Introduction
The amygdala is a complex structure composed of a heterogenous group of nuclei and subnuclei, which are primarily defined by distinct cytoarchitectonics and differing connectivity patterns (Freese and Amaral, 2005, 2006, 2009; Alheid, 2003; Price et al., 1987; Aggleton, 2000; Gloor, 1972, 1978, 1997; McDonald, 1998). Although the names and boundaries of these nuclei remain disputed, they are commonly grouped into four main divisions: lateral (LA), basal and accessory basal (BA), medial and cortical (ME), and central (CE) (e.g. (LeDoux, 1998). These structures are also functionally distinct. For example, LA is involved in learning new stimulus-affect associations (Johansen et al., 2010), whereas ME is involved in olfactory associations and sexual behavior (Lehman et al., 1980; Bian et al., 2008). These functions are likely determined by the afferent and efferent connectivity patterns to each region (LeDoux, 1996; Swanson and Petrovich, 1998; Pitkanen et al., 1997). For example, LA and BA are engaged in updating current stimulus value associations, primarily through connections with orbitofrontal regions (Baxter and Murray, 2002), whereas CE is believed to mediate behavioral responses to potentially harmful stimuli through its connectivity with hypothalamus, basal forebrain, and the brainstem (Kalin et al., 2004).
The distinct functions of the amygdala nucleus groups are not well-understood in the human brain, however, because the nuclei cannot be differentiated in standard magnetic resonance imaging. This is regrettable, because multiple studies suggest amygdalar involvement in psychopathology, such as mood (Phillips et al., 2003), anxiety (Rauch et al., 2003), and developmental disorders (Baron-Cohen et al., 2000). Some attempts have been made to segment the amygdala, either manually through visual approximation based on a single-subject histological atlas (Etkin et al., 2004), or automatically by normalizing the subject’s brain to a template brain and applying a thresholded probabilistic atlas (Amunts et al., 2005). The former approach is labor intensive and susceptible to human error, whereas the latter approach is prey to normalization errors. Further, the use of any atlas necessarily disregards individual differences in nucleic anatomy. Without an easily accessible and robust technique with which to compartmentalize the amygdala, it is difficult to elucidate the separate roles of the human amygdaloid nuclei, as well as the impact of individual differences in nucleus structure and function. Moreover, progress towards mechanistic theories of dysfunction and abnormal development will remain hindered until these structures can be explored in vivo.
Given the unique set of extrinsic connections for each nucleus, it may be possible to differentiate the distinct nuclei by their anatomic connectivity patterns. A metric of structural connectivity can be acquired non-invasively through diffusion weighted imaging (DWI), an MRI method that utilizes the propensity of water to travel along myelinated axons. Fibers can then be reconstructed using a variety of methods collectively termed tractography.
We adapted and extended methods that used probabilistic tractography (Behrens et al., 2003a) to divide each subject’s set of amygdaloid voxels into logical subsets, using Boolean expressions. Boolean logic has several properties that make it potentially advantageous for segmenting regions with highly overlapping connectivity patterns such as the amygdaloid nuclei. First, Boolean expressions can define precise combinations of connectivity patterns through specifically defined sets of unions, intersections, and negations. This should be an effective approach in disambiguating the similar connectivity profiles among amygdaloid nuclei. Second, we expected that this would be particularly useful when combining several smaller nuclei or subnuclei with distinct connectivity patterns. For example, LA is composed of dorsal, dorsal intermediate, ventral intermediate, and ventral subnuclei (Pitkanen and Amaral, 1998; Price et al., 1987), but these subdivisions are too small for typical scan resolutions and so are combined here for practical purposes. Boolean logic can easily combine connectivity patterns of these small subnuclei into a single unit. Finally, Boolean logic is especially appropriate when connectivity patterns are known a priori and are well-explored; a single expression can then be directly constructed from actual anatomical data.
Here we present a novel method, TractSeg (Tractography-based Segmentation), that localizes the four main nucleus groups in the living human amygdala (BA, LA, CE, and ME) using probabilistic tractography on DWI scans that take less than ten minutes to acquire. We hypothesized that it was possible to delineate subregions in the human amygdala based on connectivity patterns derived mainly from animal studies. To validate this method, we compared these subregions with the known topography of their corresponding nuclei, and tested how well they mapped on to the nucleic boundaries observable with a high-resolution scan. In addition, we assessed the across-subject consistency of TractSeg by measuring the spatial overlap between subjects’ nuclei, in a reference frame produced by rigid-body rotation based on each subject’s own amygdalae.
Methods
Subjects
Thirty-six subjects were recruited from the greater Boston area between the ages of 19 and 42 (mean age=25.7±0.2, 19 female). Subjects were screened for history of mental illness and were compensated at $30/hr. The diffusion sequences and anatomical sequences took approximately 20 minutes. The study was approved by the Massachusetts Institute of Technology and Massachusetts General Hospital ethics committees.
Acquisition
Diffusion-weighted data were acquired using echo planar imaging (64 slices, voxel size 2×2×2mm, 128×128 base resolution, diffusion weighting isotropically distributed along 60 directions, b-value 700s/mm2) on a 3T Siemens scanner with a 32 channel head-coil (Reese et al., 2003). A high resolution (1mm3) 3D magnetization-prepared rapid acquisition with gradient echo (MPRAGE) scan was also acquired on these subjects. An additional higher-resolution scan, which was optimized to differentiate amygdala nuclei in-vivo, was obtained on one of the subjects (dual-echo TE0=5ms, TE1=12ms, TR=20ms, 20° flip angle, 600μm × 600 μm × 600 μm, 8 runs registered and averaged). All analyses were performed on subject-specific anatomy, rather than extrapolation from a template brain.
Tractography
Automated cortical and subcortical parcellation was performed (Fischl et al., 2002, 2004) to define specific cortical and subcortical regions in each individual’s T1 scan. Automated segmentation results were reviewed for quality control, and were then registered to each individual’s diffusion images, and used as the seed and target regions for fiber tracking. The resulting cortical and subcortical targets (including the amygdala) were then checked, and corrected for parcellation errors if necessary. The principal diffusion directions were calculated per voxel, and probabilistic diffusion tractography was carried out using FSL-FDT (Behrens et al., 2003b, 2007) with 25000 streamline samples in each seed voxel to create a connectivity distribution to each of the target regions, while avoiding a mask consisting of the ventricles.
Classification
In each subject, we calculated the connection probability (using FSL-FDT’s probtrackX) from each amygdala voxel (seed) to all bilateral cortical and subcortical regions (targets), and normalized the distribution of probabilities for each seed voxel to [0,1] by dividing by the maximum probability. We then thresholded and binarized these results to exclude values below 0.1, such that every amygdaloid voxel contained a 0 or 1 for each target.
