Abstract
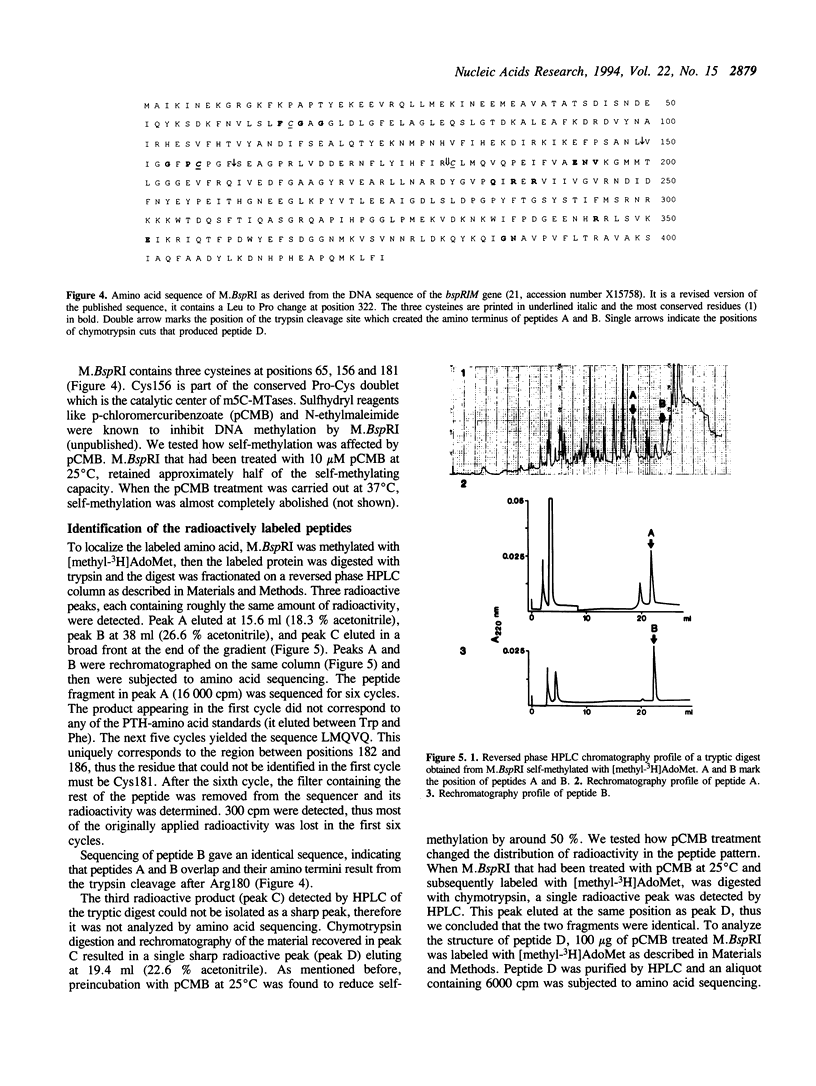
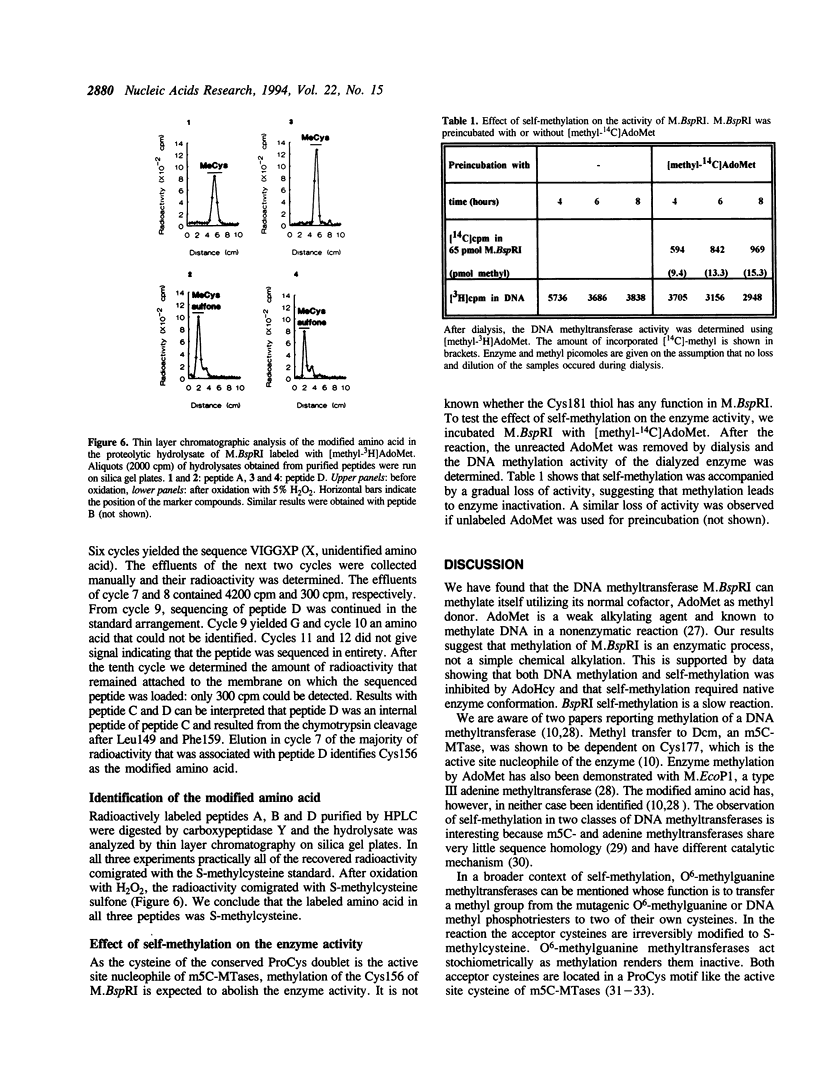
The DNA (cytosine-5)-methyltransferase (m5C-MTase) M.BspRI is able to accept the methyl group from the methyl donor S-adenosyl-L-methionine (AdoMet) in the absence of DNA. Transfer of the methyl group to the enzyme is a slow reaction relative to DNA methylation. Self-methylation is dependent on the native conformation of the enzyme and is inhibited by S-adenosyl-L-homocysteine, DNA and sulfhydryl reagents. Amino acid sequencing of proteolytic peptides obtained from M.BspRI, which had been methylated with [methyl-3H]AdoMet, and thin layer chromatography of the modified amino acid identified two cysteines, Cys156 and Cys181 that bind the methyl group in form of S-methylcysteine. One of the acceptor residues, Cys156 is the highly conserved cysteine which plays the role of the catalytic nucleophile of m5C-MTases.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balganesh T. S., Reiners L., Lauster R., Noyer-Weidner M., Wilke K., Trautner T. A. Construction and use of chimeric SPR/phi 3T DNA methyltransferases in the definition of sequence recognizing enzyme regions. EMBO J. 1987 Nov;6(11):3543–3549. doi: 10.1002/j.1460-2075.1987.tb02681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor T., Laudano A., Mattaliano R., Ingram V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J Mol Biol. 1988 Oct 20;203(4):971–983. doi: 10.1016/0022-2836(88)90122-2. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Chen L., MacMillan A. M., Chang W., Ezaz-Nikpay K., Lane W. S., Verdine G. L. Direct identification of the active-site nucleophile in a DNA (cytosine-5)-methyltransferase. Biochemistry. 1991 Nov 19;30(46):11018–11025. doi: 10.1021/bi00110a002. [DOI] [PubMed] [Google Scholar]
- Cheng X., Kumar S., Posfai J., Pflugrath J. W., Roberts R. J. Crystal structure of the HhaI DNA methyltransferase complexed with S-adenosyl-L-methionine. Cell. 1993 Jul 30;74(2):299–307. doi: 10.1016/0092-8674(93)90421-l. [DOI] [PubMed] [Google Scholar]
