Abstract
Quantum dots (QDs) were prepared in genetically engineered Escherichia coli (E. coli) through the introduction of foreign genes encoding a CdS binding peptide. The CdS QDs were successfully separated from the bacteria through two methods, lysis and freezing–thawing of cells, and purified with an anion-exchange resin. High-resolution transmission electron microscopy, X-ray diffraction, luminescence spectroscopy, and energy dispersive X-ray spectroscopy were applied to characterize the as-prepared CdS QDs. The effects of reactant concentrations, bacteria incubation times, and reaction times on QD growth were systematically investigated. Our work demonstrates that genetically engineered bacteria can be used to synthesize QDs. The biologically synthesized QDs are expected to be more biocompatible probes in bio-labeling and imaging.
Keywords: Biosynthesis, Genetic engineering, Escherichia coli, CdS
1. Introduction
As a kind of promising bioprobe, QDs have been widely used in the field of clinical medicine, biology and pharmacology, due to their unique optical properties (Bruchez et al., 1998; Chan and Nie, 1998; Lee et al., 2008; Yang et al., 2009). Although QDs have been generally acknowledged as a good biological marker, its toxicity and non-specific problems still exist and cannot be ignored (Clift et al., 2008; Goldman et al., 2004; Liu et al., 2008). Microbial synthesis carried out in living biological systems can effectively resolve this problem under the inherent capabilities of organisms. QDs have attracted broad attention in the field of analytical chemistry and life sciences due to their unique spectral properties as a new type of fluorescent nano-materials (Ho et al., 2006; Yang et al., 2007; Zhang et al., 2007). They have been shown to possess promising applications in life science with a higher fluorescence yield, narrow half-peak width and symmetrical peak-shape, which enable simultaneous multiplex labeling and detection (Deka et al., 2009; Yang et al., 2008; Zhao et al., 2009). In the last few years, there has been considerable progress in the application of QDs (Shan et al., 2008; Zaman et al., 2009). However, the existence of toxicity and non-specific problems can no longer be ignored. Moreover, in the further study of nano-materials, the potential negative biological effects of these ultra-fine particles have received increasing attention (Ma et al., 2006a). One promising alternative is the use of biological templates for the synthesis of nanocrystals. Currently, some biological templates, including carbohydrate (Dong and Qian, 2008), peptides (Peelle et al., 2005a), nucleotides (Ma et al., 2006b), and fusion proteins (Nam et al., 2006) have been used to make materials. Biological templates not only guide the nucleation of inorganic materials, but also control the crystal structure and size, under aqueous and ambient conditions. The bionic approaches to the synthesis of nanocrystals can also be extended to living biological systems (Anshup et al., 2005; Du et al., 2007; Mao et al., 2003; Yoo et al., 2006), which are ideal nano-structured templates for the design and synthesis of nanomaterials because of their endogenous ability of molecular recognition and self-assembly (Huang et al., 2005; Ngweniform et al., 2009).
Recently, the microbial synthesis of inorganic materials including noble metals (Seghal et al., 2010), alloys (Lee et al., 2006; Williams et al., 1996), binary oxides, and ternary oxides (Jha et al., 2009) using fungi (Bai et al., 2009; Kumar et al., 2007), yeast (Cui et al., 2009), and bacteria (Sweeney et al., 2004) has been reported. However, similar to any other synthesis route, the microbial synthesis meets several challenges which are yet to be explored and solved such as better control over size and shape, and scaling up of the synthesis to get large amount of nanomaterials. Chemical synthesis of nanomaterials, on the other hand, has been already developed to be able to provide excellent control over size and shape. Another challenge, in the case of the microbial technique, is to fully understand the synthesis mechanism at the molecular level, which eventually may help in providing better control over size and shapes as well as crystallinity in the future. More recently, Bao et al. (2010) demonstrate a simple and efficient biosynthesis method to prepare easily harvested biocompatible cadmium telluride (CdTe) QDs with tunable fluorescence emission using yeast cells. In particular, the CdTe QDs with uniform size (2–3.6 nm) are protein-capped, which makes them highly soluble in water, and in situ bio-imaging in yeast cells indicates that the biosynthesized QDs have good biocompatibility.
In this work, CdS QDs were prepared with genetically engineered E. coli as the carrier to establish a new method for synthesis of fluorescent QDs. A peptide reported to be able to bind to CdS, termed CDS 7 (N-GDVHHHGRHGAEHADI-C) (Peelle et al., 2005b), was expressed in the bacteria and the engineered bacteria were allowed to interact with CdS precursors to induce the formation of CdS QDs. It is expected that CdS-binding peptide, CDS 7, will cap the CdS QDs synthesized by the bacteria, and a series of parameters including reactant concentration, incubation time of bacteria and reaction time were systematically studied. The experiment results showed that the emission wavelength of as-prepared CdS ODs was of reactant concentration-dependent, varied from 445 nm to 513 nm.
2. Materials and methods
2.1. Materials
Yeast extract, tryptone, sodium chloride, isopropylthio-β-d-galactoside (IPTG), kanamycin, glycerol, DEAE-Sephadex A-50 (DEAE = diethylaminoethyl) were purchased from Shanghai Sangon Biological Engineering Technology & Services CO., Ltd of China. Cadmium chloride, sodium sulfide, trishydroxymethylaminomethanetris (Tris), potassium chloride were obtained from National Medicines Corporation Ltd. of China. All chemicals used in the experiments were of high purity and used without further purification. Triple-distilled water was used throughout the experiments.
2.2. Genetic engineering of bacteria
The DNA sequence for peptide CDS 7 (N-GDVHHHGRHGAEHADI-C) was optimized for expression in E. coli. The Nco I and BamH I restriction enzyme digestion sites were added to the sequence for ligation into the corresponding site in the pET-28b(+) vector, and a GC sequence was added to the 5′ of Nco I to keep the peptide in frame for expression. His6 tag was added at the 5′ end of the peptide. The translation start codon ATG was contained in Nco I restriction site and translation termination code TAA was added at the 3′ end of the peptide. The 5′ of the primers were phosphorylated for efficient ligation. The primer sequences include sense primer: PCATGGGCCATCATCATCATCATCACGGCGATGTGCATCATCATGGCCGCCACGGCGCGGAACATGCGGATATTTAAG, anti-sense primer: P-GATCCTTAA ATATCCGCATGTTCCGCGCCGTGGCGGCCATGATGATGCACATCGCCGTGATGATGATGATGATGGCC. The primers were annealed and cloned into pET-28b(+) vector, transfected in E. coli BL21 (DE3), and the resulting pET-28b(+)CDS 7 expression plasmid was verified by DNA sequencing.