Since many of the targets are connected to more than one amygdaloid nucleus (as are nuclei connected to more than one target), we built four Boolean expressions describing the ipsilateral targets that putatively connect with four amygdala nuclei a) LA b) BA c) ME and d) CE. We derived these expressions from histological tracing studies of animal amygdalae, such that each expression reflects known connectivity patterns of the individual nuclei (Table 1). We then applied these expressions to the connectivity distribution of each amygdala voxel. Those that fit an expression were classified as belonging to the corresponding nucleus, whereas voxels that did not match any expression remained unclassified.
Table 1.
Definition of nucleus groups as based on summary of histological tracer studies in rats, nonhuman primates, and humans.
| Target combinations | Putative nucleus |
|---|---|
| ~(Superior parietal | Post-central1-4 | Medial orbitofrontal1,2,5 | Lateral occipital | Pericalcarine | Cuneus6) & (Temporal pole | Fusiform | Lateral orbitofrontal & (Superior temporal | Inferior Temporal1-3,7-9,26))) | Lateral |
|
| |
| (Parahippocampus6 & (Hippocampus15,16 | Rostral anterior cingulate6,17 | Lateral orbitofrontal | Medial orbitofrontal18,2,3,5 | Caudal middle-frontal | Lateral occipital | Pericalcarine | Cuneus | Lingual6,19,20, 26)) | (Insula & (Accumbens | Superior frontal6,21-23)) | Basal |
|
| |
| ~(Brain Stem10,11 & Ventral Diencephalon6,12,13 & Thalamus Proper14) & (Ventral Diencephalon24,25 & (Striatum5 | Hippocampus15,16)) | Medial |
|
| |
| Brain Stem10,11 & Ventral Diencephalon6,12,13 & Thalamus Proper14 | Central |
|
| |
| Table References | |
| 1. (Aggleton et al., 1980) | 14.(Amaral et al., 1992) |
| 2. (Stefanacci and Amaral, 2000) | 15. (Aggleton, 1986) |
| 3. (Stefanacci and Amaral, 2002) | 16. (Amaral, 1986) |
| 4. (Turner et al., 1980) | 17. (Vogt and Pandya, 1987) |
| 5. (Gloor, 1994) | 18. (Carmichael and Price, 1995) |
| 6. (Amaral and Price, 1984) | 19. (Amaral et al., 2003) |
| 7. (Kosmal et al., 1997) | 20. (Freese and Amaral, 2005) |
| 8. (Yukie, 2002) | 21. (Barbas and De Olmos, 1990) |
| 9. (Bachevalier et al., 1997) | 22. (Ghashghaei and Barbas, 2002) |
| 10. (Price and Amaral, 1981) | 23. (Russchen et al., 1985) |
| 11. (Price, 1981) | 24. (Price, 1986) |
| 12. (Amaral et al., 1982) | 25. (Price et al., 1987) |
| 13. (Mehler, 1980) | 26. (Herzog and Van Hoesen, 1976) |
Legend
~ NOT | OR & AND
For example, the LA is the primary recipient of high-level sensory input, mainly from anterior temporal regions, and does not connect with lower-level visual regions; it also receives specific, but sparse, input from the lateral orbitofrontal cortex, but not from medial orbitofrontal cortex. Moreover, there is little evidence of parietal connectivity with the amygdala in general, and specifically none with LA. In order to encapsulate this connectivity pattern in a single expression, we began by negating any voxel that connects with parietal, occipital, or medial orbitofrontal cortices. The LA was defined as the intersection between the remaining subset of voxels and those that connect with anterior temporal cortices, namely temporal pole or fusiform gyrus, or lateral orbitofrontal cortex when also accompanied by connections with other anterior temporal cortices such as the inferior or superior temporal gyri, since BA also connects with lOFC. The three other expressions were also constructed in a similar manner to reflect specific connectivity patterns. The BA projects to all components of the ventral visual system and is reciprocally connected with frontal cortices, mainly mOFC and lOFC. In addition, it is also heavily connected with the hippocampus and related structures. The ME and CE are both highly connected with midbrain targets, but are distinct in their connections to brainstem, in addition to other targets. We therefore used the intersection of ventral diencephalon with the union of caudate and hippocampus to reflect CE connectivity, whereas ME connectivity was characterized by the brainstem, thalamus, and ventral diencephalon targets.
The resulting images were spatially smoothed per nucleus in 3-dimensions, based on the number of neighboring voxels of the same nucleus. Voxels with 6 or more neighbors were classified as the nucleus in question. In order to retain mutual exclusivity between nuclei, any overlapping voxels were classified as belonging to the smaller nucleus. This was implemented in order to preserve boundaries between the nuclei while overcoming the inherent problems of thresholding by number of neighbors: smaller nuclei are more prone to lose voxels, while larger ones are more prone to gain voxels. In the case of no voxels surviving the threshold (which was infrequent: left central in one subject, left medial in another subject, and right central in a third subject), the original un-smoothed nucleus was used. They were then transformed from diffusion space back into each subject’s anatomical coordinates, interpolated based on nearest neighbors, and overlaid on their anatomical MPRAGE scan for figures and quantitative analyses.
Comparison to manual segmentation
We optimized and acquired an additional high-resolution anatomical scan from one subject (see Acquisition). These high-resolution images were manually labeled based on visible boundaries between the four nuclei, and compared to the segmentation derived from tractography. Both the manual and tractographic segmentation images were registered to this subject’s MPRAGE scan (down-sampled from 600um to 1mm and up-sampled from 2mm to 1mm respectively). Performance of the tractographic segmentations was assessed by the voxel-by-voxel correspondence between these two images. The accuracy for each nucleus was measured as the proportion of matching voxels in both segmentations. We also calculated d′ for each nucleus in order to penalize false positives:
where norminv(x) is the inverse of the cumulative Gaussian distribution.
Measures of consistency between subjects
In order to compare the outcome of the connectivity-based segmentation between subjects and still preserve subject-specific anatomy (keeping the images in native-space rather than in normalized-space), we rotated the amygdalae of each subject along an axis drawn from the center-of-masses of the amygdala and the fourth ventricle, correcting for inter-subject differences in pitch (yaw and roll were consistent across subjects). After correcting for head rotation, we placed each amygdala into a common reference frame (with each subject’s amygdala centroid at the origin) by mean-shifting (subtracting the rotated coordinates from the amygdala center-of-mass). A conventional whole-brain approach would not have been practical or informative for comparing subjects or generating a probability map due to low cross-subject alignment of the entire amygdala; when we aligned subjects to the template T1 image provided by SPM8, we found that only 57.58% of the subjects were consistent in the spatial location of the right amygdala, and 60.61% for the left (as compared to our method of alignment for which there was a 97.06% overlap for the right amygdala, and 100% for the left).
