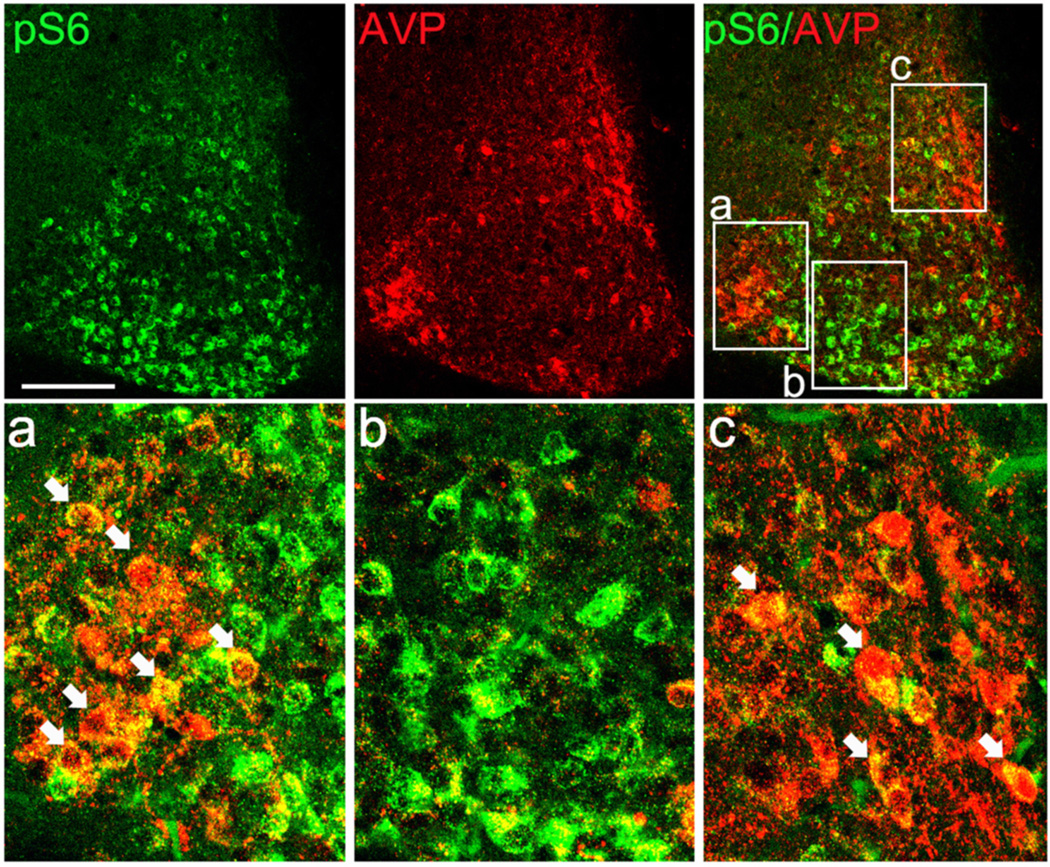
As shown in Figure 1A, at CT12 pS6 was not localized to a distinct neuroanatomical subregion of the SCN, such as the core or shell, thus suggesting that mTOR activation was not specific to distinct peptidergic neuronal populations (e.g., arginine vasopressin [AVP], vasoactive intestinal peptide, calbindin). To test this assertion, SCN tissue isolated at CT12 was double-labeled for pS6 and AVP. As expected, AVP was robustly expressed in dorsal and lateral regions of central SCN (Fig. 2), whereas pS6 was expressed in a much broader region of the SCN. Moreover, merging the immunofluorescence signals revealed that only a limited number of pS6-positive cells expressed AVP (Fig. 2a–c). Together these data support the idea that pS6 is expressed in diverse subsets of SCN neurons.
Figure 2. Dual immunolabeling for pS6 and arginine vasopressin (AVP) in the SCN.
Animals were dark-adapted for 2 d and killed at CT12. Upper row: representative confocal immunofluorescent images of pS6 (green, left panel) and AVP (red, middle panel). The merged image (right panel) reveals largely non-overlapping expression patterns. Framed regions a, b and c are presented as magnified panels in the lower row. Arrows indicate cellular-level colocalization (denoted by a yellow hue) of the two antigens. Scale bars: 100 microns.
Rhythmic mTOR activity and per1 expression
From both a neuroanatomical and temporal perspective, the circadian oscillation in pS6 activity in the SCN parallels the expression of the core clock protein PER1, thus raising the possibility that rhythmic mTOR activity is restricted to the pool of SCN neurons that exhibit robust clock gene rhythms.