As the molecular weight of CDS 7 peptide expressed in E. coli is too small to be detected on a regular SDS–PAGE gel, we further cloned CDS peptide into the pHUE expression vector for high yield and soluble expression of proteins (Baker et al., 2005; Catanzariti et al., 2004). The CDS 7 peptide sequence was first amplified by PCR from pET-28b(+)CDS 7 and cloned into the pHUE vector. The primers for the PCR reaction were, sense: 5′-TCCCTCCGCGGTGGCGGCGATGTGCATCATCATGGC-3′, anti-sense: 5′-CCCAAGCTTTTAA ATA TCCGCATGTTCCGC-3′. The PCR product was digested with Sac II and Hind III and inserted into the corresponding site in pHUE vector. The resulting expression product was His6-Ubiquitin-CDS 7. The pHUE-His6-Ubiquitin-CDS 7 plasmid was transfected and expressed in E. coli BL21.
The bacterial cells were sub-cultured (1:100) into 200 ml LB-broth and grown to OD600 = 0.6 at 37 °C with shaking. Then culture was induced by 0.5 mM IPTG for 4 h. Cells were isolated by centrifugation and lysed by mild sonication. The supernatant containing the His6-Ubiquitin-CDS 7 was applied to a Ni-Sepharose column. The fusion protein was eluted with an elution buffer (50 mM Na2HPO4/NaH2PO4 at pH 8.0, 300 mM NaCl, 150 mM imidazole). The purified His6-Ubiquitin-CDS 7 fusion protein was digested by adding 1 mM dithiothreitol (DTT) and incubating with deubiquitinase Usp2-cc at 1:100 of enzyme to substrate molar ratio for 2 h at 37 °C, pH 8.0. After incubation, the cleaved product was analyzed with 12% SDS–PAGE and silver staining.
2.3. Synthesis of CdS QDs
The E. coli cells genetically engineered according to Section 2.2 were first incubated in 20 ml of LB medium containing kanamycin. When the OD600 value reached 0.5, the expression of engineered gene was induced with 0.5 mM IPTG, and cadmium chloride was added to a final concentration of 1 mM. After incubation of the samples for 3 h, freshly prepared sodium sulfide solution was added slowly to a final concentration of 1 mM. The samples were incubated at room temperature with end-over-end rotation for 1.5 h. Finally, the samples were washed with water for 3 times and characterized with UV–vis and fluorescence spectrometry.
2.4. Isolation and purification of CdS QDs
The CdS QDs obtained initially were associated with E. coli cells. For the next use, the synthesized CdS QDs need to be separated and further purified. Here, two methods i.e. lysis and freezing–thawing of cells were used for this step. Firstly, the cells harvested in Section 2.3 were washed with water to remove extraneous medium; next, the washed cell paste was re-suspended in distilled water to form a homogenous cell suspension, and treated by one of the following two methods.
2.4.1. Lysis of cells
The homogenous cell suspension was treated with hyperacoustic lysis by a JY 92-II hyperacoustic cell grinder (Ningbo Scientz Biotechnology CO., Ltd) for several times with each time lasting 30 min. When the cell suspension finally became clear, it was centrifuged at 8000 rpm for 5 min, the supernatant was filtered with a 0.2 μm syringe filter, then stored at 4 °C for purification.
2.4.2. Freezing–thawing of cells
The resuspended cells were frozen at −70 °C for two weeks and then thawed at room temperature. Then, the cell suspension was centrifuged and filtered just as before, and the obtained supernatant was stored at 4 °C for next purification.
2.4.3. Purification of CdS QDs
An anion-exchange resin column, packed with DEAE-Sephadex A-50, was used in this step for the purification of CdS nanoparticles. The resin was first dipped in distilled water over night, then loaded and equilibrated with Tris buffer (pH 7.6) containing KCl (125 mM). Suspension containing CdS QDs were concentrated to 2 ml and kept at 4 °C or for immediate use. The concentrated supernatant (1 ml) was loaded onto the desalting gel-filtration column at a flow rate of 2mlmin−1 and eluted with Tris buffer (pH 7.6) containing KCl (150, 200, 300, 400 mM) to fractionate the cells materials and purify the CdS QDs. After purification, the column was washed with water (pH 7.0) at a flow rate of 2 ml min−1 for 60 min. Fractions were collected and used for next determination.