Each subject was then iteratively compared with every other subject, and both accuracy and d′ were calculated per subject as the average overlap across the other subjects. We also calculated the mean volumes per nucleus across the subjects, and performed a two-sample Student’s t-test across hemispheres. A cut-off of p≤0.0125 (Bonferonni corrected for multiple comparisons) was used for determining the significance of these tests.
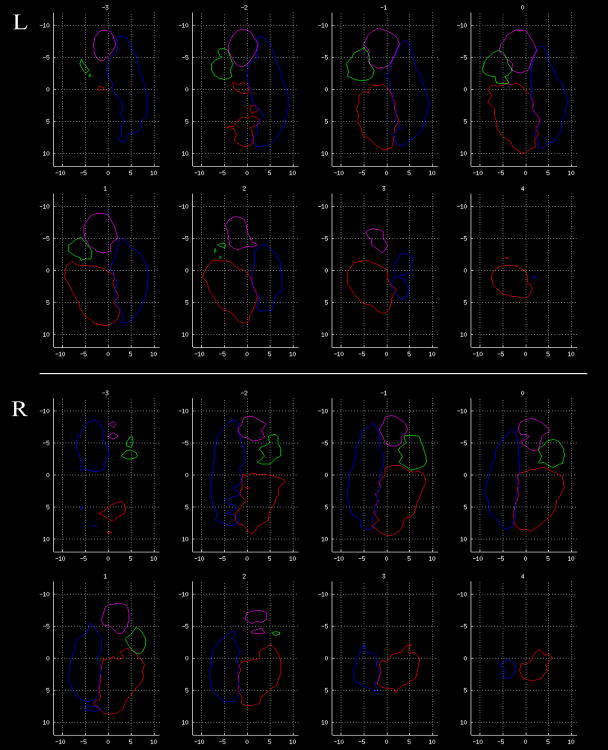
We also generated a probability atlas (Figure 4) of the amygdaloid nuclei by aligning subjects’ amygdalae as above, and calculating the proportion of subjects that share nucleus classification for each voxel. For the sake of visualization, Figure 4 displays the atlas thresholded at 15/35 subjects.
Figure 4. Amygdala tractographic segmentation atlas.
Coronal slices through the population’s left (top panels) and right (bottom panels) amygdalae, from posterior to anterior, in rotated space. The edges of the group probability maps, thresholded at >= 15/35 subjects, are shown for each subregion (color-coded as in Figure 1). Units are in millimeters, and with respect to the amygdala centroid. The reference point for rotation (4th ventricle) is posterior and normal to the plane for all subjects.
Results
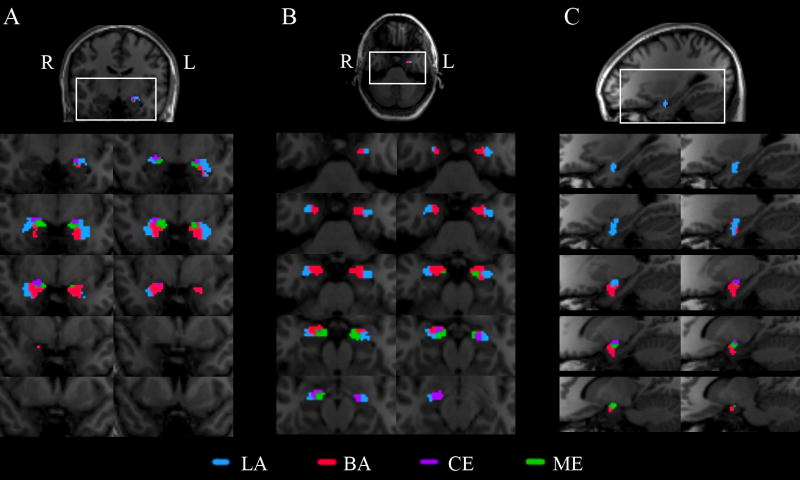
We defined four Boolean expressions that correspond with known connectivity patterns of the four major nucleus groups of the amygdala: LA, BA, CE, and ME (Table 1). The combination of target regions for LA defined the most ventrolateral subregion of the amygdala (as shown on an example subject, Figure 1b-c). This was present along the full rostrocaudal extent (Figure 1a) of the amygdala, which is morphologically and spatially characteristic of LA (Gloor, 1997; Aggleton, 2000; Freese and Amaral, 2009). A similar, but distinct, pattern of connectivity (see Methods and Table 1) identified a more ventral amygdaloid region immediately medial to the LA, corresponding to the known location of the BA (Gloor, 1997; Aggleton, 2000; Freese and Amaral, 2009) (Figure 1a-c). These two subregions were the largest of the tractographic classification, and indeed are the largest nuclei of the amygdala. The third expression defined an oblique subregion of the dorsomedial amygdala, a distinguishing feature of the ME (Figure 1a-c), whereas the last expression classified voxels that were present in the dorsal amygdala and appeared in the caudal-most region, much like the CE (Figure 1a). The four nuclei were also comparable between hemispheres (Figure 1a-b).
Figure 1. Tractographic segmentation in an example subject.
Right- and left-amygdala tractographic segmentation are resampled into anatomical coordinates, and overlaid on the same subject’s MPRAGE images. Nuclei are color-coded as: BA (red), LA (blue), ME (green), CE (purple). a, Coronal sections from posterior to anterior extents of the amygdalae demonstrate the comparable segmentations for both hemispheres, and also illustrate that the LA and BA occupy the most rostral extents, while CE and ME appear more caudally. b, Axial sections from inferior to superior further describe the nuclei, where LA and BA are more ventral than CE or ME. c, Right sagittal sections from lateral to medial show the most lateral (LA) and medial (ME) nuclei in relation to the other nuclei.