- Demple B., Sedgwick B., Robins P., Totty N., Waterfield M. D., Lindahl T. Active site and complete sequence of the suicidal methyltransferase that counters alkylation mutagenesis. Proc Natl Acad Sci U S A. 1985 May;82(9):2688–2692. doi: 10.1073/pnas.82.9.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman S., Ansari N. Binding of the EcoRII methyltransferase to 5-fluorocytosine-containing DNA. Isolation of a bound peptide. Nucleic Acids Res. 1992 Jun 25;20(12):3241–3248. doi: 10.1093/nar/20.12.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene P. H., Poonian M. S., Nussbaum A. L., Tobias L., Garfin D. E., Boyer H. W., Goodman H. M. Restriction and modification of a self-complementary octanucleotide containing the EcoRI substrate. J Mol Biol. 1975 Dec 5;99(2):237–261. doi: 10.1016/s0022-2836(75)80143-4. [DOI] [PubMed] [Google Scholar]
- Hanck T., Schmidt S., Fritz H. J. Sequence-specific and mechanism-based crosslinking of Dcm DNA cytosine-C5 methyltransferase of E. coli K-12 to synthetic oligonucleotides containing 5-fluoro-2'-deoxycytidine. Nucleic Acids Res. 1993 Jan 25;21(2):303–309. doi: 10.1093/nar/21.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho D. K., Wu J. C., Santi D. V., Floss H. G. Stereochemical studies of the C-methylation of deoxycytidine catalyzed by HhaI methylase and the N-methylation of deoxyadenosine catalyzed by EcoRI methylase. Arch Biochem Biophys. 1991 Feb 1;284(2):264–269. doi: 10.1016/0003-9861(91)90294-s. [DOI] [PubMed] [Google Scholar]
- Hornby D. P., Müller M., Bickle T. A. High level expression of the EcoP1 modification methylase gene and characterisation of the gene product. Gene. 1987;54(2-3):239–245. doi: 10.1016/0378-1119(87)90492-6. [DOI] [PubMed] [Google Scholar]
- Hümbelin M., Suri B., Rao D. N., Hornby D. P., Eberle H., Pripfl T., Kenel S., Bickle T. A. Type III DNA restriction and modification systems EcoP1 and EcoP15. Nucleotide sequence of the EcoP1 operon, the EcoP15 mod gene and some EcoP1 mod mutants. J Mol Biol. 1988 Mar 5;200(1):23–29. doi: 10.1016/0022-2836(88)90330-0. [DOI] [PubMed] [Google Scholar]
- Kiss A., Posfai G., Keller C. C., Venetianer P., Roberts R. J. Nucleotide sequence of the BsuRI restriction-modification system. Nucleic Acids Res. 1985 Sep 25;13(18):6403–6421. doi: 10.1093/nar/13.18.6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimasauskas S., Kumar S., Roberts R. J., Cheng X. