Abstract
The genome sequence for Schistosoma mansoni has been determined, allowing the complete protein complement to be predicted. However, few functional genomics techniques have been developed for use in S. mansoni, limiting the usefulness of the sequence data. Here we describe a whole mount in situ hybridization (WISH) method that can be used to identify the tissue-specific expression of transcripts in S. mansoni. Using this protocol we determine the tissue-specific expression of tetraspanin 2, a female-enriched tetraspanin, phenol oxidase, the secretory Cu/Zn superoxide dismutase, and an Argonaute family member. The localization of these transcripts by WISH correlates with prior studies performed using immunohistochemistry and/or in situ hybridization on tissue sections. WISH can be adapted to screen multiple transcripts, thus identifying novel targets for drugs or vaccines.
Keywords: functional genomic tools, Schistosoma, phenol oxidase, tetraspanin, argonaute, superoxide dismutase
Schistosomiasis, or bilharzia, affects more than 200 million people, making it a major cause of morbidity and mortality worldwide. Although the genome sequence for Schistosoma mansoni has been determined [1], our understanding of schistosomes at the molecular level is still quite rudimentary and the function of most predicted genes/proteins is unknown. Bioinformatics can predict the function of some proteins based solely on their sequence, but many schistosome genes have no homologs outside of the genus [1], posing problems for understanding the function of many schistosome-specific proteins. Since many of the techniques used in other model systems to characterize gene function have not yet been adapted for use in S. mansoni, new methodologies need to be developed. Because schistosomes have multiple tissues and organs systems involved in processes such as digestion, neural function, and reproduction, understanding the tissue and/or cellular expression pattern of a gene may provide useful information about the function of its encoded protein. In situ hybridization (ISH) experiments on histological sections are used routinely for adult schistosomes, but they are labor intensive, requiring the analysis of many sections to determine the localization of a transcript [2]. Immunohistochemistry is another useful methodology, but requires a great deal of time and resources (e.g., recombinant protein production, purification and antibody production), thus rendering it less than optimal as a screening tool to analyze multiple genes. Whole mount in situ hybridization (WISH) experiments are routinely performed to characterize gene expression patterns in numerous animal models, from hydra to higher vertebrates [3, 4]. Because of the complex tissue architecture of schistosomes, WISH would be a valuable method to determine the expression patterns of schistosome-specific gene products. Although a WISH protocol has been described for schistosomes [5], we have developed an alternative method that provides high-resolution transcript localization while maintaining the structural integrity of worms during the fixation and incubation steps. Here we present this streamlined WISH protocol that will be useful as a moderate-throughput screening tool to localize transcripts. To demonstrate the functionality of this WISH protocol we have examined the tissue expression of several transcripts: phenol oxidase (po), tetraspanin 2 (tsp2), a female-specific tetraspanin (fs-tsp), an Argonaute (Ago) family member, ago2, and a secretory form of Cu/Zn superoxide dismutase (sp-sod).
A Puerto Rican strain of S. mansoni was maintained in Biomphalaria glabrata snails and NIH Swiss mice according to previously described methods [6]. Mice were infected through tail exposure to cercariae and adult parasites were harvested through portal vein perfusion at 49 days post infection. Maintenance and experiments using vertebrate animals were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) at Rush University Medical Center (IACUC number 08-058; DHHS animal welfare assurance number A3120-01).
Transcripts were amplified from a mixed adult male and female cDNA library using primers shown in Table 1. PCR products were cloned into the pCRII vector using the TOPO TA Cloning Kit (Invitrogen) or into the pJC53.2 vector [7] through standard cloning methods. Using these plasmids we generated digoxigenin-labeled riboprobes using the Riboprobe System (Promega) with SP6, T7, or T3 RNA polymerases and digoxigenin-11-UTP (Roche).
Table 1.