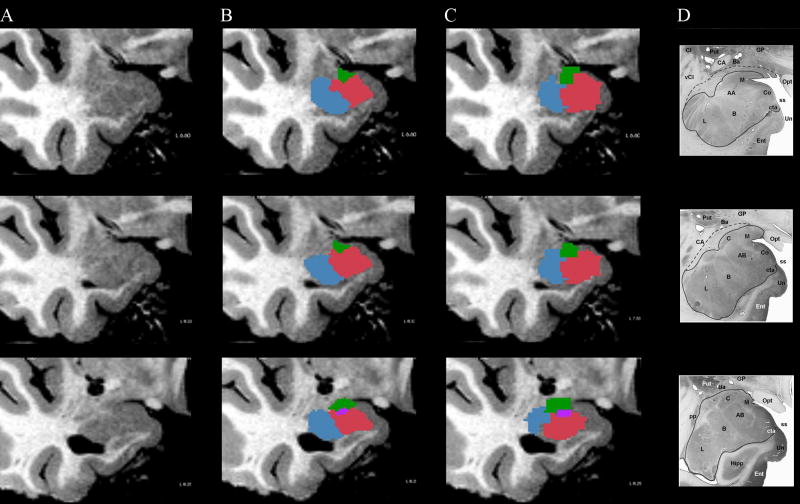
High-resolution validation
In order to visualize the boundaries between the nuclei, we acquired an additional high-resolution anatomical scan from one subject (Figure 2a). This scan, averaged over 8 runs totaling approximately two hours, gave us the resolution and contrast-to-noise ratio (CNR) needed to visualize the boundaries between the nuclei in vivo. These images were manually labeled to segment the amygdala into the 4 nuclei (Figure 2b), and compared to the connectivity-based segmentation based on the ten-minute diffusion-weighted sequence (Figure 2c) in the same subject. The size, shape, and location of the LA, BA, CE, and ME were markedly similar between the manually-labeled amygdala and the tractographic segmentation. Since both manual and tractographic segmentation were performed on the same individual, we were able to overlay and directly compare them. For each nucleus, we calculated accuracy as the proportion of matching voxels, and d′ as the difference between standardized hit rates and false alarm rates, wherein values of 0 or <0 imply an overlap at or worse than chance, and d′≥1 indicating high sensitivity. The tractographic segmentation was very similar to the manual segmentation, with high accuracy rates in both hemispheres for the LA (R:0.86; L:0.71), BA (R:0.80; L:0.66), CE (R:0.89; L:0.85), ME (R:0.93; L:0.95) and high d′ values LA (R:2.13; L:1.27), BA (R:1.76; L:1.19), CE (R:1.16; L:2.40), ME (R:2.30; L:2.14).
Figure 2. Manual vs. tractographic segmentation.
a, Coronal images of the right amgydala from a high-resolution scan (dual-echo 20° flip angle TE0 / TE1 / TR = 5ms/ 12ms/ 20ms 600μm isotropic) averaged over 8 runs. b, Boundaries visible from this scan were used to manually segment the amygdala into four nuclei, color-coded as in Figure1. c, Tractographic segmentation on the same individual was registered and overlaid on the same coronal slices as Figure 2a and b. The nuclei are visually similar to those based on a high-resolution scan, as well as to d, a coronal section based on a histological specimen of the human amygdala (reprinted with permission from Brabec et al., 2010).
Consistency across individuals
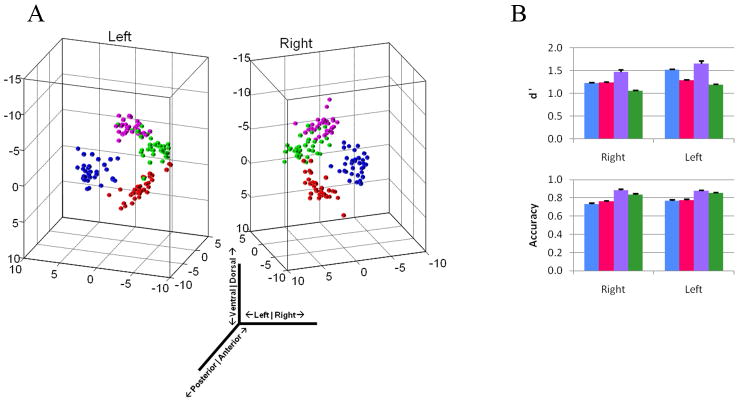
These subregions were also consistent in size, shape, and location across an additional 35 subjects, with lateral and basal occupying the largest volumes, central and medial the smallest (Table 2), and no between-hemisphere differences (LA: p=0.89; BA: p=0.24; CE: p=0.02; ME: p=0.34). We placed each subject’s amygdala into a common reference frame via rigid body rotation, free of any spatial warping (see Methods). We were then able to visualize the consistency of nucleic location in three dimensions across individuals in both the right and left amygdalae (Figure 3a).
Table 2.
Nucleus volumes in proportion to the whole amygdala across subjects.
| Lateral | Basal | Central | Medial | |
|---|---|---|---|---|
| Left | 0.40±0.03 | 0.32±0.02 | 0.16±0.01 | 0.12±0.02 |
| Right | 0.40±0.02 | 0.35±0.02 | 0.11±0.01 | 0.14±0.02 |
Values are reported in mean ± standard error.
Figure 3. Overlap of tractographic segmentation-based nuclei between subjects.
a, Resulting centroid locations, after alignment (see methods), of the segmented nuclei are plotted from 35 subjects, demonstrating the similarity of nucleus location, in three dimensions. Nuclei are color-coded as in Figure 1. b, Right- and left-amygdala tractographic segmentation was consistent among 35 subjects, as indicated by high d′ and accuracy values.
Accuracy and d′ measures of overlap between subjects were calculated by iteratively using each subject as a reference in comparison to every other subject. As observed qualitatively, the degree of overlap was high across subjects for nuclei in both hemispheres, with high average accuracy rates (Figure 3b): LA (R:0.73±0.01; L:0.77±0.01), BA (R:0.76±0.01; L:0.78±0.01), CE (R:0.89±0.01; L:0.88±0.01), ME (R:0.84±0.01; L:0.85±0.01) and high average d′ values: LA (R:1.23±0.05; L:1.52±0.06), BA (R:1.25±0.05; L:1.29±0.06), CE (R:1.47±0.05; L:1.66±0.05), ME (R:1.06±0.06; L:1.19±0.05). We used these amygdala subdivisions from our 35 subjects to generate a population-based atlas of the human amygdala, thresholded by overlap of at least 15 out of 35 subjects (Figure 4).
Discussion
By exploiting the differential connectivity patterns of four amygdaloid nuclei, we generated logical statements that anatomically define four subregions in the amygdala. These expressions were based on connectivity patterns from non-human amygdalae, since there are few human tracer studies. Nonetheless, when these expressions were applied to tractographic reconstructions of the human amygdala, they generated spatially-distinct clusters that map well to their known locations. These subregions were spatially consistent across individuals, and were validated by a high-resolution image in one subject.
