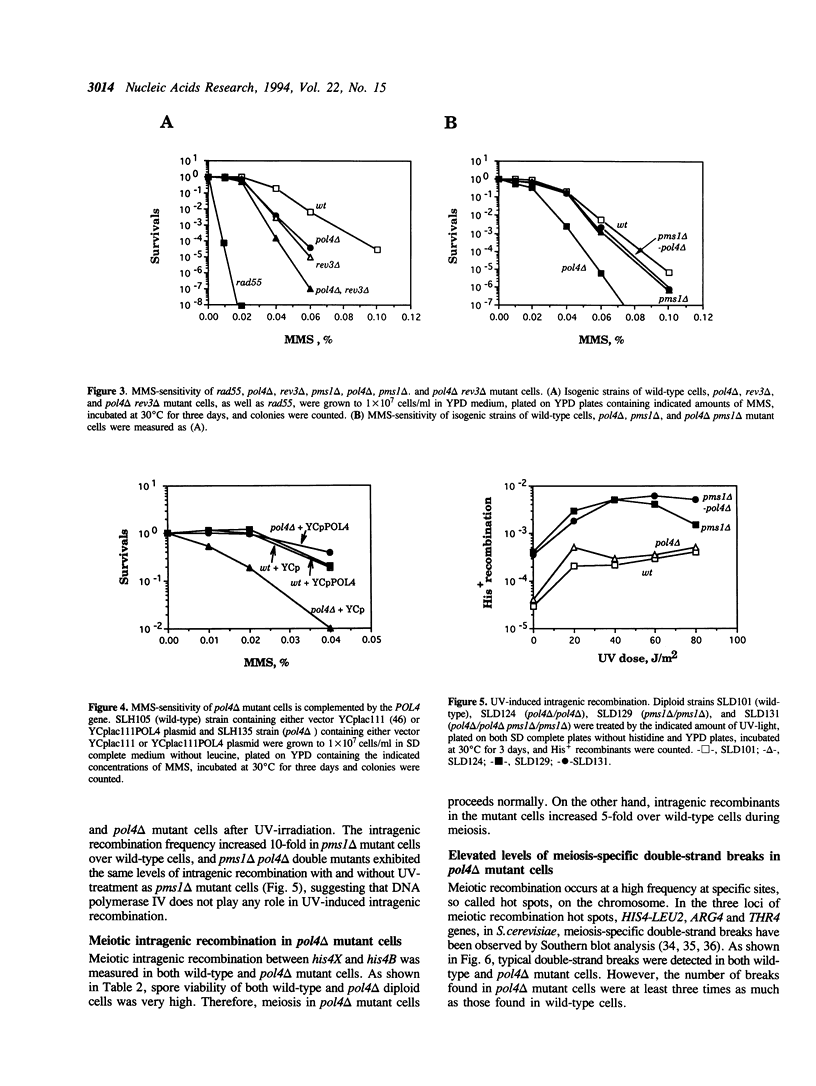
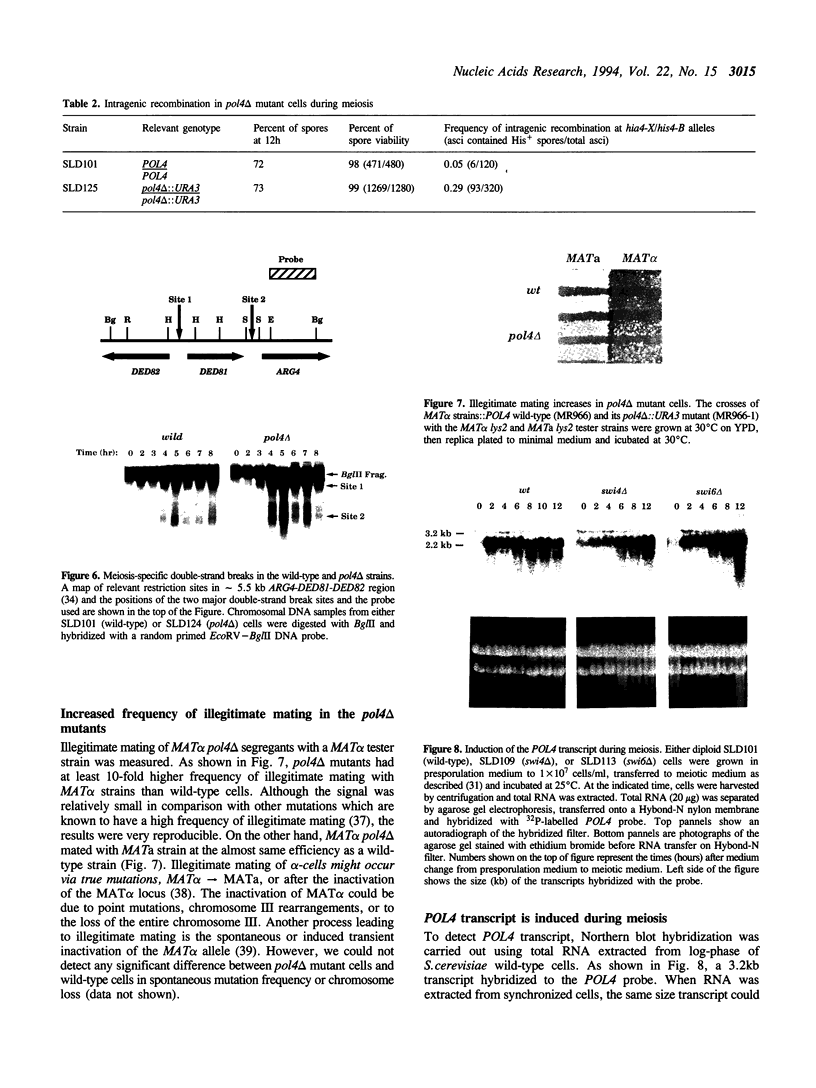
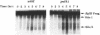Abstract
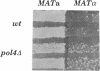
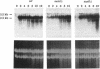
We identified and purified a new DNA polymerase (DNA polymerase IV), which is similar to mammalian DNA polymerase beta, from Saccharomyces cerevisiae and suggested that it is encoded by YCR14C (POLX) on chromosome III. Here, we provided a direct evidence that the purified DNA polymerase IV is indeed encoded by POLX. Strains harboring a pol4 deletion mutation exhibit neither mitotic growth defect nor a meiosis defect, suggesting that DNA polymerase IV participates in nonessential functions in DNA metabolism. The deletion strains did not exhibit UV-sensitivity. However, they did show weak sensitivity to MMS-treatment and exhibited a hyper-recombination phenotype when intragenic recombination was measured during meiosis. Furthermore, MAT alpha pol4 delta segregants had a higher frequency of illegitimate mating with a MAT alpha tester strain than that of wild-type cells. These results suggest that DNA polymerase IV participates in a double-strand break repair pathway. A 3.2kb of the POL4 transcript was weakly expressed in mitotically growing cells. During meiosis, a 2.2 kb POL4 transcript was greatly induced, while the 3.2 kb transcript stayed at constant levels. This induction was delayed in a swi4 delta strain during meiosis, while no effect was observed in a swi6 delta strain.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Araki H., Hamatake R. K., Johnston L. H., Sugino A. DPB2, the gene encoding DNA polymerase II subunit B, is required for chromosome replication in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):4601–4605. doi: 10.1073/pnas.88.11.4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki H., Ropp P. A., Johnson A. L., Johnston L. H., Morrison A., Sugino A. DNA polymerase II, the probable homolog of mammalian DNA polymerase epsilon, replicates chromosomal DNA in the yeast Saccharomyces cerevisiae. EMBO J. 1992 Feb;11(2):733–740. doi: 10.1002/j.1460-2075.1992.tb05106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baril E. F., Scheiner C., Pederson T. A beta-like DNA polymerase activity in the slime mold Dictyostelium discoideum. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3317–3321. doi: 10.1073/pnas.77.6.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P., Ouzounis C., Sander C., Scharf M., Schneider R., Sonnhammer E. Comprehensive sequence analysis of the 182 predicted open reading frames of yeast chromosome III. Protein Sci. 1992 Dec;1(12):1677–1690. doi: 10.1002/pro.5560011216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulet A., Simon M., Faye G., Bauer G. A., Burgers P. M. Structure and function of the Saccharomyces cerevisiae CDC2 gene encoding the large subunit of DNA polymerase III. EMBO J. 1989 Jun;8(6):1849–1854. doi: 10.1002/j.1460-2075.1989.tb03580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd M., Campbell J. L. Temperature-sensitive mutations in the yeast DNA polymerase I gene. Proc Natl Acad Sci U S A. 1987 May;84(9):2838–2842. doi: 10.1073/pnas.84.9.