Primers used to generate probes for this study. Primers used to generate probes for phenol oxidase (po), tetraspanin 2 (tsp2), female specific tetraspanin (fs-tsp), argonaute 2 (ago2) and secretory Cu/Zn superoxide dismutase (sp-sod) and the size of the probe generated are shown. The GenBank™ accession number (Acc #) for each gene is given.
| Transcript | Size (bp) | Acc # | Forward primer (5′ -> 3′) | Reverse primer (5′ -> 3′) |
|---|---|---|---|---|
| po | 464 | XP_002576328 | GTTTCAGCATGTGATGAATG | CAATATGTTAAACCAGTCCAATC |
| tsp2 | 390 | XP_002581393 | CTCTTCGTTGTGGGTATAAG | CATGTTCGTCATTACGGTAC |
| fs-tsp | 479 | XP_002570007 | CATAGTACTCGGATGTTTCC | CTCGGATAACACAGACAGC |
| ago2 | 1195 | XP_002581078 | CCAGTGAAAGTCGTTGCAGA | ACTTGCGGACTTGCTGAGTT |
| sp-sod | 325 | XP_002580485 | ATGACAGTATATTCCTATTTAG | GTGTCTGGGGTATCCATG |
The method for S. mansoni WISH described here has been adapted from a method developed for Schmidtea mediterranea [8]. Adult parasite pairs were separated by incubating worm pairs on ice and then fixing them in 4% paraformaldehyde (PFA) (Ted Pella) diluted in PBS with 0.3% Triton-X 100 (PBSTx) for 15 min. To enhance probe penetration adult males were reduced in 50 mM DTT, 1% NP-40, and 0.1% SDS for 10 min at 37°C and then both males and females were dehydrated in a methanol series (100% PBSTx, 50% methanol, and 100% methanol) for 5 minutes each and stored at -20°C for up to one month. Prior to use, worms were bleached in 6% hydrogen peroxide diluted from a 30% stock in methanol under light for up to 20 hr; this procedure was found to remove color from the gut and prevent tanning (browning) of the female vitellaria upon exposure to heat. Following bleaching, worms were rinsed in methanol, rehydrated through a methanol series to PBSTx and then incubated for 10 minutes in PBSTx with 0.1% SDS. Worms were permeabilized with proteinase K (Invitrogen) at 1 μg/mL diluted in PBS. The permeabilization time depends on the sex of the worms; male worms were permeabilized for 15 to 20 min and female worms were permeabilized for 10 min. After permeabilization, worms were post-fixed in 4% PFA for 10 min and rinsed with PBSTx. Prehybridization began with incubation for 10 min in prehybridization solution (50% deionized formamide, 5× SSC (diluted from 20× SSC from Sigma), 1 mg/mL torula yeast RNA, 1% Tween 20) mixed with PBSTx at a 1:1 ratio, followed by transfer to prehybridization solution and incubation at 56°C for 2 hr in a hybridization oven (Thermo Scientific Hybaid Shake and Stack). Prehybridization solution was then removed and the worms were incubated in hybridization buffer (prehybridization solution with 10% dextran sulfate and 2 ng/mL of the riboprobe) for 18 to 20 hr at 56°C. The following day, the riboprobe mix