To the best of our knowledge, this is the first time that the amygdala has been noninvasively segmented into four putative nucleus groups based on structural connectivity patterns. Previous research used other methods, such as visual approximation to distinguish the dorsal vs. ventral amygdala (Etkin et al., 2004; Dolan, 2002, 2007; Dolan and Vuilleumier, 2003; Dolan et al., 2001, 2006) and posit functional roles for these subregions with fMRI, which can be further explored now at a single-subject level and with more subdivisions with which to predict and test models of amygdalar function. Fiber orientations (based on DWI scans) within the amygdala have been used to divide the structure into two subregions, centromedial and basolateral (Solano-Castiella et al., 2010). However, this method, like others before it, performed analyses on images normalized to a template brain, and were also restricted to two subdivisions. Visual approximation or normalization methods may be susceptible to errors which the current method circumvents. We used native-space analyses (Fischl et al., 2008) to generate target regions which were specific to individuals’ anatomy, and performed all subsequent analyses in native-space as well, such that the resulting amygdala subdivisions were also unique to the individual’s own anatomy.
Native-space analyses better accommodate individual variation in subcortical volume (Di Martino et al., 2008; Pujol et al., 2010), and are thus best-suited for volumetric analyses and studies of clinical populations that have smaller or larger average amygdala volumes (e.g. Nacewicz et al., 2006; Brambilla et al., 2003; Chance et al., 2002). The current method could be implemented to explore differences in amygdaloid nucleus volumes and their relative contributions to the size of the whole amygdala. Furthermore, volumetric differences between populations, elucidated via TractSeg, could indeed be due to either nucleic variation or to connectivity differences between populations. This can be further explored by applying the probability atlas to the pathological population, and analyzing connectivity differences between the atlas-based segmentation and the subject-specific TractSeg-based segmentation. Additionally, future studies can investigate the relative contribution of connectivity versus actual amygdala subdivision differences by generating a database of nucleus volumes (based on histology and/or high-resolution imaging) in order to probe normal variations, and relate them to connectivity differences.
We also extended other efforts to segment the amygdala or other gray matter structures through connectivity by validating the connectivity-based subdivisions using a high-resolution structural scan, similar to a previous approach for localizing lateral geniculate bodies (Devlin et al., 2006). High-resolution scans averaged across multiple runs, such as the one developed for the purposes of this study, allow for dramatically better visualization than standard resolution images. However they are currently too long to be commonly used in conjunction with other types of scans (such as functional MRI) and too strenuous for many subjects who cannot remain motionless throughout the scan; in our case, the high-resolution image took two hours to acquire. Such long scan durations are especially impractical for clinical and developmental applications. The present study used a DWI scan lasting less than ten minutes to segment the amygdala, and produced results that converged substantially with those of the optimized high-resolution acquisition.
Although we employed many of the basic principles and pre-processing steps of pioneering probabilistic tractographic studies, e.g. (Behrens et al., 2003a), our use of native-space analyses (discussed above) and Boolean logic extends these in ways that will facilitate future research at the single-subject level. This method can be applied to not only the amygdala, but to any gray matter structure. Furthermore, it is particularly effective because it allows for a combination of target regions and thus can be robust across individuals and noisy MR signals. These Boolean expressions can disambiguate the highly overlapping patterns of connectivity among gray matter nuclei with specifically defined sets of unions, intersections, and negations. It is particularly appropriate when connectivity patterns are known a priori in order to construct expressions that should theoretically define the nuclei in question. Future methods might also benefit from logical solutions that can handle continuous probabilities, as opposed to binarization, such as fuzzy logic, e.g. (McNeill and Freiberger, 1993).
One possible limitation of DWI in general is that the polarity of connections is unknown; future studies employing this method should keep this in consideration when building the sets of connectivity profiles. Also, since all cortical and subcortical regions were used to create target regions and connectivity distributions to the amygdala, whole-brain coverage during DWI acquisition is necessary to use this method.
We suggest that this method has applications in exploring the functions of distinct nuclei, exploring structural and functional networks, and can be used to segment other gray matter regions. The regions-of-interest (ROIs) generated from this method (which remain in native space and are true to the individual’s own anatomy in shape, size, and location) can be used as independently localized ROIs for fMRI analyses. This could be useful in elucidating the specific roles of distinct nuclei within the human amygdala, both in healthy controls, and in clinical populations. By expanding the seed region to encompass a larger region than what is typically defined as the amygdala, TractSeg can also be used to explore specific hypotheses of the function and structural organization of the extended amygdala (Cassell et al., 1999). Furthermore, the nuclei can be used as seed regions for functional connectivity analyses, and thus for exploring differences in functional networks between populations or across development. The nuclei might also be definable by these Boolean expressions but from functional rather than structural connectivity. This will broaden our understanding of the similarities or differences of structural vs. functional networks.
Conclusions
In the present paper, we proposed a new method of using known structural connectivity patterns to define subject-specific amygdaloid subregions. We have shown that these regions correspond to the known locations of the nuclei based on histology, as well as to a high-resolution MR scan on which nucleic boundaries are visible. The subregions were also spatially consistent across 35 individuals. Future studies can explore the specific roles of distinct nuclei within the human amygdala, especially since this method relies on rapid diffusion sequences that can be easily introduced into experimental paradigms and for studying clinical populations. We demonstrate a specific application to the amygdala here, but TractSeg can be applied to any gray matter structure. To facilitate future applications, we use software packages that are easily available, documented and supported, and free. Finally, the method will be released both as a stand-alone program (http://gablab.mit.edu/software/ArchiTract/), and as a component of FreeSurfer (http://surfer.nmr.mgh.harvard.edu/) and NyPype, a free, open-source framework capable of integrating analysis software from a variety of sources (http://nipy.org/nipype).