3. Results and discussion
3.1. Expression of QDs binding peptide in E. coli
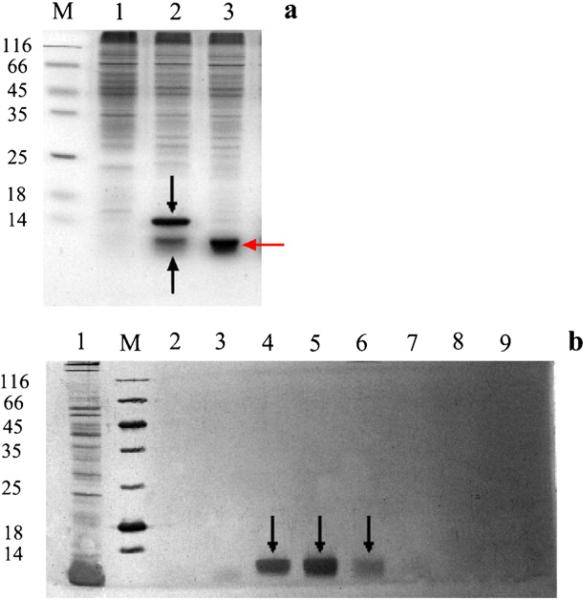
A CdS QDs binding histidine-rich peptide CDS 7 obtained by yeast surface display has been reported to catalyze the aqueous synthesis of fluorescent CdS QDs at room temperature (Peelle et al., 2005b). We were interested in testing the in vivo synthesis ability of CdS QDs by bacteria expressing this peptide. We firstly cloned CDS 7 peptide with a His6 tag into the pET-28b(+) vector and transfected E. coli strain BL21 (DE3). As the small size of the peptide (2.57 kDa) could not be distinguished by the SDS–PAGE and visualized as a clear band, we employed pHUE expression vector for high yield and high-resolution expression of proteins (Baker et al., 2005; Catanzariti et al., 2004). The CDS 7 peptide was expressed as fusion protein with His6-Ubiquitin. The resulting expression product His6-Ubiquitin-CDS 7 (12.5 kDa) was readily detected by 15% SDS gel and purified by Ni-Sepharose column, the results are shown in Fig. 1(a) and (b), respectively. In Fig. 1(a), Lane 1, supernatant of the cell lysate of pHUE-His6-Ubiquitin-CDS 7 plasmid transfected E. coli BL21 (DE3) without IPTG induction. Lane 2, supernatant of the cell lysate of pHUE-His6-Ubiquitin-CDS 7 plasmid transfected E. coli with IPTG induction. The downward arrow indicates the expression product corresponding to the molecular weight of His6-Ubiquitin-CDS 7. The upward arrow indicates a band corresponding to the molecular weight of His6-Ubiquitin. Lane 3, supernatant of the cell lysate of pHUE-His6-Ubiquitin-CDS 7 plasmid transfected E. coli incubated with deubiquitinase Usp2-cc at 1:100 of enzyme to substrate molar ratio for 2 h at 37 °C, pH 8.0. The left-pointing arrow indicates a band corresponding to the molecular weight of His6-Ubiquitin. For Fig. 1(b), the supernatant of the cell lysate of pHUE-His6-Ubiquitin-CDS 7 plasmid transfected E. coli with IPTG induction. Lanes 1 and 2, eluted fractions of 20 mM imidazole. Lanes 3 and 4, eluted fractions of 100 mM imidazole. Lanes 5 and 6, eluted fractions of 200 mM imidazole. Lanes 7 and 8, eluted fractions of 300 mM imidazole. The downward arrow indicates the purified product corresponding to the molecular weight of His6-Ubiquitin-CDS 7. Furthermore, the fusion protein can be cleaved by deubiquitinase at the C-terminus of ubiquitin and release the His6-Ubiquitin tag (Fig. 1(a)).
Fig. 1.
Expression of QDs binding peptide in E. coli. (a) 15% SDS–PAGE of E. coli. expressing QDs binding peptide. (b) Purification of His6-Ubiquitin-CDS 7 by Ni-Sepharose column and 15% SDS–PAGE of the imidazole eluted fractions.
3.2. Characterization of the CdS QDs synthesized by bacteria
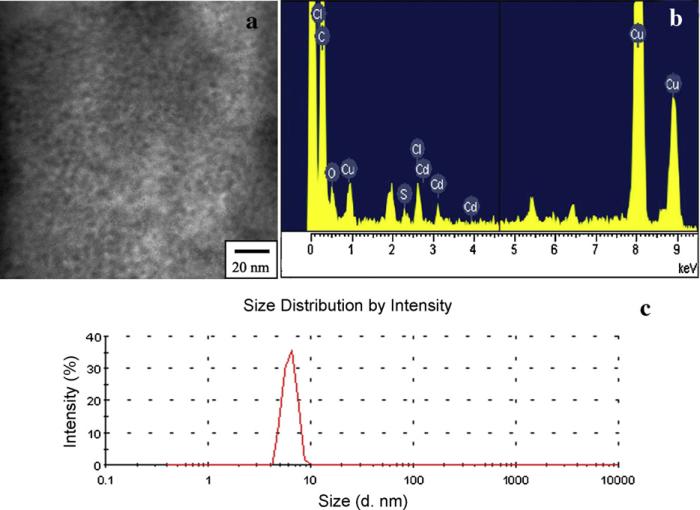
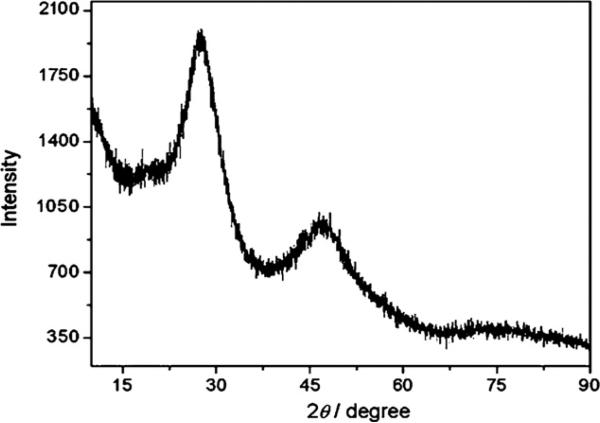
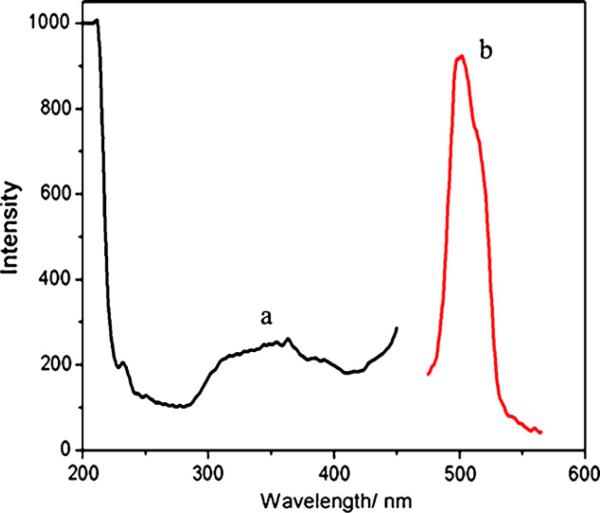
To further confirm the formation of CdS QDs, the morphology, crystalline structure and optical properties of the products were characterized respectively. High-resolution transmission electron microscopy (HRTEM) and JEM-2100HR transmission electron microscope (TEM, JEOL Ltd., Japan), using an accelerating voltage of 200 kV were used to probe the CdS QDs. Fig. 2(a) and (c) shows the TEM image and size distribution of the CdS QDs, respectively. It can be seen that the water-soluble CdS QDs were fairly monodisperse with an average size of 6 nm. EDS spectrum in Fig. 2(b) taken from nanoparticle presented in Fig. 2(a) shows the signals from Cd and S elements present in the nanoparticles. It should be noted that Cu signal is from the carbon-coated copper TEM grid. An analysis of the EDS spectrum indicates a Cd/S molar ratio of nearly 1:1 just as bold values presented in Table 1. This result confirmed that the products were CdS QDs. Powder XRD measurements were also performed at room temperature (pXRD, PW3040/60 X’Pert Pro MDP, Holland Panalytical B.V.) by using Cu Kα (1.5418 Å) radiation. As shown in Fig. 3, the diffraction peaks of the product match well with the main peaks in the standard pattern of a crystalline cubic structure of CdS nanocrystal (JCPDS No. 00-021-0829). Considering their further application as bioprobes in the field of biology, the optical properties of the synthesized CdS QDs were studied. The fluorescence spectra were measured on a LS-55 Luminescence Spectrometer (Perkin-Elmer, USA) with 350 nm excitation. As shown in Fig. 4, the as-prepared CdS QDs still possess excellent optical properties just as QDs synthesized with other chemical synthesis methods, such as broad and continuous excitation spectra, narrow and symmetric emission spectra.