was removed and worms were transferred to incubation baskets. These containers were made by cutting the bottom off of microfuge tubes and melting a nylon mesh (100 μm, Millipore) to the bottoms to create a basket that can be moved through solutions in 24-well plates used throughout the remainder of the protocol. Stringency washes were performed with solutions preheated to 56°C in the following order: Prehybridization solution mixed with 2 × SSC at a 1:1 ratio, 2 × SSC with 0.1% Triton-X 100, 0.2 × SSC with 0.1% Triton-X 100 in the hybridization oven with agitation. All washes were carried out twice for 30 min each at 56°C. After stringency washes specimens were cooled to room temperature and washed twice with maleic acid buffer (100 mM maleic acid, 150 mM NaCl, 0.1% Tween-20, pH 7.5, MABT). Specimens were then incubated in blocking solution (10% horse serum in MABT) for 2 hr followed by an overnight incubation at 4°C with an anti-digoxigenin-alkaline phosphatase-conjugated antibody (Roche) diluted 1:2000 in blocking solution. The following day specimens were washed for 2 hr in MABT, changing buffer every 20 min. After washing, specimens were incubated in AP buffer (100 mM Tris, pH 9.5; 100 mM NaCl; 50 mM MgCl2; 0.1% Tween-20 brought up to volume with 10% polyvinylalcohol solution). Hybridization signals were detected by adding a nitro blue tetrazolium (NBT)/5-bromo-4-chloro-3-indolyl phosphate (BCIP) solution (AP buffer with 450 μg/mL NBT (Roche) and 175 μg/mL BCIP (Roche)). Samples were developed for 20 min to 24 hr depending on the probe. Development was halted by replacing the development buffer with PBSTx and washing thoroughly. Specimens were then post-fixed in 4% PFA for 10 min, washed with PBSTx, and incubated in 100% ethanol for 20 min. Specimens were rinsed in 50% ethanol and then PBSTx before being cleared in 80% glycerol. Specimens were mounted in 80% glycerol under glass coverslips and imaged. Images were obtained from at least two independent WISH experiments using at least 20 worms of each sex in each experiment.
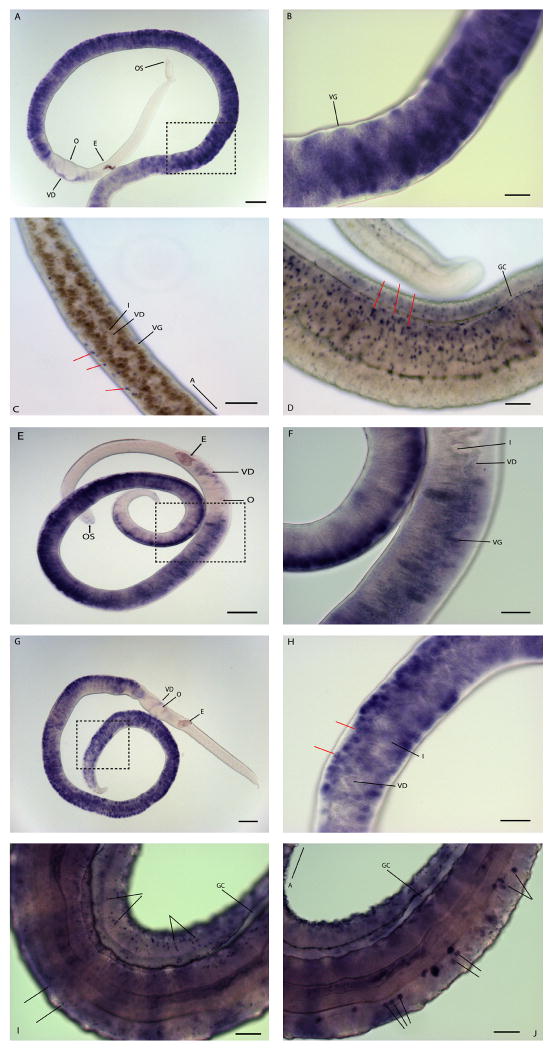
Phenol oxidase (PO), or tyrosinase, is known to function in the cross-linking of eggshell precursor proteins and the protein has been localized to the vitellaria of S. mansoni [9-11]. Using WISH we find that po mRNA is expressed in the vitellaria of adult female schistosomes (Fig 1A-B). Specifically, mature vitelline cells (proximal to the vitelline duct) are labeled more intensely than the immature vitelline cells located distally to the vitelline duct. In addition, we find that po probes also label cells within the entire vitelline duct; however, po transcripts are not detected in the ovary or other female reproductive tissues such as the oviduct, ootype, and uterus.