Acknowledgments
We are grateful to S. Ghosh and R. R. Saxe for valuable discussions and revisions, A. Kouwe, F. Polli, S. Sabhlok, and G. Reynolds for help with acquisition, and M. Reuter for help with high-resolution registration methods. This work was supported by PHS grant DA023427 (Z.M.S) and by the Poitras Center for Affective Disorders Research (J.D.E.G.). Support was also provided in part by: the National Center for Research Resources (P41-RR14075, and the NCRR BIRN Morphometric Project BIRN002, U24 RR021382), the National Institute for Biomedical Imaging and Bioengineering (R01EB006758), the National Institute on Aging (AG022381), the National Institute for Neurological Disorders and Stroke (R01 NS052585-01, 1R21NS072652-01), Shared Instrumentation Grants (1S10RR023401, 1S10RR019, and 1S10RR023043), and the Ellison Medical Foundation (B.F.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- AGGLETON JP. A description of the amygdalo-hippocampal interconnections in the macaque monkey. Exp Brain Res. 1986;64:515–26. doi: 10.1007/BF00340489. [DOI] [PubMed] [Google Scholar]
- AGGLETON JP, BURTON MJ, PASSINGHAM RE. Cortical and subcortical afferents to the amygdala of the rhesus monkey (Macaca mulatta) Brain Res. 1980;190:347–68. doi: 10.1016/0006-8993(80)90279-6. [DOI] [PubMed] [Google Scholar]
- AGGLETON JPE. The Amygdala. Oxford, United Kingdom: Oxford University Press; 2000. [Google Scholar]
- ALHEID GF. Extended amygdala and basal forebrain. Ann N Y Acad Sci. 2003;985:185–205. doi: 10.1111/j.1749-6632.2003.tb07082.x. [DOI] [PubMed] [Google Scholar]
- AMARAL DG. Amygdalohippocampal and amygdalocortical projections in the primate brain. Adv Exp Med Biol. 1986;203:3–17. doi: 10.1007/978-1-4684-7971-3_1. [DOI] [PubMed] [Google Scholar]
- AMARAL DG, BEHNIEA H, KELLY JL. Topographic organization of projections from the amygdala to the visual cortex in the macaque monkey. Neuroscience. 2003;118:1099–120. doi: 10.1016/s0306-4522(02)01001-1. [DOI] [PubMed] [Google Scholar]
- AMARAL DG, PRICE JL. Amygdalo-cortical projections in the monkey (Macaca fascicularis) J Comp Neurol. 1984;230:465–96. doi: 10.1002/cne.902300402. [DOI] [PubMed] [Google Scholar]
- AMARAL DG, PRICE JL, PITKANEN A, CARMICHAEL ST. Anatomical organization of the primate amygdaloid complex. In: AGGLETON JP, editor. The Amygdala Neurobiological Aspects of Emotion, Memory, and Mental Dysfunction. New York: Wiley-Liss; 1992. [Google Scholar]
- AMARAL DG, VEAZEY RB, COWAN WM. Some observations on hypothalamo-amygdaloid connections in the monkey. Brain Res. 1982;252:13–27. doi: 10.1016/0006-8993(82)90974-x. [DOI] [PubMed] [Google Scholar]
- AMUNTS K, KEDO O, KINDLER M, PIEPERHOFF P, MOHLBERG H, SHAH NJ, HABEL U, SCHNEIDER F, ZILLES K. Cytoarchitectonic mapping of the human amygdala, hippocampal region and entorhinal cortex: intersubject variability and probability maps. Anat Embryol (Berl) 2005;210:343–52. doi: 10.1007/s00429-005-0025-5. [DOI] [PubMed] [Google Scholar]
- BACHEVALIER J, MEUNIER M, LU MX, UNGERLEIDER LG. Thalamic and temporal cortex input to medial prefrontal cortex in rhesus monkeys. Exp Brain Res. 1997;115:430–44. doi: 10.1007/pl00005713. [DOI] [PubMed] [Google Scholar]
- BARBAS H, DE OLMOS J. Projections from the amygdala to basoventral and mediodorsal prefrontal regions in the rhesus monkey. J Comp Neurol. 1990;300:549–71. doi: 10.1002/cne.903000409. [DOI] [PubMed] [Google Scholar]
- BARON-COHEN S, RING HA, BULLMORE ET, WHEELWRIGHT S, ASHWIN C, WILLIAMS SC. The amygdala theory of autism. Neurosci Biobehav Rev. 2000;24:355–64. doi: 10.1016/s0149-7634(00)00011-7. [DOI] [PubMed] [Google Scholar]
- BAXTER MG, MURRAY EA. The amygdala and reward. Nat Rev Neurosci. 2002;3:563–73. doi: 10.1038/nrn875. [DOI] [PubMed] [Google Scholar]
- BEHRENS TE, BERG HJ, JBABDI S, RUSHWORTH MF, WOOLRICH MW. Probabilistic diffusion tractography with multiple fibre orientations: What can we gain? Neuroimage. 2007;34:144–55. doi: 10.1016/j.neuroimage.2006.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BEHRENS TE, JOHANSEN-BERG H, WOOLRICH MW, SMITH SM, WHEELER-KINGSHOTT CA, BOULBY PA, BARKER GJ, SILLERY EL, SHEEHAN K, CICCARELLI O, THOMPSON AJ, BRADY JM, MATTHEWS PM. Non-invasive mapping of connections between human thalamus and cortex using diffusion imaging. Nat Neurosci. 2003a;6:750–7. doi: 10.1038/nn1075. [DOI] [PubMed] [Google Scholar]
- BEHRENS TE, WOOLRICH MW, JENKINSON M, JOHANSEN-BERG H, NUNES RG, CLARE S, MATTHEWS PM, BRADY JM, SMITH SM. Characterization and propagation of uncertainty in diffusion-weighted MR imaging. Magn Reson Med. 2003b;50:1077–88. doi: 10.1002/mrm.10609. [DOI] [PubMed] [Google Scholar]
- BIAN X, YANAGAWA Y, CHEN WR, LUO M. Cortical-like functional organization of the pheromone-processing circuits in the medial amygdala. J Neurophysiol. 2008;99:77–86. doi: 10.1152/jn.00902.2007. [DOI] [PubMed] [Google Scholar]
- BRAMBILLA P, HARDAN A, DI NEMI SU, PEREZ J, SOARES JC, BARALE F. Brain anatomy and development in autism: review of structural MRI studies. Brain Res Bull. 2003;61:557–69. doi: 10.1016/j.brainresbull.2003.06.001. [DOI] [PubMed] [Google Scholar]
- CARMICHAEL ST, PRICE JL. Limbic connections of the orbital and medial prefrontal cortex in macaque monkeys. J Comp Neurol. 1995;363:615–641. doi: 10.1002/cne.903630408. [DOI] [PubMed] [Google Scholar]
- CASSELL MD, FREEDMAN LJ, SHI C. The intrinsic organization of the central extended amygdala. Ann N Y Acad Sci. 1999;877:217–41. doi: 10.1111/j.1749-6632.1999.tb09270.x. [DOI] [PubMed] [Google Scholar]