Fig. 2.
TEM image (a), EDS analysis (b) and size distribution (c) of CdS QDs. In (b), the Cu signal is from the TEM grid.
Table 1.
Elementary analysis of CdS QDs.
| Element | Weight percentage | Atomic percent |
|---|---|---|
| C K | 27.00 | 66.19 |
| O K | 1.85 | 3.40 |
| S K | 0.52 | 0.47 |
| Cl K | 1.79 | 1.49 |
| Cu K | 57.08 | 26.45 |
| Cd L | 1.73 | 0.45 |
| Cu L | 10.03 | 1.55 |
| Total | 100.00 |
Fig. 3.
XRD pattern of CdS QDs.
Fig. 4.
Fluorescence excitation (a) and emission (b) spectra of CdS QDs.
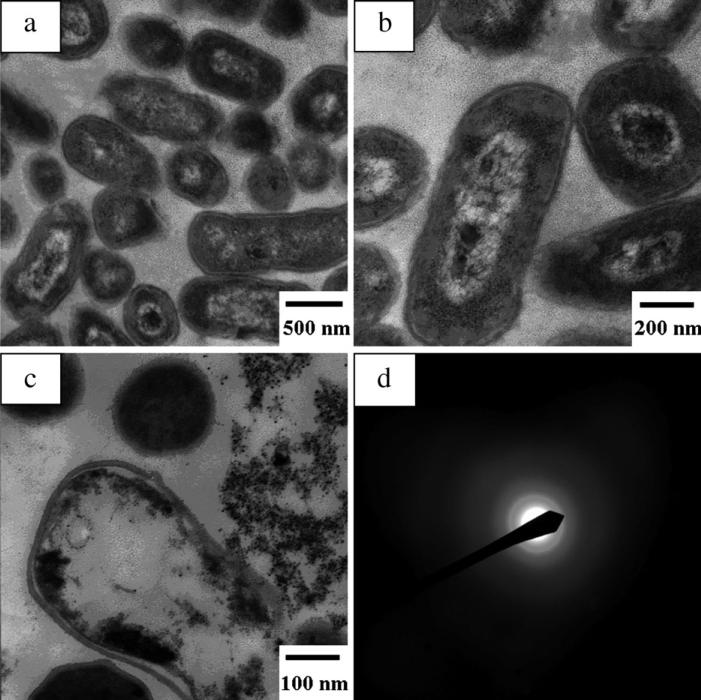
To further verify the synthesis of CdS QDs in E. coli, the cells harvested by centrifugation were first resuspended in resin, hardened at 60 °C overnight, then the hard pellets were cut into 60 nm thin slices and the slices were floated on water and placed on a carbon-coated copper TEM grid for TEM characterization. The results of the microtome treated specimens with different magnifications are presented in Fig. 5(a)–(c), which clearly indicate the presence of QDs within bacterial cells. It can be seen that the CdS QDs are all present over the cytoplasm. In Fig. 5(c), the bacterial cells might be lysed, resulting in some aggregation. However, as shown in the image, many monodisperse QDs appear around the lysed cell, indicating that most of the QDs had been released from the cells. The diffraction rings in electron diffraction (ED) pattern in Fig. 5(d) indicated the synthesized CdS QDs had a cubic crystal structure.
Fig. 5.
TEM images of microtome treated specimens (a–c) and ED pattern of the QDs (d). The diffraction rings in ED pattern indicated that the CdS QDs had a cubic microcrystal structure.
3.3. Optimization of synthesis parameters
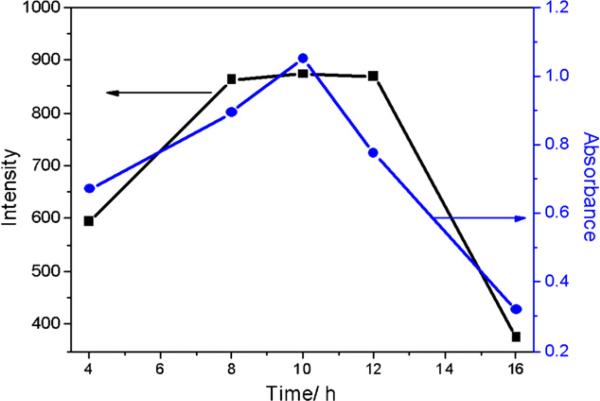
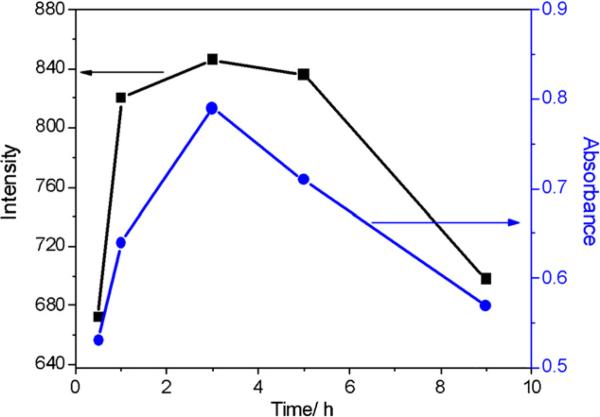
Nanocrystal formation was found to vary dramatically with the parameters, such as incubation time of bacterial cells, reaction time and temperature, and molar ratio of cadmium chloride and sodium sulfide. The E. coli cells gradually increased at the beginning of incubation, and reached saturation after 10 h of growth. At the same time, cells harvested after 10 h of growth are most active, also beneficial for the formation of QDs. Fig. 6 shows the best incubation time for the reaction according to absorbance at 400 nm and emission intensity at 510 nm of CdS QDs. Besides the incubation time, reaction time is also an important factor. After the QDs growth was studied for the reaction time of 0.5–9 h, the experimental results showed that QDs synthesized at 3 h exhibit the best optical properties (Fig. 7). The data were also measured under the same condition as in Fig. 6.