Figure 1.
Localization of po, tsp-2, fs-tsp, and sp-sod transcripts by whole mount in situ hybridization. (A–B) po localized by WISH (3 hr development). (A and B) Female worms showing staining in the vitellaria but not in the other reproductive organs; (B) is a higher magnification view of the boxed image in (A). (C-D) tsp2 localized by WISH, (4 hr development). tsp2 localizes to sub-tegumental cells in females (C) and males (D). Red arrows indicate punctate blue staining representing the localization of tsp2 transcripts. (E-F) Localization of fs-tsp, (3 hr development). fs-tsp localizes to the vitelline duct and vitellaria but does not localize to subtegumental cells; (F) is a higher magnification image of the boxed area in (E). (G-I) localization of sp-sod, (3 hr and 22 hr development time for panels G-H and I-J, respectively). A female enriched sp-sod localizes to the immature vitellaria of females (indicated by red arrows) and the vitelline duct downstream of the vitellaria (G and H) but is not detected in the oviduct, ovary, or uterus in the female worm; (H) is a higher magnification view of the image shown in (G). (I) Localization of sp-sod in the intestine and sub-tegumental cells of a male worm. (J) Localization of sp-sod in a subset of cells in the parenchyma that appear to be associated with the intestinal epithelium. (A-B) and (E-I) were visualized using a Zeiss Axio Star plus microscope, (C-D) were visualized using a Zeiss Axio Observer Z1 microscope. Abbreviations: A, anterior end of worm; E, egg; GC, gynecophoral canal; I, intestine; OS, oral sucker; O, ovary; T, testes; VG, vitelline gland; VD, vitelline duct. Scale Bars: (B-D, F, H, I, and J) 50 μm; (A, E, and G) 150 μm.
Tetraspanin 2 (Tsp2) is a promising vaccine target against S. mansoni [12]. Immunofluorescence studies show that the Tsp2 protein is localized to the tegument of adult schistosomes where it is thought to function in immune evasion [12]. In agreement with the antibody localization studies, we find that tsp-2 mRNA localizes to sub-tegumental cells of both adult male and female schistosomes (Fig 1C-D). Internal focal planes (Fig. 1C) and more superficial focal planes (Fig. 1D) reveal tsp2 signal at the periphery of the worm (Fig. 1C), consistent with expression below the tegument. Because there are 28 tetraspanin genes in the schistosome genome [1], which should be expressed in many different tissues, the localization of tsp-2 transcripts to the sub-tegumental cells demonstrates the specificity of this WISH protocol. Although the expression patterns of all 28 genes have not been reported, we have localized another tetraspanin transcript (fs-tsp), which has been shown by SAGE to be enriched in female worms [13]. fs-tsp, like po, specifically labels the vitellaria and cells in the vitelline duct. fs-tsp localizes to the vitelline duct both near the vitellaria and the area of the duct near the ovary. Like po, fs-tsp mRNAs are not detected in the ovary or uterus; unlike tsp-2, fs-tsp mRNAs are not detected in the parenchyma or sub-tegumental cells in the male (not shown) or female (Fig 1E-F). The female-specific tetraspanin protein has four membrane spanning segments typical of tetraspanins (not shown) and is therefore expected to be localized to the cell surface. Although the exact function of this tetraspanin has not been defined, its expression on the surface of vitellocytes suggests it plays a role in vitellocyte-vitellocyte or vitellocyte-oocyte interactions during egg formation.