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Alani E., Kleckner N. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell. 1990 Jun 15;61(6):1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- Chang L. M. DNA polymerases from bakers' yeast. J Biol Chem. 1977 Mar 25;252(6):1873–1880. [PubMed] [Google Scholar]
- Goldway M., Sherman A., Zenvirth D., Arbel T., Simchen G. A short chromosomal region with major roles in yeast chromosome III meiotic disjunction, recombination and double strand breaks. Genetics. 1993 Feb;133(2):159–169. doi: 10.1093/genetics/133.2.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordenin D. A., Proscyavichus Y. Y., Malkova A. L., Trofimova M. V., Peterzen A. Yeast mutants with increased bacterial transposon Tn5 excision. Yeast. 1991 Jan;7(1):37–50. doi: 10.1002/yea.320070105. [DOI] [PubMed] [Google Scholar]
- Haber J. E., Weiffenbach B., Rogers D. T., McCusker J., Rowe L. B. Chromosomal rearrangements accompanying yeast mating-type switching: evidence for a gene-conversion model. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 2):991–1002. doi: 10.1101/sqb.1981.045.01.115. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H., Mortimer R. K., Culotti J., Culotti M. Genetic Control of the Cell Division Cycle in Yeast: V. Genetic Analysis of cdc Mutants. Genetics. 1973 Jun;74(2):267–286. doi: 10.1093/genetics/74.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose F., Hotta Y., Yamaguchi M., Matsukage A. Difference in the expression level of DNA polymerase beta among mouse tissues: high expression in the pachytene spermatocyte. Exp Cell Res. 1989 Mar;181(1):169–180. doi: 10.1016/0014-4827(89)90191-2. [DOI] [PubMed] [Google Scholar]
- Klar A. J., Strathern J. N., Abraham J. A. Involvement of double-strand chromosomal breaks for mating-type switching in Saccharomyces cerevisiae. Cold Spring Harb Symp Quant Biol. 1984;49:77–88. doi: 10.1101/sqb.1984.049.01.011. [DOI] [PubMed] [Google Scholar]
- Kolodkin A. L., Klar A. J., Stahl F. W. Double-strand breaks can initiate meiotic recombination in S. cerevisiae. Cell. 1986 Aug 29;46(5):733–740. doi: 10.1016/0092-8674(86)90349-1. [DOI] [PubMed] [Google Scholar]
- Lee S. H., Eki T., Hurwitz J. Synthesis of DNA containing the simian virus 40 origin of replication by the combined action of DNA polymerases alpha and delta. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7361–7365. doi: 10.1073/pnas.86.19.7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leem S. H., Ogawa H. The MRE4 gene encodes a novel protein kinase homologue required for meiotic recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 1992 Feb 11;20(3):449–457. doi: 10.1093/nar/20.3.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linn S. How many pols does it take to replicate nuclear DNA? Cell. 1991 Jul 26;66(2):185–187. doi: 10.1016/0092-8674(91)90608-2. [DOI] [PubMed] [Google Scholar]