Fig. 6.
Effect of incubation time on the fluorescence intensity (black curve) and absorbance (blue curve) of CdS QDs. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
Fig. 7.
Effect of reaction time on the fluorescence intensity (black curve) and absorbance (blue curve) of CdS QDs. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
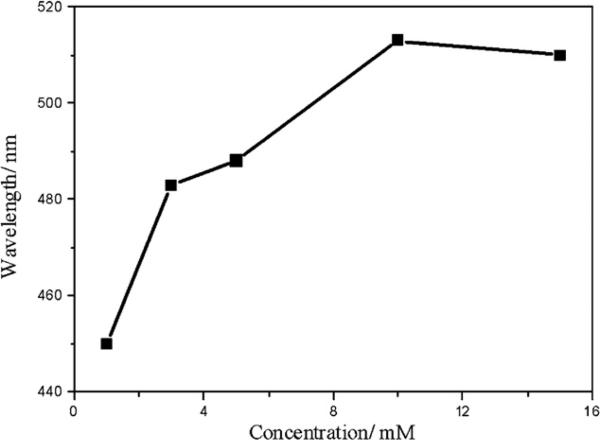
In chemical synthesis process of QDs, concentration and molar ratio of reactants were also reported as important parameters for control of products. It is generally accepted that the concentration of reactants can directly affect the products in chemical synthesis. Thus we studied the relationship between the concentration of reactants (Cd2+ and S2−) and fluorescence emission spectra of CdS QDs. From Fig. 8 it can be seen that the fluorescence emission exhibits an obvious red-shift with the increase of the concentration of reactant within the range of 0.5–10 mM. It is possible that the increase of reactant concentrations weakened the control over CdS nanocrystals nucleation by foreign proteins in the bacterial cells, which resulted in the increase of average particle size and the consequent red shift in emission wavelength.
Fig. 8.
Effect of reactant concentration on the fluorescence wavelength of CdS QDs.
3.4. Isolation and purification of the CdS QDs from the cells
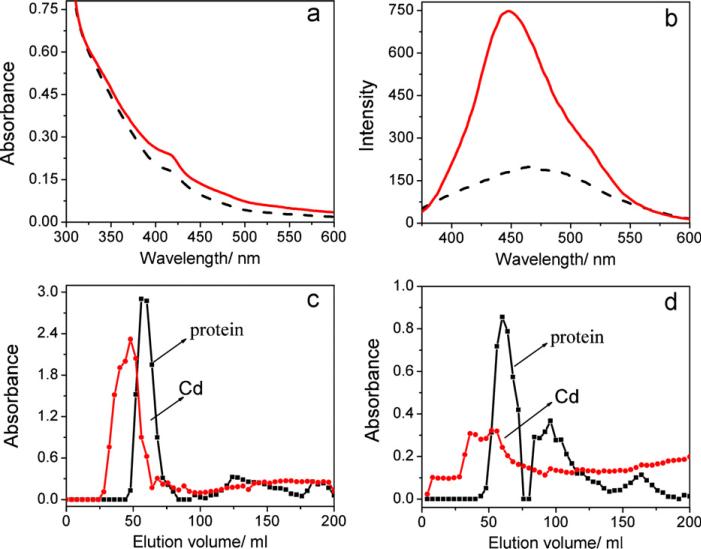
To further validate the formation of CdS QDs, two methods, cell lysis by hyperacoustic cell grinder and freezing–thawing of cells, were used to release the CdS QDs from the water-soluble fraction, and the UV–vis and fluorescence spectra of the obtained CdS samples were presented in Fig. 9(a) and (b). The released QDs were then separated from other cellular components by anion-exchange chromatography. Successive fractions eluted from the column were collected and analyzed by atomic adsorption spectroscopy for cadmium at 228.8 nm and UV–vis adsorption spectroscopy for protein at 260 nm as shown in Fig. 9(c) and (d), respectively. The products obtained from lysed cells possess much better optical properties compared with those from frozen–thawed cells in Fig. 9(a) and (b). Meanwhile, according to the elution curves shown in Fig. 9(c) and (d), the total content of Cd released from lysed cells at the entire volume range was twice more than that from the frozen–thawed cells. It can be concluded that the isolation and purification of CdS QDs by cell lysis method was much better than that by the freezing–thawing of cells.
Fig. 9.
(a) UV–vis and (b) fluorescence spectra of the concentrated CdS QDs samples after treated by lysis (solid line) and freezing–thawing (dash line) methods as well as the absorbances of cadmium and protein in the eluates treated with lysis (c) and freezing–thawing (d).