- CHANCE SA, ESIRI MM, CROW TJ. Amygdala volume in schizophrenia: post-mortem study and review of magnetic resonance imaging findings. Br J Psychiatry. 2002;180:331–8. doi: 10.1192/bjp.180.4.331. [DOI] [PubMed] [Google Scholar]
- DEVLIN JT, SILLERY EL, HALL DA, HOBDEN P, BEHRENS TEJ, NUNES RG, CLARE S, MATTHEWS PM, MOORE DR, JOHANSEN-BERG H. Reliable identification of the auditory thalamus using multi-modal structural analyses. Neuroimage. 2006;30(4):1112–1120. doi: 10.1016/j.neuroimage.2005.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DI MARTINO A, SCHERES A, MARGULIES DS, KELLY AM, UDDIN LQ, SHEHZAD Z, BISWAL B, WALTERS JR, CASTELLANOS FX, MILHAM MP. Functional connectivity of human striatum: a resting state FMRI study. Cereb Cortex. 2008;18:2735–47. doi: 10.1093/cercor/bhn041. [DOI] [PubMed] [Google Scholar]
- DOLAN RJ. Emotion, cognition, and behavior. Science. 2002;298:1191–4. doi: 10.1126/science.1076358. [DOI] [PubMed] [Google Scholar]
- DOLAN RJ. The human amygdala and orbital prefrontal cortex in behavioural regulation. Philos Trans R Soc Lond B Biol Sci. 2007;362:787–99. doi: 10.1098/rstb.2007.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOLAN RJ, HEINZE HJ, HURLEMANN R, HINRICHS H. Magnetoencephalography (MEG) determined temporal modulation of visual and auditory sensory processing in the context of classical conditioning to faces. Neuroimage. 2006;32:778–89. doi: 10.1016/j.neuroimage.2006.04.206. [DOI] [PubMed] [Google Scholar]
- DOLAN RJ, MORRIS JS, DE GELDER B. Crossmodal binding of fear in voice and face. Proc Natl Acad Sci U S A. 2001;98:10006–10. doi: 10.1073/pnas.171288598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOLAN RJ, VUILLEUMIER P. Amygdala automaticity in emotional processing. Ann N Y Acad Sci. 2003;985:348–55. doi: 10.1111/j.1749-6632.2003.tb07093.x. [DOI] [PubMed] [Google Scholar]
- ETKIN A, KLEMENHAGEN KC, DUDMAN JT, ROGAN MT, HEN R, KANDEL ER, HIRSCH J. Individual differences in trait anxiety predict the response of the basolateral amygdala to unconsciously processed fearful faces. Neuron. 2004;44:1043–55. doi: 10.1016/j.neuron.2004.12.006. [DOI] [PubMed] [Google Scholar]
- FISCHL B, RAJENDRAN N, BUSA E, AUGUSTINACK J, HINDS O, YEO BT, MOHLBERG H, AMUNTS K, ZILLES K. Cortical folding patterns and predicting cytoarchitecture. Cereb Cortex. 2008;18:1973–80. doi: 10.1093/cercor/bhm225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FISCHL B, SALAT DH, BUSA E, ALBERT M, DIETERICH M, HASELGROVE C, VAN DER KOUWE A, KILLIANY R, KENNEDY D, KLAVENESS S, MONTILLO A, MAKRIS N, ROSEN B, DALE AM. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–55. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- FISCHL B, VAN DER KOUWE A, DESTRIEUX C, HALGREN E, SEGONNE F, SALAT DH, BUSA E, SEIDMAN LJ, GOLDSTEIN J, KENNEDY D, CAVINESS V, MAKRIS N, ROSEN B, DALE AM. Automatically parcellating the human cerebral cortex. Cereb Cortex. 2004;14:11–22. doi: 10.1093/cercor/bhg087. [DOI] [PubMed] [Google Scholar]
- FREESE JL, AMARAL DG. The organization of projections from the amygdala to visual cortical areas TE and V1 in the macaque monkey. J Comp Neurol. 2005;486:295–317. doi: 10.1002/cne.20520. [DOI] [PubMed] [Google Scholar]
- FREESE JL, AMARAL DG. Synaptic organization of projections from the amygdala to visual cortical areas TE and V1 in the macaque monkey. J Comp Neurol. 2006;496:655–67. doi: 10.1002/cne.20945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FREESE JL, AMARAL DG. Neuroanatomy of the Primate Amygdala. In: WHALEN PJ, PHELPS EA, editors. The Human Amygdala. New York: The Guilford Press; 2009. [Google Scholar]
- GHASHGHAEI HT, BARBAS H. Pathways for emotion: interactions of prefrontal and anterior temporal pathways in the amygdala of the rhesus monkey. Neuroscience. 2002;115:1261–79. doi: 10.1016/s0306-4522(02)00446-3. [DOI] [PubMed] [Google Scholar]
- GLOOR P. Temporal lobe epilepsy: its possible contribution to the understanding of the functional significance of the amygdala and of its interaction with neocortical-temporal mechanisms. In: ELEFTHERION BE, editor. The Neurobiology of the Amygdala. New York: Plenum Press; 1972. [Google Scholar]
- GLOOR P. Inputs and outputs of the amygdala: what the amygdala is trying to tell the rest of the brain. In: LIVINGSTON KE, HORNIKIEWICZ O, editors. Limbic Mechanisms The Continuing Evolution of the Limbic System Concept. New York and London: Plenum Press; 1978. [Google Scholar]
- GLOOR P. Mesial temporal lobe structures. In: SHORVON SD, FISH DR, ANDERMANN F, BYDDER GM, STEFAN H, editors. Magnetic Resonance Scanning and Epilepsy. New York/London: Plenum Press; 1994. [Google Scholar]
- GLOOR P. The Temporal Lobe and Limbic System. New York, New York: Oxford University Press, Inc; 1997. [Google Scholar]
- HERZOG AG, VAN HOESEN GW. Temporal neocortical afferent connections to the amygdala in the rhesus monkey. Brain Res. 1976;115(1):57–69. doi: 10.1016/0006-8993(76)90822-2. [DOI] [PubMed] [Google Scholar]
- JOHANSEN JP, HAMANAKA H, MONFILS MH, BEHNIA R, DEISSEROTH K, BLAIR HT, LEDOUX JE. Optical activation of lateral amygdala pyramidal cells instructs associative fear learning. Proc Natl Acad Sci U S A. 2010;107:12692–7. doi: 10.1073/pnas.1002418107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KALIN NH, SHELTON SE, DAVIDSON RJ. The role of the central nucleus of the amygdala in mediating fear and anxiety in the primate. J Neurosci. 2004;24:5506–15. doi: 10.1523/JNEUROSCI.0292-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOSMAL A, MALINOWSKA M, KOWALSKA DM. Thalamic and amygdaloid connections of the auditory association cortex of the superior temporal gyrus in rhesus monkey (Macaca mulatta) Acta Neurobiol Exp (Wars) 1997;57:165–88. doi: 10.55782/ane-1997-1224. [DOI] [PubMed] [Google Scholar]