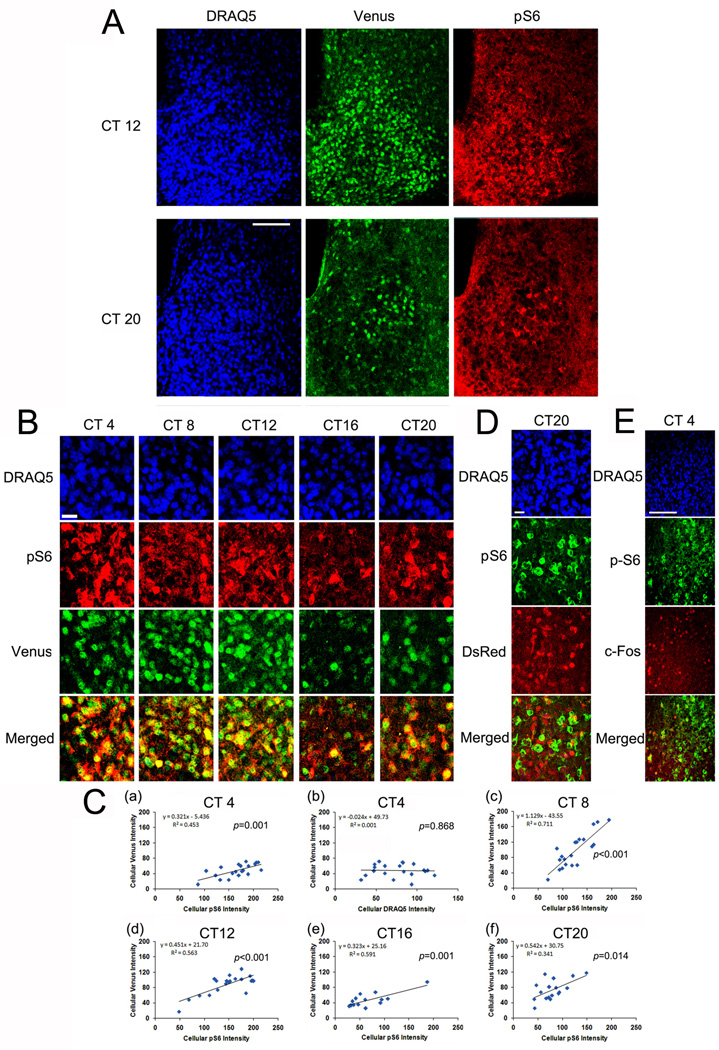
To begin to address this question, we profiled per1 expression and mTOR activity as a function of circadian time. To monitor rhythms in per1 gene expression, we employed a transgenic mouse line in which the per1 promoter-drives the expression of the GFP derivative Venus (Cheng et al., 2009). Of note, in these transgenic animals, the Venus coding sequence was modified to allow for rapid protein turnover and nuclear localization of the reporter protein, thus providing a dynamic signal that closely matches the transcriptional profile of the endogenous gene (Cheng et al., 2009). As shown in Figure 3A and 3B, representative confocal images of triple fluorescent labeling for pS6, Venus and the nuclear marker DRAQ5 reveal a cellular-level colocalization of pS6 and Venus over the circadian cycle. As expected, Venus was expressed within the cell nuclei while pS6 was expressed within the cytoplasm. To quantify the cellular-level relationship between pS6 and Venus expression, linear correlation analysis between pS6 intensity and Venus expression was examined across the circadian cycle. Interestingly, at all time points, a significant linear correlation was detected between pS6 and Venus expressions (p<0.05, Fig. 3C). To test the veracity of this quantitation technique, we also performed correlation analysis between Venus and DRAQ5. Importantly, no significant correlation between these two markers was noted [Fig. 3C(b)]. Together these data suggest that the rhythmic actuation of mTOR activity is tightly linked to expression of the core clock gene per1.
Figure 3. Colocalized expression of pS6 and per1-Venus in the SCN.
Entrained animals were dark-adapted for 2 d and then killed under dim red light at 4 h intervals over a 24 h cycle. A, Representative confocal microscopic images of triple immunofluorescence labeling for pS6 (red), Venus (green) and DRAQ5 (blue) at CT12 and CT20. B, High magnification images of the central SCN as a function of circadian time: the pS6 and Venus channels are merged (bottom panels). Note the cellular level correlation between pS6 and Venus expression (revealed by a yellow cellular-level hue). Scale bar: 15 microns. C, Linear correlation analysis of cellular Venus and pS6 expressions at CT4 (a), CT8 (c), CT12 (d), CT16 (e) and CT20 (f). The Y-axis denotes normalized fluorescence intensity (0–255 scale) of Venus and the X-axis denotes normalized fluorescent intensity of pS6. As a control, the relationship between Venus (Y-axis) expression and DRAQ5 (X-axis) labeling at CT4 is shown in C(b). Each dot represents a single cell. The significance (p value) for Pearson's correlation analysis is shown in each panel. D, Central SCN tissue isolated at CT 20 was immunolabeling for pS6 and per2-DsRed. Of note, the merged panel (bottom) reveals little cellular colocalization between the two antigens. This lack of concordant labeling is in contrast to the colocalized expression between pS6 and Venus. Scale bar: 15 microns. E, Central SCN tissue isolated at CT 4 was immunolabeled for pS6 and c-FOS. Of note, the merged panel (bottom) reveals that the majority of pS6-postive cells do not express c-FOS. Scale bar: 100 microns.