3.5. Control experiment
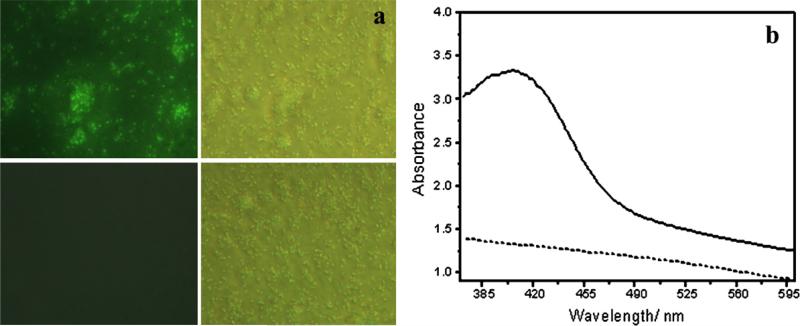
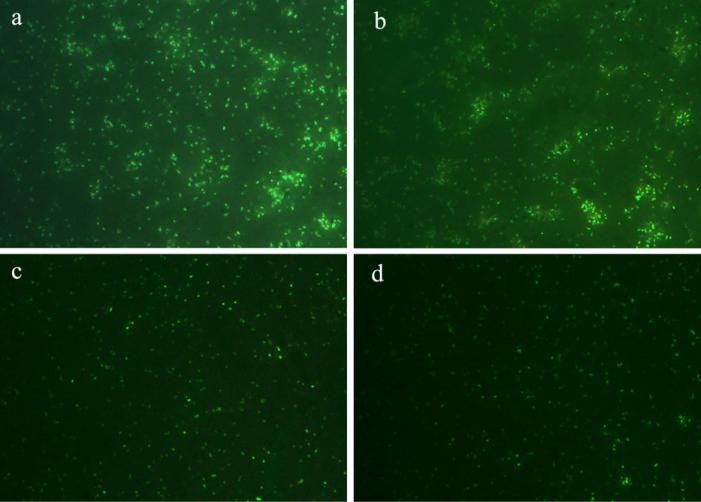
Bio-transformation process of the reaction was observed by visual inspection, imaging on a Leica DMIL inverted fluorescence microscope as well as UV–vis absorption spectroscopic measurements (Fig. 10). The E. coli cells without incubation with CdCl2 and Na2S do not show any fluorescence under the 350 nm excitation, whereas E. coli cells incubated with CdCl2 and Na2S show bright green fluorescence in Fig. 10(a). UV–vis spectrum for the resus-pended E. coli bacteria solution (dash line) shows no evidence of absorption peak in the region of 300–600 nm, while the absorption spectrum of BL 21 cells suspension incubated with CdCl2 and Na2S, shows an obvious absorption peak at about 410 nm (solid line), as shown in Fig. 10(b). After characterization, the residual bacterial cells were stored at 4 °C, and imaged on the microscope every day. As shown in Fig. 11, after 4 days the synthesized CdS quantum dots in E. coli cells still emit bright green fluorescence under 350 nm excitation, indicating their good stability.
Fig. 10.
(a) Images of E. coli incubated with (upper) and without (bottom) Cd2+ and S2−. The left rows represent fluorescent images, and the right rows represent phase contrast image. (b) UV–vis absorption spectra of E. coli incubated with (solid line) and without (dash line) Cd2+ and S2−.
Fig. 11.
Fluorescence images of E. coli cells stored at 4 °C for (a) 1, (b) 2, (c) 3 and (d) 4 days after incubated with Cd2+ and S2−.
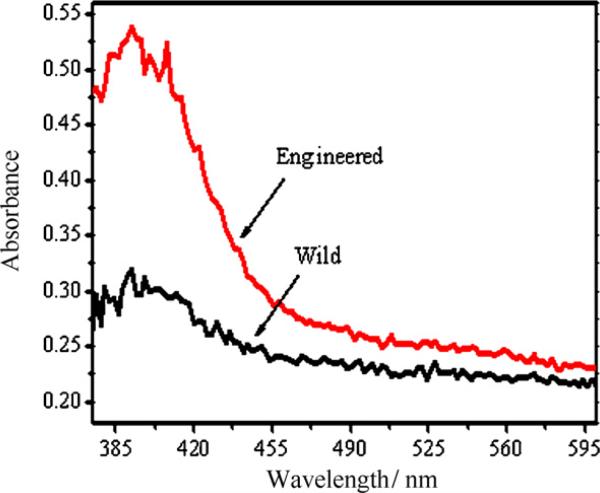
Biomolecules such as protein can bind with inorganic materials, the CdS binding peptide expressed in bacterial cells in this work was identified by yeast surface display technique and has proved to have high CdS binding affinity. Here, we used BL 21 as carrier to synthesize CdS QDs through introduction of foreign gene encoding the CdS binding peptide, and found that the genetically engineered bacteria can be successfully used to form CdS QDs. For further comparison, the wild type bacterial BL21 was taken as control. As shown in Fig. 12, it is obvious that the engineered E. coli BL21 is more beneficial for the formation of CdS QDs than the wild type bacteria. These results indicate that CdS binding peptide expressed inside the cells can favor the formation of CdS QDs and cap them to stabilize them.
Fig. 12.
Absorption spectra of CdS QDs synthesized with engineered and wild type bacteria.
Biological systems can possess highly specific molecular recognition capabilities after genetic engineering, and can control nucleation, phase formation and assembly of materials precisely. Here, we demonstrate the formation of CdS nanocrystals utilizing engineered E. coli cells as carriers. We firstly cloned CDS 7 peptide with a His6 tag into the pET-28b(+) vector and transfected E. coli strain BL21 (DE3). After induced with IPTG, the foreign proteins were expressed and could bind Cd2+ due to the addition of the cadmium chloride. When sodium sulfide solution was added, the nucleation of CdS nanocrystals occurred. We took E. coli cells that were not incubated with Cd2+ and S2− as the control. The control experiment, along with the XRD and EDS measurements, proved the formation of CdS QDs in the bacteria. What's more, we also found that the CdS QDs possessed a narrow size distribution and good stability.
4. Conclusions
CdS QDs were prepared via a bio-mimetic method, using engineered E. coli bacteria cells as carriers. Through the control experiment, it was proved that during the reaction, foreign protein expressed in E. coli cells under induction of IPTG, can well control the size of CdS QDs. The synthesis parameters were investigated. It was observed that both the incubation time of bacterial cells and the reaction time dramatically affect the formation of the CdS QDs. The experimental results showed that the fluorescence emission exhibit an obvious red-shift with the increase of the reactant concentration within the range of 0.5–10 mM, making the emission wavelength vary in the range of 445–510 nm. Through the release of the as-prepared CdS QDs by the hyperacoustic lysis and freezing–thawing of cells and the isolation with DEAE-Sephadex A-50, an anion-exchange resin, the synthesized CdS QDs can be separated from the cells and purified effectively. The biologically synthesized CdS QDs are coated by the CdS binding peptides and expected to be biocompatible in biolabeling.