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994 Jan 28;76(2):357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- Klimasauskas S., Nelson J. L., Roberts R. J. The sequence specificity domain of cytosine-C5 methylases. Nucleic Acids Res. 1991 Nov 25;19(22):6183–6190. doi: 10.1093/nar/19.22.6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimasauskas S., Timinskas A., Menkevicius S., Butkienè D., Butkus V., Janulaitis A. Sequence motifs characteristic of DNA[cytosine-N4]methyltransferases: similarity to adenine and cytosine-C5 DNA-methylases. Nucleic Acids Res. 1989 Dec 11;17(23):9823–9832. doi: 10.1093/nar/17.23.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Cheng X., Klimasauskas S., Mi S., Posfai J., Roberts R. J., Wilson G. G. The DNA (cytosine-5) methyltransferases. Nucleic Acids Res. 1994 Jan 11;22(1):1–10. doi: 10.1093/nar/22.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mi S., Roberts R. J. The DNA binding affinity of HhaI methylase is increased by a single amino acid substitution in the catalytic center. Nucleic Acids Res. 1993 May 25;21(10):2459–2464. doi: 10.1093/nar/21.10.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M. H., Gulbis J. M., Dodson E. J., Demple B., Moody P. C. Crystal structure of a suicidal DNA repair protein: the Ada O6-methylguanine-DNA methyltransferase from E. coli. EMBO J. 1994 Apr 1;13(7):1495–1501. doi: 10.1002/j.1460-2075.1994.tb06410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers L. C., Verdine G. L., Wagner G. Solution structure of the DNA methyl phosphotriester repair domain of Escherichia coli Ada. Biochemistry. 1993 Dec 28;32(51):14089–14094. doi: 10.1021/bi00214a003. [DOI] [PubMed] [Google Scholar]
- Pogolotti A. L., Jr, Ono A., Subramaniam R., Santi D. V. On the mechanism of DNA-adenine methylase. J Biol Chem. 1988 Jun 5;263(16):7461–7464. [PubMed] [Google Scholar]
- Pósfai G., Kiss A., Erdei S., Pósfai J., Venetianer P. Structure of the Bacillus sphaericus R modification methylase gene. J Mol Biol. 1983 Nov 5;170(3):597–610. doi: 10.1016/s0022-2836(83)80123-5. [DOI] [PubMed] [Google Scholar]