These findings led us to question whether mTOR activity was a feature of all oscillatory neurons in the SCN. In an attempt to address this question, we examined whether pS6 is detected in the subset of SCN neurons that exhibit per2 oscillations. Here, it is worth noting that the neuroanatomical pattern of the per2 oscillation is distinct from the pattern of per1 rhythmicity in the SCN. Along these lines, PER2 protein rhythms are largely observed in the dorsal and lateral regions of the SCN (Cheng et al., 2007), whereas, PER1 protein oscillations are observed throughout the SCN (Hastings et al., 1999). Simply based on the published distribution pattern of rhythmic per2 cells, it would appear that the pS6 and per2 oscillations occur in distinct cell populations. To test this idea, we employed a per2-DsRED transgenic mouse strain (Cheng et al., 2009), to profile per2 transcription and pS6 expression. Similar to the per1-Venus transgenic mouse strain, the per2 transgenic mouse strain drives the expression of a nuclear-localized, destabilized reporter (DsRED) transgene, thus allowing for single cell resolution of cells in which per2 transcription occurs. Double immunolabeling of CT20 tissue revealed a divergent cellular-level expression pattern of DsRED and pS6: hence, the majority of pS6-positive cells did not express the per2 transgene (Fig. 3D). These data contrast with the high degree of colocalization between Venus and pS6. We also tested whether pS6 is expressed in SCN neurons that exhibit a circadian-gated increase in c-Fos expression during the early subjective day (Guido et al., 1999). Double labeling analysis of tissue collected at CT4 revealed that the majority of pS6-positive cells in the SCN did not express c-Fos (Fig. 3E). Together, these data suggest that mTOR activity is not a marker of all rhythmic cells, but rather, is localized to a subset of per1-expressing clock neurons.
Light-induced per1 expression and mTOR activation
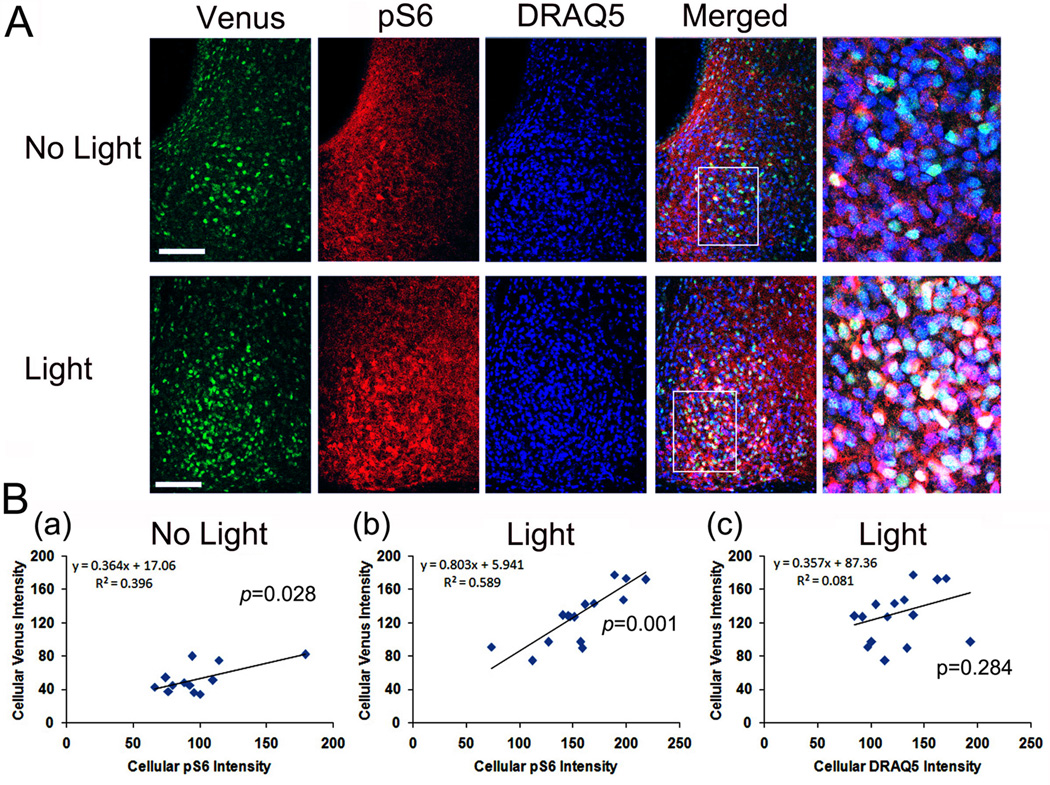
Our previous work found that mTOR activity facilitates light-induced PER1 expression in the SCN (Cao et al., 2010). Here we tested for a cellular level relationship between light-evoked mper1 transcription of S6 activation. To this end, Venus and pS6 expression was examined 4h after an early night (CT 15) light pulse (100 lux). Paralleling our previous results (Cheng et al., 2009; Cao et al., 2008), light triggered a marked increase in Venus expression and S6 phosphorylation relative to control, no light, animals (Figure 4A). Further, quantitative cellular level analysis of the control and light-treated groups revealed a significant positive correlation between pS6 and Venus expression (p<0.05, Figure 4B). As a control, analysis of DRAQ5 and Venus following light treatment revealed that there was no correlation between the two labels. These data lend further support to the idea that mTOR activity is largely restricted to cells that exhibit a high degree of per1 gene expression.