SODs are important redox proteins, defending cells against reactive oxygen species by converting the superoxide radical to molecular oxygen and hydrogen peroxide [14]. Using WISH we find that sp-sod localizes to the vitellaria of females, the sub-tegumental cells, and a subset of cells in the parenchyma that appear to be associated with the intestine of male worms (Fig 1G-J). Although sp-sod labels the vitellaria of females it does not have the same pattern as either fs-tsp or po. sp-sod labels the immature vitelline cells strongly and only weakly labels mature vitelline cells in the vitelline lobes or in the vitelline duct (Fig 1G-H). Immature vitelline cells, found at the periphery of the gland, have a large nuclear-to-cytoplasmic ratio [15], giving them a doughnut-like appearance. In addition, the transcript is not found in the ovo-vitelline duct, the ovary, or other female reproductive tissues. Transcriptome analysis determined that sp-sod was expressed almost exclusively (>20 fold enriched) and at high levels in reproductively active adult female worms (0.7 % of SAGE tags sequenced) compared to adult paired males [13]. In addition this transcript was found at very low levels in females or males from single sex infections or liver stage worms. A similar, female-enriched expression of another antioxidant protein, glutathione peroxidase, has been reported [16], suggesting a redox system enriched in female reproductive tissues. Other studies found that sp-SOD was much less abundant than the cytosolic form of SOD and localized to the sub-tegument of adult male worms [17]; however, no localization in female worms was reported. sp-sod specific mRNA was detected in the female within 3 hr of incubation in detection solution but sp-sod mRNA in the male was not detected until 22 hr of incubation, reflecting the difference in sp-sod transcript abundance between males and females. The detection of low levels of sp-sod mRNA in male worms illustrates the utility of WISH in detecting both abundant and rare transcripts. We generated the sp-sod probe from a region that is poorly conserved between the three SOD genes and includes the secretory leader sequence of sp-sod; therefore, there should be no cross-hybridization with the cytosolic Cu/Zn SOD or the mitochondrial Mn SOD.
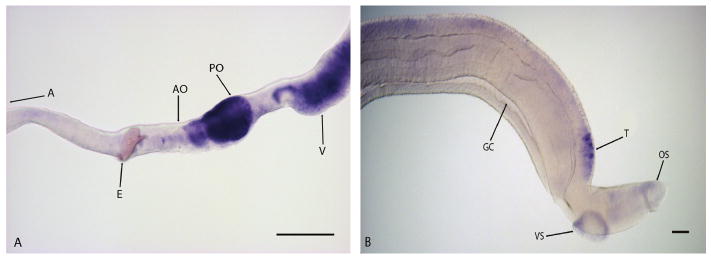
Argonaute (Ago) proteins participate in post-transcriptional regulatory processes through gene silencing mediated by small RNAs. Mature small RNAs generated by Dicer bind Ago proteins in the RNA-induced silencing complex (RISC) and guide them to their gene targets where they function in gene silencing via direct mRNA degradation, mRNA deadenylation, or blocking translation [18,19]. Ago proteins are critical for germline cell maintenance in fruit flies and mice [20] and knockdown of ago2 by RNAi leads to the loss of neoblasts and tissue regeneration in the planarian Dugesia japonica [21]. S. mansoni contains 4 Ago proteins expressed in all life cycle stages but enriched in adult worms, cercaria, and eggs [22]. These proteins contain the conserved PAZ and PIWI domains present in other Ago proteins. In WISH experiments S. mansoni ago2 transcripts are found in the ovary, especially the posterior ovary segments containing mature oocytes (Fig 2A). ago2 transcripts are also found in vitelline glands and testes of adult worms (Fig 2). These data suggest that ago2 may play a role in the regulation of germ cell maintenance in the gonads and vitelline cells of S. mansoni. The ago2 probe used in this study is designed to the PAZ domain and part of the PIWI domain but does not have a high degree of nucleotide identity with S. mansoni ago1 or ago3 (not shown).
Figure 2.
argonaute localization in adult female (A) and male (B) Schistosoma mansoni worms. ago2 was developed for 30 min in males and 3 hours in females. argonaute is localized to the gonads (testes, ovary) and vitellaria of S. mansoni, with the highest expression in the posterior ovary (PO). Abbreviations: A, anterior end of worm; AO, anterior ovary; E, egg; GC, gynecophoral canal; OS, oral sucker; PO, posterior ovary; T, testes; V, vitellaria; VS, ventral sucker. Images obtained with a Zeiss Axio Star plus microscope. Scale Bars: 100 μm.