- LEDOUX J. Emotional networks and motor control: a fearful view. Prog Brain Res. 1996;107:437–46. doi: 10.1016/s0079-6123(08)61880-4. [DOI] [PubMed] [Google Scholar]
- LEDOUX J. Fear and the brain: where have we been, and where are we going? Biol Psychiatry. 1998;44:1229–38. doi: 10.1016/s0006-3223(98)00282-0. [DOI] [PubMed] [Google Scholar]
- LEHMAN MN, WINANS SS, POWERS JB. Medial nucleus of the amygdala mediates chemosensory control of male hamster sexual behavior. Science. 1980;210:557–60. doi: 10.1126/science.7423209. [DOI] [PubMed] [Google Scholar]
- MCDONALD AJ. Cortical pathways to the mammalian amygdala. Prog Neurobiol. 1998;55:257–332. doi: 10.1016/s0301-0082(98)00003-3. [DOI] [PubMed] [Google Scholar]
- MCNEILL D, FREIBERGER P. Fuzzy logic. New York: Simon & Schuster; 1993. [Google Scholar]
- MEHLER WR. Subcortical afferent connections of the amygdala in the monkey. J Comp Neurol. 1980;190:733–62. doi: 10.1002/cne.901900407. [DOI] [PubMed] [Google Scholar]
- NACEWICZ BM, DALTON KM, JOHNSTONE T, LONG MT, MCAULIFF EM, OAKES TR, ALEXANDER AL, DAVIDSON RJ. Amygdala volume and nonverbal social impairment in adolescent and adult males with autism. Arch Gen Psychiatry. 2006;63:1417–28. doi: 10.1001/archpsyc.63.12.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PHILLIPS ML, DREVETS WC, RAUCH SL, LANE R. Neurobiology of emotion perception II: Implications for major psychiatric disorders. Biol Psychiatry. 2003;54:515–28. doi: 10.1016/s0006-3223(03)00171-9. [DOI] [PubMed] [Google Scholar]
- PITKANEN A, AMARAL DG. Organization of the intrinsic connections of the monkey amygdaloid complex: projections originating in the lateral nucleus. J Comp Neurol. 1998;398:431–58. doi: 10.1002/(sici)1096-9861(19980831)398:3<431::aid-cne9>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- PITKANEN A, SAVANDER V, LEDOUX JE. Organization of intra-amygdaloid circuitries in the rat: an emerging framework for understanding functions of the amygdala. Trends Neurosci. 1997;20:517–23. doi: 10.1016/s0166-2236(97)01125-9. [DOI] [PubMed] [Google Scholar]
- PRICE JL. The efferent projections of the amygdaloid complex in the rat, cat and monkey. In: BEN-ARI Y, editor. The Amygdaloid Complex. Amsterdam: Elsevier/North Holland Biomedical Press; 1981. [Google Scholar]
- PRICE JL. Subcortical projections from the amygdaloid complex. Adv Exp Med Biol. 1986;203:19–33. doi: 10.1007/978-1-4684-7971-3_2. [DOI] [PubMed] [Google Scholar]
- PRICE JL, AMARAL DG. An autoradiographic study of the projections of the central nucleus of the monkey amygdala. J Neurosci. 1981;1:1242–59. doi: 10.1523/JNEUROSCI.01-11-01242.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PRICE JL, RUSSCHEN FT, AMARAL DG. The limbic region. II. The amygdaloid complex. In: BJÖRKLUND A, HÖKFELT T, SWANSON LW, editors. Handbook of Chemical Neuroanatomy Integrated Systems of the CNS. Amsterdam: Elsevier Science Publishers BV; 1987. [Google Scholar]
- PUJOL J, SORIANO-MAS C, GISPERT JD, BOSSA M, REIG S, ORTIZ H, ALONSO P, CARDONER N, LOPEZ-SOLA M, HARRISON BJ, DEUS J, MENCHON JM, DESCO M, OLMOS S. Variations in the shape of the frontobasal brain region in obsessive-compulsive disorder. Hum Brain Mapp. 2010 doi: 10.1002/hbm.21094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAUCH SL, SHIN LM, WRIGHT CI. Neuroimaging studies of amygdala function in anxiety disorders. Ann N Y Acad Sci. 2003;985:389–410. doi: 10.1111/j.1749-6632.2003.tb07096.x. [DOI] [PubMed] [Google Scholar]
- REESE TG, HEID O, WEISSKOFF RM, WEDEEN VJ. Reduction of eddy-current-induced distortion in diffusion MRI using a twice-refocused spin echo. Magn Reson Med. 2003;49:177–82. doi: 10.1002/mrm.10308. [DOI] [PubMed] [Google Scholar]
- RUSSCHEN FT, BAKST I, AMARAL DG, PRICE JL. The amygdalostriatal projections in the monkey. An anterograde tracing study. Brain Res. 1985;329:241–57. doi: 10.1016/0006-8993(85)90530-x. [DOI] [PubMed] [Google Scholar]
- SOLANO-CASTIELLA E, ANWANDER A, LOHMANN G, WEISS M, DOCHERTY C, GEYER S, REIMER E, FRIEDERICI AD, TURNER R. Diffusion tensor imaging segments the human amygdala in vivo. Neuroimage. 2010;49:2958–65. doi: 10.1016/j.neuroimage.2009.11.027. [DOI] [PubMed] [Google Scholar]
- STEFANACCI L, AMARAL DG. Topographic organization of cortical inputs to the lateral nucleus of the macaque monkey amygdala: a retrograde tracing study. J Comp Neurol. 2000;421:52–79. doi: 10.1002/(sici)1096-9861(20000522)421:1<52::aid-cne4>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- STEFANACCI L, AMARAL DG. Some observations on cortical inputs to the macaque monkey amygdala: an anterograde tracing study. J Comp Neurol. 2002;451:301–23. doi: 10.1002/cne.10339. [DOI] [PubMed] [Google Scholar]
- SWANSON LW, PETROVICH GD. What is the amygdala? Trends Neurosci. 1998;21:323–31. doi: 10.1016/s0166-2236(98)01265-x. [DOI] [PubMed] [Google Scholar]
- TURNER BH, MISHKIN M, KNAPP M. Organization of the amygdalopetal projections from modality-specific cortical association areas in the monkey. J Comp Neurol. 1980;191:515–43. doi: 10.1002/cne.901910402. [DOI] [PubMed] [Google Scholar]
- VOGT BA, PANDYA DN. Cingulate cortex of the rhesus monkey: II. Cortical afferents. J Comp Neurol. 1987;262:271–89. doi: 10.1002/cne.902620208. [DOI] [PubMed] [Google Scholar]
- YUKIE M. Connections between the amygdala and auditory cortical areas in the macaque monkey. Neurosci Res. 2002;42:219–29. doi: 10.1016/s0168-0102(01)00325-x. [DOI] [PubMed] [Google Scholar]