Abstract
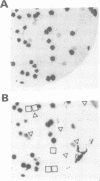
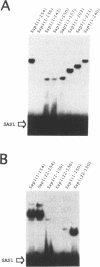
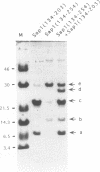
Mating type switching in fission yeast, Schizosaccharomyces pombe, is initiated by a site-specific double-strand break (DSB) at the mat1 locus. The DSB is controlled from a distance by cis- and trans-acting elements. The switch-activating protein, Sap1 binds to the SAS1 cis-acting element which controls the frequency of the DSB at the mat1 locus and, consequently the efficiency of mating type switching. We developed a general method for screening randomly mutagenized expression libraries of DNA-binding protein in E.coli. Sap1 gene was mutagenized by PCR under conditions of reduced Taq polymerase fidelity. The mutated DNA was expressed in E.coli and screened for SAS1-recognition. This method was used to isolated 16 point mutations that abolished SAS1 interaction together with 18 mutations that did not affect binding. The position of these point mutations allowed the identification of three protein domains located in the N-terminal part of Sap1 that are essential for DNA-binding. Deletions and biochemical analysis showed that Sap1 is a dimer both in solution and when bound to SAS1 sequence. The dimerization domain was localized C-terminally to the three domains described above and when used in exess it inhibited DNA binding.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arcangioli B., Copeland T. D., Klar A. J. Sap1, a protein that binds to sequences required for mating-type switching, is essential for viability in Schizosaccharomyces pombe. Mol Cell Biol. 1994 Mar;14(3):2058–2065. doi: 10.1128/mcb.14.3.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcangioli B., Klar A. J. A novel switch-activating site (SAS1) and its cognate binding factor (SAP1) required for efficient mat1 switching in Schizosaccharomyces pombe. EMBO J. 1991 Oct;10(10):3025–3032. doi: 10.1002/j.1460-2075.1991.tb07853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcangioli B., Lescure B. Identification of proteins involved in the regulation of yeast iso- 1-cytochrome C expression by oxygen. EMBO J. 1985 Oct;4(10):2627–2633. doi: 10.1002/j.1460-2075.1985.tb03980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach D. H., Klar A. J. Rearrangements of the transposable mating-type cassettes of fission yeast. EMBO J. 1984 Mar;3(3):603–610. doi: 10.1002/j.1460-2075.1984.tb01855.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr C. M., Kim P. S. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993 May 21;73(4):823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Cohen C., Parry D. A. Alpha-helical coiled coils and bundles: how to design an alpha-helical protein. Proteins. 1990;7(1):1–15. doi: 10.1002/prot.340070102. [DOI] [PubMed] [Google Scholar]
- Egel R., Beach D. H., Klar A. J. Genes required for initiation and resolution steps of mating-type switching in fission yeast. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3481–3485. doi: 10.1073/pnas.81.11.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S. C. A structural taxonomy of DNA-binding domains. Nature. 1991 Oct 24;353(6346):715–719. doi: 10.1038/353715a0. [DOI] [PubMed] [Google Scholar]
- Hoffmann A., Roeder R. G. Purification of his-tagged proteins in non-denaturing conditions suggests a convenient method for protein interaction studies. Nucleic Acids Res. 1991 Nov 25;19(22):6337–6338. doi: 10.1093/nar/19.22.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. GCN4, a eukaryotic transcriptional activator protein, binds as a dimer to target DNA. EMBO J. 1987 Sep;6(9):2781–2784. doi: 10.1002/j.1460-2075.1987.tb02573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly M., Burke J., Smith M., Klar A., Beach D. Four mating-type genes control sexual differentiation in the fission yeast. EMBO J. 1988 May;7(5):1537–1547. doi: 10.1002/j.1460-2075.1988.tb02973.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar A. J., Bonaduce M. J., Cafferkey R. The mechanism of fission yeast mating type interconversion: seal/replicate/cleave model of replication across the double-stranded break site at mat1. Genetics. 1991 Mar;127(3):489–496. doi: 10.1093/genetics/127.3.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar A. J. Differentiated parental DNA strands confer developmental asymmetry on daughter cells in fission yeast. Nature. 1987 Apr 2;326(6112):466–470. doi: 10.1038/326466a0. [DOI] [PubMed] [Google Scholar]
- Klar A. J., Miglio L. M. Initiation of meiotic recombination by double-strand DNA breaks in S. pombe. Cell. 1986 Aug 29;46(5):725–731. doi: 10.1016/0092-8674(86)90348-x. [DOI] [PubMed] [Google Scholar]
- Klar A. J. The developmental fate of fission yeast cells is determined by the pattern of inheritance of parental and grandparental DNA strands. EMBO J. 1990 May;9(5):1407–1415. doi: 10.1002/j.1460-2075.1990.tb08256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988 Jun 24;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Nielsen O., Egel R. Mapping the double-strand breaks at the mating-type locus in fission yeast by genomic sequencing. EMBO J. 1989 Jan;8(1):269–276. doi: 10.1002/j.1460-2075.1989.tb03373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perisic O., Xiao H., Lis J. T. Stable binding of Drosophila heat shock factor to head-to-head and tail-to-tail repeats of a conserved 5 bp recognition unit. Cell. 1989 Dec 1;59(5):797–806. doi: 10.1016/0092-8674(89)90603-x. [DOI] [PubMed] [Google Scholar]
- Presta L. G., Rose G. D. Helix signals in proteins. Science. 1988 Jun 17;240(4859):1632–1641. doi: 10.1126/science.2837824. [DOI] [PubMed] [Google Scholar]
- Rosenberg A. H., Lade B. N., Chui D. S., Lin S. W., Dunn J. J., Studier F. W. Vectors for selective expression of cloned DNAs by T7 RNA polymerase. Gene. 1987;56(1):125–135. doi: 10.1016/0378-1119(87)90165-x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. B., Johnson K. S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988 Jul 15;67(1):31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Sorger P. K., Nelson H. C. Trimerization of a yeast transcriptional activator via a coiled-coil motif. Cell. 1989 Dec 1;59(5):807–813. doi: 10.1016/0092-8674(89)90604-1. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Yang-Yen H. F., Chambard J. C., Sun Y. L., Smeal T., Schmidt T. J., Drouin J., Karin M. Transcriptional interference between c-Jun and the glucocorticoid receptor: mutual inhibition of DNA binding due to direct protein-protein interaction. Cell. 1990 Sep 21;62(6):1205–1215. doi: 10.1016/0092-8674(90)90396-v. [DOI] [PubMed] [Google Scholar]