This study shows that WISH is a powerful tool for characterizing the expression of S. mansoni transcripts. WISH can be used to screen transcripts with little prior information. As there are still many predicted genes in the S. mansoni genome that remain uncharacterized or have no functional data, analysis of their cell-type and tissue-specific expression patterns will be useful to inform potential roles for these genes in the biology of the parasite, leading to more productive drug and vaccine candidate screening programs.
Acknowledgments
We would like to thank Dr. Fred Lewis at the Biomedical Research Institute (Rockville Maryland, United States) for parasite material through NIH-NIAID contract N01-AI-55270; Dr. Sasha Shafikhani, RUMC, for the use of his Zeiss Axio Observer Z1 microscope; Dr. Philip LoVerde for the adult S. mansoni cDNA library; and Dr. Ryan King, the University of Illinois Urbana-Champaign, for advice and encouragement.
Funding: This work was supported in part by the National Institute of Allergy and Infectious Diseases grants AI065622 and AI081107 (DLW), by The Eunice Kennedy Schriver National Institute of Child Health and Human Development grant F32 HD062124 (JJC), and by a 2010 Travel Award from the Burroughs Wellcome Fund to AAC. PAN is an Investigator of the Howard Hughes Medical Institute (www.hhmi.org).
Footnotes
Author contributions: AAC, DLW, JC, and PN conceived and designed the experiments and analyzed the data. AAC wrote the paper.
References
- 1.Berriman M, Haas BJ, LoVerde PT, Wilson RA, Dillon GP, Cerqueira GC, et al. The genome of the blood fluke Schistosoma mansoni. Nature. 2009;460:352–8. doi: 10.1038/nature08160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beckmann S, Buro C, Dissous C, Hirzmann J, Grevelding C. The syk kinase SmTK4 of Schistosoma mansoni is involved in the regulation of spermatogenesis and oogenesis. PLoS Pathog. 2010;6:e1000769. doi: 10.1371/journal.ppat.1000769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grens A, Gee L, Fisher DA, Bode HR. CnNK-2 homeobox gene, has a role in patterning the basal end of the axis in hydra. Dev Biol. 1996;180:473–88. doi: 10.1006/dbio.1996.0321. [DOI] [PubMed] [Google Scholar]
- 4.Rosen B, Beddington RS. Whole-mount in situ hybridization in the mouse embryo: Gene expression in three dimensions. Trends Genet. 1993;9:162–7. doi: 10.1016/0168-9525(93)90162-b. [DOI] [PubMed] [Google Scholar]
- 5.Dillon GP, Illes JC, Isaacs HV, Wilson RA. Patterns of gene expression in schistosomes: Localization by whole mount in situ hybridization. Parasitology. 2007;134:1589–97. doi: 10.1017/S0031182007002995. [DOI] [PubMed] [Google Scholar]
- 6.Lewis F. Schistosomiasis. In: Coligan JE, Kruisbeek AM, Margulies DH, Shevach EM, Strober W, editors. Current Protocols in Immunology. John Wiley & Sons; New York: 1998. pp. 19.1.1–19.1.28. [Google Scholar]
- 7.Collins JJ, 3rd, Hou X, Romanova EV, Lambrus BG, Miller CM, Saberi A, Sweedler JV, Newmark PA. Genome-wide analyses reveal a role for peptide hormones in planarian germline development. PLoS Biol. 2010;8:e1000509. doi: 10.1371/journal.pbio.1000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pearson BJ, Eisenhoffer GT, Gurley KA, Rink JC, Miller DE, Sanchez Alvarado A. Formaldehyde-based whole-mount in situ hybridization method for planarians. Dev Dyn. 2009;238:443–50. doi: 10.1002/dvdy.21849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seed JL, Bennett JL. Schistosoma mansoni: Phenol oxidase's role in eggshell formation. Exp Parasitol. 1980;49:430–1. doi: 10.1016/0014-4894(80)90077-6. [DOI] [PubMed] [Google Scholar]