Figure 4. Light-evoked pS6 and per1-Venus expressions in the SCN.
Dark-adapted (2d) mice were exposed to a light (100 lux, 15 min) at CT 15, killed at CT19, and coronal SCN-containing sections were processed for Venus (green) and pS6 (red) expression and nuclei were visualized with DRAQ5 (blue). A, Representative confocal microscopic of “no light” and “light” groups. Framed regions on Merged images are magnified to the right. B, Linear correlation analysis of cellular Venus and pS6 expressions in the No Light (a) and Light treated (b) group. The Y-axis denotes normalized cellular-level Venus fluorescence intensity (0–255 scale) and the X-axis denotes normalized fluorescence intensity of pS6. Each dot represents a single cell. As a control, the correlation between Venus (Y-axis) and DRAQ5 (X-axis) expressions after light is shown (c). The significance (p value) for Pearson's correlation analysis is shown for each panel.
p42/44 MAPK activity and mTOR oscillation
Next, we turned to an examination of potential upstream regulatory pathways that drive mTOR activity in the SCN. Given their well-recognized role as regulators of mTOR, two pathways were of particular interest: the p42/44 ERK/mitogen-activated protein kinase (ERK/MAPK) pathway and the PI3 kinase/AKT pathway. Initially, emphasis was placed on the MAPK pathway, since we previously reported that activation this pathway exhibits robust circadian oscillation with a similar temporal profile to that of mTOR activity (Obrietan et al., 1998), and because the MAPK pathway couples light to mTOR in the SCN (Cao et al., 2008). To examine this question, we first profiled the circadian regulation of MAPK activity by using an antibody that detects the dual phosphorylated (Thr 202 and Tyr 204) form of ERK (here referred to as pERK). Circadian profiling revealed that pERK has a similar pattern as pS6: high during the circadian day and low during the circadian night (Figure 5A and 5B). Given the similarities in the temporal expression pattern of mTOR and MAPK signaling, we examined whether there was a parallel cellular-level expression pattern of pERK and pS6. Double immunofluorescent labeling at both CT8 and CT20 did not detect marked cellular colabeling between pERK with pS6. Hence, cells with high levels of pS6 typically did not show concordant elevated level of pERK (Fig. 5C). Although enhanced MAPK signaling was not specific to pS6-expressing cells, basal levels of ERK activity could contribute to mTOR activity. Likewise, a lack of cellular-level colocalization could be explained by different enzymatic kinetics of ERK and S6 dephosphorylation. Hence, to directly test whether MAPK signaling regulates S6 phosphorylation, we employed a pharmacological approach to disrupt ERK activation and assess the effects on S6 phosphorylation. Microinfusion of the MEK inhibitor U0126 (10 mM, 2µl) into the lateral ventricle 30 min before sacrifice led to a moderate, yet significant, decrease (Fig. 5D and 5E) in pS6 expression during both the subjective day (CT 8) and the subjective night (CT16). As a validation of the efficacy of pharmacological approach to abrogate MAPK signaling, we also provide data showing that the infusion led to a potent inhibition of ERK phosphorylation (Fig. 5D). Together, these data suggest that circadian-regulated mTOR activity is modulated by the MAPK pathway. As a further control, animals were infused with the mTOR inhibitor rapamycin (100 µM, 2µl) at CT8. In line with our prior work (Cao et al., 2008), rapamycin inhibited pS6 (Fig. 5D and 5E), thus indicating that S6 phosphorylation is driven by mTOR activity.
We also tested the potential contribution of the PI3K/Akt pathway to mTOR signaling in the SCN. As a first step, we profiled Akt phosphorylation (pAKT) at Ser 308, a marker of AKT activity. Interestingly, only sparse non-rhythmic pAkt expression was detected in the SCN (data not shown). Further, light-induced (CT 15, 100 lux, 30 min) Akt phosphorylation was not detected in the SCN (data not shown). These findings indicate a minimal level of Akt activity in the SCN, and thus suggest that PI3K/Akt does not contribute to rhythmic mTOR activity. To directly test this question, the Akt inhibitor Akti-1,2 (20 mM 2 uL) was infused into the lateral ventricle, and the effects of pS6 expression were examined. This approximate concentration of inhibitor has been used in other models to effectively suppress Akt activity (Logie et al., 2007; Xu et al., 2008). Notably, relative to vehicle-infused mice, S6 phosphorylation in the SCN was not affected by Akti-1/2 infusion (Fig. 5E), thus indicating that signaling via the PI3K/Akt pathway does not markedly contribute to mTOR activity in the SCN. Of note, we were not able to directly assess the contribution of PI3K signaling, given that the potent, and commonly used PI3K antagonist LY294002, is also an inhibitor of mTOR (Ballou et al., 2007).