Acknowledgements
We are grateful for the support from the National Science Foundation of China (Grant No. 20875011), National High-tech R&D Program (863 Program, No. 2009AA03Z309), as well as the support from Northeastern University on PhD students. CBM would like to thank the financial support from the US National Science Foundation (DMR-0847758, CBET-0854414, CBET-0854465), National Institutes of Health (R21EB009909-01A1, R03AR056848-01, R01HL092526-01A2), and Oklahoma Center for the Advancement of Science and Technology.
Footnotes
Publisher's Disclaimer: This article appeared in a journal published by Elsevier. The attached copy is furnished to the author for internal non-commercial research and education use, including for instruction at the authors institution and sharing with colleagues. Other uses, including reproduction and distribution, or selling or licensing copies, or posting to personal, institutional or third party websites are prohibited. In most cases authors are permitted to post their version of the article (e.g. in Word or Tex form) to their personal website or institutional repository. Authors requiring further information regarding Elsevier's archiving and manuscript policies are encouraged to visit: http://www.elsevier.com/copyright
References
- Anshup A, Venkataraman JS, Subramaniam C, Kumar RR, Priya S, Kumar TRS, Omkumar RV, John A, Pradeep T. Growth of gold nanoparticles in human cells. Langmuir. 2005;21:11562–11567. doi: 10.1021/la0519249. [DOI] [PubMed] [Google Scholar]
- Bai HJ, Zhang ZM, Guo Y, Yang GE. Biosynthesis of cadmium sulfide nanoparticles by photosynthetic bacteria Rhodopseudomonas palustris. Colloid Surf. B. 2009;70:142–146. doi: 10.1016/j.colsurfb.2008.12.025. [DOI] [PubMed] [Google Scholar]
- Baker RT, Catanzariti AM, Karunasekara Y, Soboleva TA, Sharwood R, Whitney S, Board PG. Using deubiquitylating enzymes as research tools. Meth. Enzymol. 2005;398:540–554. doi: 10.1016/S0076-6879(05)98044-0. [DOI] [PubMed] [Google Scholar]
- Bao HF, Hao N, Yang YX, Zhao DY. Biosynthesis of biocompatible cadmium telluride quantum dots using yeast cells. Nano Res. 2010;3:481–489. [Google Scholar]
- Bruchez M, Moronne M, Gin P, Weiss S, Alivisatos AP. Semiconductor nanocrystals as fluorescent biological labels. Science. 1998;281:2013–2016. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- Catanzariti AM, Soboleva TA, Jans DA, Board PG, Baker RT. An efficient system for high-level expression and easy purification of authentic recombinant proteins. Protein Sci. 2004;13:1331–1339. doi: 10.1110/ps.04618904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan WCW, Nie SM. Quantum dot bioconjugates for ultrasensitive non-isotopic detection. Science. 1998;281:2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- Clift MJD, Rutishauser BR, Brown DM, Duffin R, Donaldson K, Proudfoot L, Guy K, Stone V. The impact of different nanoparticle surface chemistry and size on uptake and toxicity in a murine macrophage cell line. Toxicol. Appl. Pharm. 2008;232:418–427. doi: 10.1016/j.taap.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Cui R, Liu HH, Xie HY, Zhang ZL, Yang YR, Pang DW, Xie ZX, Chen BB, Hu B, Shen P. Living yeast cells as a controllable biosynthesizer for fluorescent quantum dots. Adv. Funct. Mater. 2009;19:2359–2364. [Google Scholar]
- Deka S, Quarta A, Lupo MG, Falqui A, Boninelli S, Giannini C, Morello G, Giorgi MD, Lanzami G, Spinella C, Cingolani R, Pellegrino T, Manna L. CdSe/CdS/ZnS double shell nanorods with high photoluminescence efficiency and their exploitation as biolabeling probes. J. Am. Chem. Soc. 2009;131:2948–2958. doi: 10.1021/ja808369e. [DOI] [PubMed] [Google Scholar]
- Dong WW, Qian WP. Facile synthesis of Ag and Au nanoparticles utilizing chitosan as a mediator agent. Colloid Surf. B. 2008;62:136–142. doi: 10.1016/j.colsurfb.2007.09.030. [DOI] [PubMed] [Google Scholar]
- Du LW, Jiang H, Liu XH, Wang EK. Biosynthesis of gold nanoparticles assisted by Escherichia coli DH5α and its application on direct electrochemistry of hemoglobin. Electrochem. Commun. 2007;9:1165–1170. [Google Scholar]
- Goldman ER, Clapp AR, Anderson GP, Uyeda HT, Mauro JM, Medintz IL, Mattoussi H. Multiplexed toxin analysis using four colors of quantum dot fluororeagents. Anal. Chem. 2004;76:684–688. doi: 10.1021/ac035083r. [DOI] [PubMed] [Google Scholar]
- Ho YP, Chen HH, Leong KW, Wang TH. Evaluating the intracellular stability and unpacking of DNA nanocomplexes by guantum dots FRET. J. Control. Release. 2006;116:83–89. doi: 10.1016/j.jconrel.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Chiang CY, Lee SK, Gao Y, Hu EL, Yoreo JD, Belcher AM. Programmable assembly of nanoarchitectures using genetically engineered viruses. Nano Lett. 2005;5:1429–1434. doi: 10.1021/nl050795d. [DOI] [PubMed] [Google Scholar]
- Jha AK, Prasad K, Prasad K. A green low-cost biosynthesis of Sb2O3 nanoparticles. Biochem. Eng. J. 2009;43:303–306. doi: 10.1002/biot.200900144. [DOI] [PubMed] [Google Scholar]