- Rydberg B., Lindahl T. Nonenzymatic methylation of DNA by the intracellular methyl group donor S-adenosyl-L-methionine is a potentially mutagenic reaction. EMBO J. 1982;1(2):211–216. doi: 10.1002/j.1460-2075.1982.tb01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scopes R. K. Measurement of protein by spectrophotometry at 205 nm. Anal Biochem. 1974 May;59(1):277–282. doi: 10.1016/0003-2697(74)90034-7. [DOI] [PubMed] [Google Scholar]
- Smith S. S., Kaplan B. E., Sowers L. C., Newman E. M. Mechanism of human methyl-directed DNA methyltransferase and the fidelity of cytosine methylation. Proc Natl Acad Sci U S A. 1992 May 15;89(10):4744–4748. doi: 10.1073/pnas.89.10.4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Som S., Friedman S. Identification of a highly conserved domain in the EcoRII methyltransferase which can be photolabeled with S-adenosyl-L-[methyl-3H]methionine. Evidence for UV-induced transmethylation of cysteine 186. J Biol Chem. 1991 Feb 15;266(5):2937–2945. [PubMed] [Google Scholar]
- Szomolányi E., Kiss A., Venetianer P. Cloning the modification methylase gene of Bacillus sphaericus R in Escherichia coli. Gene. 1980 Aug;10(3):219–225. doi: 10.1016/0378-1119(80)90051-7. [DOI] [PubMed] [Google Scholar]
- Verdine G. L. The flip side of DNA methylation. Cell. 1994 Jan 28;76(2):197–200. doi: 10.1016/0092-8674(94)90326-3. [DOI] [PubMed] [Google Scholar]
- Wilke K., Rauhut E., Noyer-Weidner M., Lauster R., Pawlek B., Behrens B., Trautner T. A. Sequential order of target-recognizing domains in multispecific DNA-methyltransferases. EMBO J. 1988 Aug;7(8):2601–2609. doi: 10.1002/j.1460-2075.1988.tb03110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. C., Santi D. V. Kinetic and catalytic mechanism of HhaI methyltransferase. J Biol Chem. 1987 Apr 5;262(10):4778–4786. [PubMed] [Google Scholar]
- Wyszynski M. W., Gabbara S., Bhagwat A. S. Substitutions of a cysteine conserved among DNA cytosine methylases result in a variety of phenotypes. Nucleic Acids Res. 1992 Jan 25;20(2):319–326. doi: 10.1093/nar/20.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyszynski M. W., Gabbara S., Kubareva E. A., Romanova E. A., Oretskaya T. S., Gromova E. S., Shabarova Z. A., Bhagwat A. S. The cysteine conserved among DNA cytosine methylases is required for methyl transfer, but not for specific DNA binding. Nucleic Acids Res. 1993 Jan 25;21(2):295–301. doi: 10.1093/nar/21.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]