DISCUSSION
We have previously identified a light-responsive mTOR signaling cascade in the SCN that modulates clock entrainment. As a logical extension of these studies, we were curious about whether mTOR signaling might also be under the control of the circadian clock. To this end, we characterized the 24 h temporal profile of mTOR signaling expression in the SCN. The data presented here reveal that 1) mTOR activity exhibits a marked circadian oscillation, 2) rhythmicity is not limited to subregions of the SCN, but rather is broadly distributed, 3) at a cellular level mTOR rhythms are highly correlated with rhythms of the circadian clock gene per1 and 4) mTOR activity is influenced by the p42/44 MAPK pathway. Together these data raise the prospect that inducible mRNA translation contributes to circadian clock physiology.
Circadian profiling revealed that mTOR activity peaked during the subjective day, and reached its nadir during the late subjective night. Interestingly, this daily mTOR activity profile fits with the rhythmic regulation of SCN cellular physiology. Along these lines, SCN neuronal activity and glucose metabolism, peak during the middle of the subjective day (Schwartz and Gainer, 1977; Shibata and Moore, 1988). Work in other model systems showing that mTOR signaling is a linker between cellular energy metabolism and neuronal excitability (Potter et al., 2010) raises the possibility that elevated mTOR activity during the daytime is required to meet the physiological demands of the elevated level of metabolic and firing activities of SCN cells. In some senses, this may be similar to the recent work from the arcuate nucleus, which showed that mTOR activity is directly and tightly regulated by energy intake (Cota et al., 2006).
Another, potentially related, explanation for the S6 phosphorylation pattern is that mTOR activity occurs in cells that exhibit robust, rhythmic, transcriptional activity. As described in the Introduction, the molecular underpinning of the cellular-level 24-hr oscillator is a transcription/translation feedback loop that is centered on the rhythmic expression of per and cry genes. Both per1 and per2 exhibit an increase in mRNA expression that is coincident with the beginning of the subjective daytime (Shearman et al., 1997). Here, it is worth noting that, at a neuroanatomical level, per1 and per2 expression appears to be quite distinct. For example, during the middle of the subjective day, per1 is detected throughout the SCN, whereas per2 expression is largely localized to the dorsal and lateral regions of the SCN, with only limited expression in the SCN “core” (Takumi et al., 1998; Yan and Okamura, 2002; Yan and Silver, 2004; Cheng et al., 2009). Given the neuroanatomical discord between the pattern of pS6 expression reported here, and published PER2 expression pattern it was not surprising to find that pS6 and per2 transgene expression were not correlated. Although this observation, combined with the c-FOS data, was a clear rejection of our hypothesis that mTOR activity occurs in cells that exhibit transcriptional rhythms, the finding that the pS6 expression pattern approximated the expression pattern of per1, raised the possibility that robust mTOR activity oscillations occur in a subset of transcriptional oscillators: per1 rhythmic cells. We pursued this question by examining the composite profile of S6 phosphorylation and per1 transcriptional rhythms. With the use of a per1-Venus transgenic mouse strain that nicely recapitulates the temporal and spatial pattern of the endogenous per1 gene, we showed that there is a statistically significant cellular-level correlation between pS6 and Venus, thus indicating that high-levels of clock-regulated mTOR activity occur in per1 oscillator cell populations. Interestingly, per1-expressing SCN neurons appear to be a distinct subclass of oscillator cells. Along these lines, work by Belle et al., (2009) showed that per1-expressing neurons exhibit markedly altered electrophysiological characteristics, relative to per1-negative SCN oscillator neurons. As further support for the unique relationship between mTOR and per1, we provided data showing that photic stimulation lead to a concordant, cellular-level, upregulation of both pS6 and Venus. Although we have yet to test the functional significance of mTOR activity within in the PER1-expressing SCN cell population, there are several possible roles that should be given consideration. Based on work showing that mTOR affects neuronal structure and synaptic communication (Tang et al., 2002; Cammalleri et al., 2003 Buckmaster et al., 2009; Li et al., 2010) it may be worthwhile examining whether mTOR influences the rhythmic reorganization of SCN neuronal networks, which is observed across the day/night cycle (Becquet et al., 2008; Girardet et al., 2010). Further, mTOR signaling may contribute to the distinct firing properties of PER1-expressing SCN neurons. Lastly, recent work has shown that the rhythmic expression of many proteins within the SCN is regulated at a post-translational level (Deery et al, 2009); clearly an examination of whether mTOR contributes to ostensibly unique phenotype of per1-expressing neurons is merited.