- Kumar SA, Ansary AA, Ahmad A, Khan MI. Extracellular biosynthesis of CdSe quantum dots by the fungus, Fusarium oxysporum. J. Biomed. Nanotechnol. 2007;3:190–194. [Google Scholar]
- Lee JI, Ha KS, Yoo HS. Quantum-dot-assisted fluorescence resonance energy transfer approach for intracellular trafficking of chitosan/DNA complex. Acta Biomater. 2008;4:791–798. doi: 10.1016/j.actbio.2008.01.010. [DOI] [PubMed] [Google Scholar]
- Lee SK, Yun DS, Belcher AM. Cobalt ion mediated self-assembly of genetically engineered bacteriophage for biomimetic Co–Pt hybrid material. Biomacromolecules. 2006;7:14–17. doi: 10.1021/bm050691x. [DOI] [PubMed] [Google Scholar]
- Liu L, Zhang J, Su X, Mason RP. In vitro and in vivo assessment of CdTe and CdHgTe toxicity and clearance. J. Biomed. Nanotechnol. 2008;4:1–5. doi: 10.1166/jbn.2008.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Chen JY, Guo J, Wang CC, Yang WL, Xu L, Wang PN. Photostability of thiol-capped CdTe quantum dots in living cells: the effect of photo-oxidation. Nanotechnology. 2006a;17:2083–2089. [Google Scholar]
- Ma N, Dooley CJ, Kelley SO. RNA-templated semiconductor nanocrystals. J. Am. Chem. Soc. 2006b;128:12598–12599. doi: 10.1021/ja0638962. [DOI] [PubMed] [Google Scholar]
- Mao CB, Flynn CE, Hayhurst A, Sweeney R, Qi JF, Georgiou G, Iverson B, Belcher AM. Viral assembly of oriented quantum dot nanowires. Proc. Natl. Acad. Sci. U. S. A. 2003;100:6946–6951. doi: 10.1073/pnas.0832310100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam KT, Kim DW, Yoo PJ, Chiang CY, Meethong N, Hammond PT, Chiang YM, Belcher AM. Virus-enabled synthesis and assembly of nanowires for lithium ion battery electrodes. Science. 2006;312:885–888. doi: 10.1126/science.1122716. [DOI] [PubMed] [Google Scholar]
- Ngweniform P, Li D, Mao CB. Self-assembly of drug-loaded liposomes on genetically engineered protein nanotubes: a potential anti-cancer drug delivery vector. Soft Matter. 2009;5:954–956. [Google Scholar]
- Peelle BR, Krauland EM, Wittrup KD, Belcher AM. Design criteria for engineering inorganic material-specific peptides. Langmuir. 2005a;21:6929–6933. doi: 10.1021/la050261s. [DOI] [PubMed] [Google Scholar]
- Peelle BR, Krauland EM, Wittrup KD, Belcher AM. Probing the interface between biomolecules and inorganic materials using yeast surface display and genetic engineering. Acta Biomater. 2005b;1:145–154. doi: 10.1016/j.actbio.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Seghal KG, Sabu A, Joseph S. Synthesis of silver nanoparticles by glycolipid biosurfactant produced from marine Brevibacterium casei MSA19. J. Biotechnol. 2010;148:221–225. doi: 10.1016/j.jbiotec.2010.06.012. [DOI] [PubMed] [Google Scholar]
- Shan YM, Wang LP, Shi YH, Zhao H, Li HM, Liu HZ, Yang B, Li TY, Fang XX, Li W. NHS-mediated QDs-peptide/protein conjugation and its application for cell labeling. Talanta. 2008;75:1008–1014. doi: 10.1016/j.talanta.2007.12.040. [DOI] [PubMed] [Google Scholar]
- Sweeney RY, Mao CB, Gao XX, Burt JL, Belcher AM, Georgiou G, Iverson BL. Bacterial biosynthesis of cadmium sulfide nanocrystals. Chem. Biol. 2004;11:1553–1559. doi: 10.1016/j.chembiol.2004.08.022. [DOI] [PubMed] [Google Scholar]
- Williams P, Keshavarz-Moore E, Dunnill P. Production of cadmium sulphide microcrystallites in batch cultivation by Schizosaccharomyces pombe. J. Biotechnol. 1996;48:259–267. doi: 10.1016/0168-1656(96)01520-9. [DOI] [PubMed] [Google Scholar]
- Yang YJ, Tao X, Hou Q, Chen JF. Fluorescent mesoporous silica nanotubes incorporating CdS quantum dots for controlled release of ibuprofen. Acta Bio-mater. 2009;5:3488–3496. doi: 10.1016/j.actbio.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Yang DZ, Chen QF, Xu SK. Synthesis of CdSe/CdS with a simple non-TOP-based route. J. Lumin. 2007;126:853–858. [Google Scholar]
- Yang DZ, Chen QF, Wang WX, Xu SK. Direct and indirect immunolabelling of HeLa cells with quantum dots. Luminescence. 2008;23:169–174. doi: 10.1002/bio.1029. [DOI] [PubMed] [Google Scholar]
- Yoo PJ, Nam KT, Qi J, Lee SK, Park J, Belcher AM, Hammond PT. Spontaneous assembly of viruses on multilayered polymer surfaces. Nat. Mater. 2006;5:234–240. doi: 10.1038/nmat1596. [DOI] [PubMed] [Google Scholar]
- Zaman MB, Baral TN, Zhang JB, Whitfield D, Yu K. Single-domain antibody functionalized CdSe/ZnS quantum dots for cellular imaging of cancer cells. J. Phys. Chem. C. 2009;113:496–499. [Google Scholar]
- Zhang Y, Deng ZT, Yue JC, Tang FQ, Wei Q. Using cadmium telluride quantum dots as a proton flux sensor and applying to detect H9 avian influenza virus. Anal. Biochem. 2007;364:122–127. doi: 10.1016/j.ab.2007.02.031. [DOI] [PubMed] [Google Scholar]
- Zhao D, He ZK, Chan WH, Choi MMF. Synthesis and characterization of high-quality water-soluble near-infrared-emitting CdTe/CdS quantum dots capped by N-acetyl-l-cysteine via hydrothermal method. J. Phys. Chem. C. 2009;113:1293–1300. [Google Scholar]