- McCusker J. H., Haber J. E. Evidence of Chromosomal Breaks near the Mating-Type Locus of SACCHAROMYCES CEREVISIAE That Accompany MATalpha xMATalpha Matings. Genetics. 1981 Nov;99(3-4):383–403. doi: 10.1093/genetics/99.3-4.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melendy T., Stillman B. Purification of DNA polymerase delta as an essential simian virus 40 DNA replication factor. J Biol Chem. 1991 Jan 25;266(3):1942–1949. [PubMed] [Google Scholar]
- Mitchell A. P. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol Rev. 1994 Mar;58(1):56–70. doi: 10.1128/mr.58.1.56-70.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A., Araki H., Clark A. B., Hamatake R. K., Sugino A. A third essential DNA polymerase in S. cerevisiae. Cell. 1990 Sep 21;62(6):1143–1151. doi: 10.1016/0092-8674(90)90391-q. [DOI] [PubMed] [Google Scholar]
- Morrison A., Christensen R. B., Alley J., Beck A. K., Bernstine E. G., Lemontt J. F., Lawrence C. W. REV3, a Saccharomyces cerevisiae gene whose function is required for induced mutagenesis, is predicted to encode a nonessential DNA polymerase. J Bacteriol. 1989 Oct;171(10):5659–5667. doi: 10.1128/jb.171.10.5659-5667.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A., Johnson A. L., Johnston L. H., Sugino A. Pathway correcting DNA replication errors in Saccharomyces cerevisiae. EMBO J. 1993 Apr;12(4):1467–1473. doi: 10.1002/j.1460-2075.1993.tb05790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver S. G., van der Aart Q. J., Agostoni-Carbone M. L., Aigle M., Alberghina L., Alexandraki D., Antoine G., Anwar R., Ballesta J. P., Benit P. The complete DNA sequence of yeast chromosome III. Nature. 1992 May 7;357(6373):38–46. doi: 10.1038/357038a0. [DOI] [PubMed] [Google Scholar]
- Pizzagalli A., Valsasnini P., Plevani P., Lucchini G. DNA polymerase I gene of Saccharomyces cerevisiae: nucleotide sequence, mapping of a temperature-sensitive mutation, and protein homology with other DNA polymerases. Proc Natl Acad Sci U S A. 1988 Jun;85(11):3772–3776. doi: 10.1073/pnas.85.11.3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R., Widen S. G., Singhal R. K., Watkins J., Prakash L., Wilson S. H. Yeast open reading frame YCR14C encodes a DNA beta-polymerase-like enzyme. Nucleic Acids Res. 1993 Nov 25;21(23):5301–5307. doi: 10.1093/nar/21.23.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sakaguchi K., Boyd J. B. Purification and characterization of a DNA polymerase beta from Drosophila*. J Biol Chem. 1985 Sep 5;260(19):10406–10411. [PubMed] [Google Scholar]