Interestingly our previous work showed that abrogation of mTOR signaling via rapamycin infusion decreased light-evoked PERIOD expression (Cao et al., 2010), but that basal (i.e., clock-regulated) PER1 expression was not affected. This reported lack of an effect on basal (clock-regulated) PER1 expression should be viewed with a bit of caution. Of note, the paradigm used in the prior study was designed to specifically test the effects of light-evoked mTOR. Further, the rapamycin infusion paradigm led to a transient (~ 90 min) repression of mTOR, which may not be of sufficient duration to reveal an effect of mTOR on circadian PER1 expression. Finally, S6 maintains activity for an extended period, following transient mTOR and p70 S6K activity (Cao et al., 2010), thus raising the possibility that long-term suppression of mTOR (which in turn would suppress S6) would be required to detect an effect on rhythmic PER1 expression or circadian clock function. Interestingly, a recent study in Drosophila used genetic approaches to show that TOR activity affects circadian period (Zheng and Sehgal, 2010). Although the effect of TOR on PER expression was not examined, the findings of Zheng and Sehgal coupled with our work showing a tight correlation between rhythmic per1 expression and mTOR activity raises the prospect that mTOR modulates clock timing via an augmentation of per1 expression in mammals.
To gain insights into the upstream signaling cascade(s) that couple the clock to mTOR, we examined the potential contributions of the p42/44 MAPK and PI3K/AKT pathways. The rationale behind our analysis of MAPK signaling was based on our previous work showing that the MAPK pathway exhibits a circadian oscillation in the SCN (Obrietan et al.,1998) and on work showing that MAPK signaling couples light to mTOR activation (Cao et al., 2008). Further, data provided here revealed a parallel temporal activation pattern for ERK and S6 in the SCN. However, an examination of kinase activation patterns within the SCN as a function of clock time revealed a good degree of neuroanatomical discord between pERK and pS6. Along these lines, during the subjective day, peak pERK expression was observed in the dorsal and lateral regions of the SCN, whereas pS6 levels were observed throughout the SCN. Further, double labeling did not detect markedly up-regulated pERK expression in pS6-immunopositive cells during either the subjective day or night. However, infusion of the MEK inhibitor during both the subjective day and night led to a modest, yet significant, suppression of mTOR activity. Given the distinct expression patterns of pERK and pS6 in the SCN, this finding was a bit surprising; however, there are several potential explanations for this effect. Along these lines, although maximal ERK activation was detected in cell populations that were distinct from those exhibiting robust pS6 levels, relatively low-level MAPK activity may be sufficient to facilitate mTOR activity. Further, the lack of colocalization could be explained by different enzymatic kinetics of ERK and S6 dephosphorylation, Hence, pERK may have been dephosphorylated (i.e., transiently-activated) in the cells showing relatively long-lasting S6 phosphorylation. Another possible explanation is that MAPK signaling may function at system-wide level in the SCN, modulating the efficacy of synaptic circuits, which drive circadian mTOR activity. In this model, cellular colocalization of MAPK and pS6 activity would not be required. Within this context, it is interesting to note that MAPK signaling has been shown to affect the robustness of SCN neuron firing (Akashi et al., 2008). It remains to be determined what kinase pathways, in addition to the MAPK cascade, contribute to mTOR rhythmicity. Signaling pathways that are responsive to changes in metabolic activity and energy levels may be good candidates for further examination (Hay and Sonenberg, 2004; Wullschleger et al., 2006).
In conclusion the data presented here reveal a prominent rhythm in the activation state of the mTOR pathway in the SCN, thus raising the prospect that inducible translation control contributes to the clock timing process.