- 10.Eshete F, LoVerde PT. Characteristics of phenol oxidase of Schistosoma mansoni and its functional implications in eggshell synthesis. J Parasitol. 1993;79:309–17. [PubMed] [Google Scholar]
- 11.Fitzpatrick JM, Hirai Y, Hirai H, Hoffmann KF. Schistosome egg production is dependent upon the activities of two developmentally regulated tyrosinases. FASEB J. 2007;21:823–35. doi: 10.1096/fj.06-7314com. [DOI] [PubMed] [Google Scholar]
- 12.Tran MH, Pearson MS, Bethony JM, Smyth DJ, Jones MK, Duke M, et al. Tetraspanins on the surface of Schistosoma mansoni are protective antigens against schistosomiasis. Nat Med. 2006;12:835–40. doi: 10.1038/nm1430. [DOI] [PubMed] [Google Scholar]
- 13.Williams DL, Sayed AA, Bernier J, Birkeland SR, Cipriano MJ, Papa AR, et al. Profiling Schistosoma mansoni development using serial analysis of gene expression (SAGE) Exp Parasitol. 2007;117:246–58. doi: 10.1016/j.exppara.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fridovich I. Superoxide dismutases. Annu Rev Biochem. 1975;44:147–59. doi: 10.1146/annurev.bi.44.070175.001051. [DOI] [PubMed] [Google Scholar]
- 15.Erasmus DA. Schistosoma mansoni: development of the vitelline cell, its role in drug sequestration, and changes induced by Astiban. Exp Parasitol. 1975;38:240–56. doi: 10.1016/0014-4894(75)90027-2. [DOI] [PubMed] [Google Scholar]
- 16.Roche C, Liu JL, LePresle T, Capron A, Pierce RJ. Tissue localization and stage-specific expression of the phospholipid hydroperoxide glutathione peroxidase of Schistosoma mansoni. Mol Biochem Parasitol. 1996;75:187–95. doi: 10.1016/0166-6851(95)02523-5. [DOI] [PubMed] [Google Scholar]
- 17.Hong Z, LoVerde PT, Thakur A, Hammarskjold ML, Rekosh D. Schistosoma mansoni: A Cu/Zn superoxide dismutase is glycosylated when expressed in mammalian cells and localizes to a subtegumental region in adult schistosomes. Exp Parasitol. 1993;7:101–14. doi: 10.1006/expr.1993.1012. [DOI] [PubMed] [Google Scholar]
- 18.Höck J, Meister G. The Argonaute protein family. Genome Biol. 2008;9:210. doi: 10.1186/gb-2008-9-2-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet. 2011;12:19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siomi MC, Kuramochi-Miyagawa S. RNA silencing in germlines: exquisite collaboration of Argonaute proteins with small RNAs for germline survival. Curr Opin Cell Bio. 2009;21:426–34. doi: 10.1016/j.ceb.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 21.Rouhana L, Shibata N, Nishimura O, Agata K. Different requirements for conserved post-transcriptional regulators in planarian regeneration and stem cell maintenance. Dev Biol. 2010;341:429–43. doi: 10.1016/j.ydbio.2010.02.037. [DOI] [PubMed] [Google Scholar]
- 22.Gomes MS, Cabral FJ, Jannotti-Passos LK, Carvallho O, Rodrigues V, Baba EH, Renata GS. Preliminary analysis of miRNA pathway in Schistosoma mansoni. Parasit Internat. 2008;58:61–8. doi: 10.1016/j.parint.2008.10.002. [DOI] [PubMed] [Google Scholar]