- Shimizu K., Santocanale C., Ropp P. A., Longhese M. P., Plevani P., Lucchini G., Sugino A. Purification and characterization of a new DNA polymerase from budding yeast Saccharomyces cerevisiae. A probable homolog of mammalian DNA polymerase beta. J Biol Chem. 1993 Dec 25;268(36):27148–27153. [PubMed] [Google Scholar]
- Shinohara A., Ogawa H., Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992 May 1;69(3):457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- Shivji K. K., Kenny M. K., Wood R. D. Proliferating cell nuclear antigen is required for DNA excision repair. Cell. 1992 Apr 17;69(2):367–374. doi: 10.1016/0092-8674(92)90416-a. [DOI] [PubMed] [Google Scholar]
- Singhal R. K., Hinkle D. C., Lawrence C. W. The REV3 gene of Saccharomyces cerevisiae is transcriptionally regulated more like a repair gene than one encoding a DNA polymerase. Mol Gen Genet. 1992 Dec;236(1):17–24. doi: 10.1007/BF00279638. [DOI] [PubMed] [Google Scholar]
- Sitney K. C., Budd M. E., Campbell J. L. DNA polymerase III, a second essential DNA polymerase, is encoded by the S. cerevisiae CDC2 gene. Cell. 1989 Feb 24;56(4):599–605. doi: 10.1016/0092-8674(89)90582-5. [DOI] [PubMed] [Google Scholar]
- Sun H., Treco D., Schultes N. P., Szostak J. W. Double-strand breaks at an initiation site for meiotic gene conversion. Nature. 1989 Mar 2;338(6210):87–90. doi: 10.1038/338087a0. [DOI] [PubMed] [Google Scholar]
- Tsurimoto T., Melendy T., Stillman B. Sequential initiation of lagging and leading strand synthesis by two different polymerase complexes at the SV40 DNA replication origin. Nature. 1990 Aug 9;346(6284):534–539. doi: 10.1038/346534a0. [DOI] [PubMed] [Google Scholar]
- Wang T. S. Eukaryotic DNA polymerases. Annu Rev Biochem. 1991;60:513–552. doi: 10.1146/annurev.bi.60.070191.002501. [DOI] [PubMed] [Google Scholar]
- Wang Z., Wu X., Friedberg E. C. DNA repair synthesis during base excision repair in vitro is catalyzed by DNA polymerase epsilon and is influenced by DNA polymerases alpha and delta in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Feb;13(2):1051–1058. doi: 10.1128/mcb.13.2.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wu X., Friedberg E. C. Excision repair of DNA in nuclear extracts from the yeast Saccharomyces cerevisiae. Biochemistry. 1992 Apr 14;31(14):3694–3702. doi: 10.1021/bi00129a019. [DOI] [PubMed] [Google Scholar]
- Weinberg D. H., Kelly T. J. Requirement for two DNA polymerases in the replication of simian virus 40 DNA in vitro. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9742–9746. doi: 10.1073/pnas.86.24.9742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintersberger E. Deoxyribonucleic acid polymerases from yeast. Further purification and characterization of DNA-dependent DNA polymerases A and B. Eur J Biochem. 1974 Dec 16;50(1):41–47. doi: 10.1111/j.1432-1033.1974.tb03871.x. [DOI] [PubMed] [Google Scholar]