Acknowledgments
We thank Victor Liu for his technical assistance. This work was supported by an NSF grant (IBN-0090974) and grants from the NIH (MH62335 and NS067409) and by the Ohio State Neuroscience Center Core grant (5P30NS045758).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Akashi M, Hayasaka N, Yamazaki S, Node K. Mitogen-activated protein kinase is a functional component of the autonomous circadian system in the suprachiasmatic nucleus. J Neurosci. 2008;28:4619–4623. doi: 10.1523/JNEUROSCI.3410-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballou LM, Selinger ES, Choi JY, Drueckhammer DG, Lin RZ. Inhibition of Mammalian Target of Rapamycin Signaling by 2-(Morpholin-1-yl)pyrimido[2,1-α]isoquinolin-4-one. J Biol Chem. 2007;282:24463–24470. doi: 10.1074/jbc.M704741200. [DOI] [PubMed] [Google Scholar]
- Belle MD, Diekman CO, Forger DB, Piggins HD. Daily electrical silencing in the mammalian circadian clock. Science. 2009;326:281–284. doi: 10.1126/science.1169657. [DOI] [PubMed] [Google Scholar]
- Becquet D, Girardet C, Guillaumond F, François-Bellan AM, Bosler O. Ultrastructural plasticity in the rat suprachiasmatic nucleus. Possible involvement in clock entrainment. Glia. 2008;56:294–305. doi: 10.1002/glia.20613. [DOI] [PubMed] [Google Scholar]
- Buckmaster PS, Ingram EA, Wen X. Inhibition of the mammalian target of rapamycin signaling pathway suppresses dentate granule cell axon sprouting in a rodent model of temporal lobe epilepsy. J Neurosci. 2009;29:8259–8269. doi: 10.1523/JNEUROSCI.4179-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammalleri M, Lütjens R, Berton F, King AR, Simpson C, Francesconi W, Sanna PP. Time-restricted role for dendritic activation of the mTOR-p70S6K pathway in the induction of late-phase long-term potentiation in the CA1. Proc Natl Acad Sci USA. 2003;100:14368–14373. doi: 10.1073/pnas.2336098100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Lee B, Cho HY, Saklayen S, Obrietan K. Photic regulation of the mTOR signaling pathway in the suprachiasmatic circadian clock. Mol Cell Neurosci. 2008;38:312–324. doi: 10.1016/j.mcn.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Li A, Cho HY, Lee B, Obrietan K. Mammalian target of rapamycin signaling modulates photic entrainment of the suprachiasmatic circadian clock. J Neurosci. 2010;30:6302–6314. doi: 10.1523/JNEUROSCI.5482-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, Nakazawa T, Shimizu K, Okamura H, Impey S, Obrietan K. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54:813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HY, Alvarez-Saavedra M, Dziema H, Choi YS, Li A, Obrietan K. Segregation of expression of mPeriod gene homologs in neurons and glia: possible divergent roles of mPeriod1 and mPeriod2 in the brain. Hum Mol Genet. 2009;18:3110–3124. doi: 10.1093/hmg/ddp252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cota D, Proulx K, Smith KA, Kozma SC, Thomas G, Woods SC, Seeley RJ. Hypothalamic mTOR signaling regulates food intake. Science. 2006;312:927–930. doi: 10.1126/science.1124147. [DOI] [PubMed] [Google Scholar]
- Deery MJ, Maywood ES, Chesham JE, Sládek M, Karp NA, Green EW, Charles PD, Reddy AB, Kyriacou CP, Lilley KS, Hastings MH. Proteomic analysis reveals the role of synaptic vesicle cycling in sustaining the suprachiasmatic circadian clock. Curr Biol. 2009;19:2031–2036. doi: 10.1016/j.cub.2009.10.024. [DOI] [PubMed] [Google Scholar]
- Douris N, Green CB. NOC out the fat: a short review of the circadian deadenylase Nocturnin. Ann Med. 2008;40:622–626. doi: 10.1080/07853890802084878. [DOI] [PubMed] [Google Scholar]
- Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- Girardet C, Blanchard MP, Ferracci G, Lévêque C, Moreno M, François-Bellan AM, Becquet D, Bosler O. Daily changes in synaptic innervation of VIP neurons in the rat suprachiasmatic nucleus: contribution of glutamatergic afferents. Eur J Neurosci. 2010;31:359–370. doi: 10.1111/j.1460-9568.2009.07071.x. [DOI] [PubMed] [Google Scholar]
- Guido M, Goguen D, De Guido L, Robertson HA, Rusak B. Circadian and photic regulation of immediate-early gene expression in the hamster suprachiasmatic nucleus. Neuroscience. 1999;90:555–571. doi: 10.1016/s0306-4522(98)00467-9. [DOI] [PubMed] [Google Scholar]
- Hastings MH, Field MD, Maywood ES, Weaver DR, Reppert SM. Differential regulation of mPER1 and mTIM proteins in the mouse suprachiasmatic nuclei: new insights into a core clock mechanism. J Neurosci. 1999;19:RC11. doi: 10.1523/JNEUROSCI.19-12-j0001.1999. 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- Ko C, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15:R271–R277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- Kojima S, Matsumoto K, Hirose M, Shimada M, Nagano M, Shigeyoshi Y, Hoshino S, Ui-Tei K, Saigo K, Green CB, Sakaki Y, Tei H. LARK activates posttranscriptional expression of an essential mammalian clock protein, PERIOD1. Proc Natl Acad Sci U S A. 2007;104:1859–1864. doi: 10.1073/pnas.0607567104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Lee B, Liu RJ, Banasr M, Dwyer JM, Iwata M, Li XY, Aghajanian G, Duman RS. mTOR-dependent synapse formation underlies the rapid antidepressant effects of NMDA antagonists. Science. 2010;329:959–964. doi: 10.1126/science.1190287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logie L, Ruiz-Alcaraz AJ, Keane M, Woods YL, Bain J, Marquez R, Alessi DR, Sutherland C. Characterization of a protein kinase B inhibitor in vitro and in insulin-treated liver cells. Diabetes. 2007;56:2218–2227. doi: 10.2337/db07-0343. [DOI] [PubMed] [Google Scholar]
- McNeil GP, Zhang X, Genova G, Jackson FR. A molecular rhythm mediating circadian clock output in Drosophila. Neuron. 1998;20:297–303. doi: 10.1016/s0896-6273(00)80457-2. [DOI] [PubMed] [Google Scholar]
- Na Y, Sung JH, Lee SC, Lee YJ, Choi YJ, Park WY, Shin HS, Kim JH. Comprehensive analysis of microRNA-mRNA co-expression in circadian rhythm. Exp Mol Med. 2009;41:638–647. doi: 10.3858/emm.2009.41.9.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel R, Clijsters L, Agami R. The miRNA-192/194 cluster regulates the Period gene family and the circadian clock. FEBS J. 2009;276:5447–5455. doi: 10.1111/j.1742-4658.2009.07229.x. [DOI] [PubMed] [Google Scholar]
- Obrietan K, Impey S, Storm DR. Light and circadian rhythmicity regulate MAP kinase activation in the suprachiasmatic nuclei. Nat Neurosci. 1998;1:693–700. doi: 10.1038/3695. [DOI] [PubMed] [Google Scholar]
- Potter W, O'Riordan KJ, Barnett D, Osting SM, Wagoner M, Burger C, Roopra A. Metabolic regulation of neuronal plasticity by the energy sensor AMPK. PLoS One. 2010;5:e8996. doi: 10.1371/journal.pone.0008996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppert S, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- Shi L, Ko ML, Ko GY. Rhythmic expression of microRNA-26a regulates the L-type voltage-gated calcium channel alpha1C subunit in chicken cone photoreceptors. J Biol Chem. 2009;284:25791–25803. doi: 10.1074/jbc.M109.033993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz W, Gainer H. Suprachiasmatic nucleus: use of 14C-labeled deoxyglucose uptake as a functional marker. Science. 1977;197:1089–1091. doi: 10.1126/science.887940. [DOI] [PubMed] [Google Scholar]
- Schwartz W, Zimmerman P. Circadian timekeeping in BALB/c and C57BL/6 inbred mouse strains. J Neurosci. 1990;10:3685–3694. doi: 10.1523/JNEUROSCI.10-11-03685.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearman L, Zylka MJ, Weaver DR, Kolakowski LF, Jr, Reppert SM. Two period homologs: circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron. 1997;19:1261–1269. doi: 10.1016/s0896-6273(00)80417-1. [DOI] [PubMed] [Google Scholar]
- Shibata S, Moore RY. Electrical and metabolic activity of suprachiasmatic nucleus neurons in hamster hypothalamic slices. Brain Res. 1988;438:374–378. doi: 10.1016/0006-8993(88)91367-4. [DOI] [PubMed] [Google Scholar]
- Tang SJ, Reis G, Kang H, Gingras AC, Sonenberg N, Schuman EM. A rapamycin-sensitive signaling pathway contributes to long-term synaptic plasticity in the hippocampus. Proc Natl Acad Sci U S A. 2002;99:467–472. doi: 10.1073/pnas.012605299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takumi T, Matsubara C, Shigeyoshi Y, Taguchi K, Yagita K, Maebayashi Y, Sakakida Y, Okumura K, Takashima N, Okamura H. A new mammalian period gene predominantly expressed in the suprachiasmatic nucleus. Genes Cells. 1998;3:167–176. doi: 10.1046/j.1365-2443.1998.00178.x. [DOI] [PubMed] [Google Scholar]
- Ukai H, Ueda HR. Systems biology of mammalian circadian clocks. Annu Rev Physiol. 2010;72:579–603. doi: 10.1146/annurev-physiol-073109-130051. [DOI] [PubMed] [Google Scholar]
- Welsh D, Takahashi JS, Kay SA. Suprachiasmatic nucleus: cell autonomy and network properties. Annu Rev Physiol. 2010;72:551–577. doi: 10.1146/annurev-physiol-021909-135919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Yan L, Okamura H. Gradients in the circadian expression of Per1 and Per2 genes in the rat suprachiasmatic nucleus. Eur J Neurosci. 2002;15:1153–1162. doi: 10.1046/j.1460-9568.2002.01955.x. [DOI] [PubMed] [Google Scholar]
- Yan L, Silver R. Resetting the brain clock: time course and localization of mPER1 and mPER2 protein expression in suprachiasmatic nuclei during phase shifts. Eur J Neurosci. 2004;19:1105–1109. doi: 10.1111/j.1460-9568.2004.03189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Chua CC, Gao J, Chua KW, Wang H, Hamdy RC, Chua BH. Neuroprotective effect of humanin on cerebral ischemia/reperfusion injury is mediated by a PI3K/Akt pathway. Brain Res. 2008;1227:12–18. doi: 10.1016/j.brainres.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Sehgal A. AKT and TOR signaling set the pace of the circadian pacemaker. Curr Biol. 2010;20:1203–1208. doi: 10.1016/j.cub.2010.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]