Abstract
Background
NKX2-3 is associated with inflammatory bowel disease (IBD). NKX2-3 is expressed in microvascular endothelial cells and the muscularis mucosa of the gastrointestinal tract. Human intestinal microvascular endothelial cells (HIMECs) are actively involved in the pathogenesis of IBD and IBD-associated microvascular dysfunction. To understand the cellular function of NKX2-3 and its potential role underlying IBD pathogenesis, we investigated the genes regulated by NKX2-3 in HIMEC using cDNA microarray.
Methodology/Principal Findings
NKX2-3 expression was suppressed by shRNA in two HIMEC lines and gene expression was profiled by cDNA microarray. Pathway Analysis was used to identify gene networks according to biological functions and associated pathways. Validation of microarray and genes expression in intestinal tissues was assessed by RT-PCR. NKX2-3 regulated genes are involved in immune and inflammatory response, cell proliferation and growth, metabolic process, and angiogenesis. Several inflammation and angiogenesis related signaling pathways that play important roles in IBD were regulated by NKX2-3, including endothelin-1 and VEGF-PI3K/AKT-eNOS. Expression levels of NKX2-3, VEGFA, PI3K, AKT, and eNOS are increased in intestinal tissues from IBD patients and expression levels of EDN1 are decreased in intestinal tissues from IBD patients. These results demonstrated the important roles of NKX2-3, VEGF, PI3K, AKT, eNOS, and EDN1 in IBD pathogenesis. Correlation analysis showed a positive correlation between mRNA expression of NKX2-3 and VEGFA and a negative correlation between mRNA expression of NKX2-3 and EDN1 in intestinal tissues from IBD patients.
Conclusion/Relevance
NKX2-3 may play an important role in IBD pathogenesis by regulating endothelin-1 and VEGF signaling in HIMECs.
Introduction
Crohn's disease (CD) and ulcerative colitis (UC), the two main subtypes of inflammatory bowel disease (IBD), are chronic, relapsing inflammatory disorders of the gastrointestinal tract. Genetics play an important role in the development of IBD [1]. NKX2-3 (NK2 transcription factor related, locus 3) has been shown to be associated with both CD and UC by recent genome-wide association studies [2], [3].
NKX2-3 is a member of the Nkx family of homeodomain transcription factors that play critical roles in regulating tissue-specific gene expression essential for determining tissue differentiation, as well as the temporal and spatial patterns of development [4], [5]. During development, NKX2-3 is primarily expressed in the midgut and hindgut mesoderm and spleen, as well as in pharyngeal endoderm [6], [7], [8]. Analysis of NKX2-3-deficient mice has revealed a critical role for this homeobox transcription factor in spleen development and organization [9], and in establishing the correct environment for normal B cell development and T cell dependent immune response [10]. NKX2-3 is also essential for normal small intestine development and function [11]. NKX2-3 is also expressed in microvascular endothelial cells within the lamina propria and submucosa of the intestine, where it is required for expression of the lymphocyte adhesion molecule MAdCAM-1 in the mouse [9].
Microvascular endothelial cells have a critical “gatekeeper” role in the inflammatory process through their ability to recruit circulating immune cells to foci of inflammation. Endothelial activation in response to cytokines and bacterial products results in cell adhesion molecule expression and chemokine production, which mediate increased binding and transmigration of leukocytes across the vascular wall. Intestinal microvascular endothelial cells are recognized as a cell population actively involved in the pathogenesis of inflammatory bowel diseases (IBD) and IBD-associated microvascular dysfunction [12].
Transcription factors can regulate the expression of downstream genes. Recently, we found that the expression of NKX2-3 is up-regulated in intestinal tissues and B cells from CD patients [13] and subsequently identified many inflammation and immune-response genes regulated by NKX2-3 in B cell lines from a CD patient. These included several genes which also have important functions in endothelial cells, such as endothelin-1 (EDN1), ASS1, and KLF2 [14]. Since endothelial cells play a key role in mucosal immune homeostasis and NKX2-3 is expressed in intestinal endothelial cells, we further performed cDNA microarray to identify genes regulated by NKX2-3 in two human intestinal microvascular endothelial cell lines (HIMEC).
Results
Suppression of NKX2-3 expression in 2 HIMEC lines
pSUPER.retro.puro.shRNA-NKX2-3 and empty vector were transfected into 21B and 432 HIMEC. RT-PCR results showed that mRNA expression levels of NKX2-3 in the two shRNA-NKX2-3 cells were significantly reduced compared with the empty vector cells (control cells) 48 hours after transfection (Fig. 1).
Figure 1. NKX2-3 mRNA expression is suppressed by shRNA in 21B and 432 HIMEC.
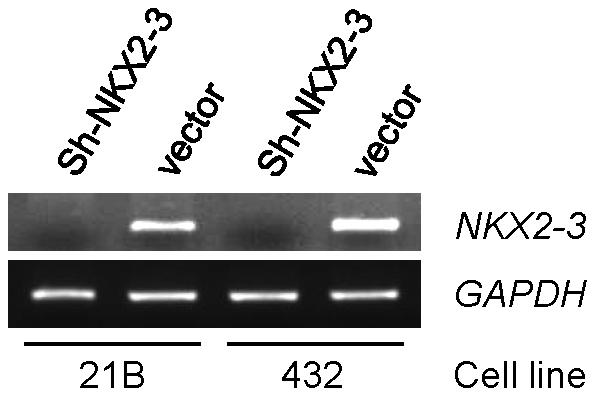
pSUPER.retro.puro.shRNA-NKX2-3 and empty vector were transfected into two HIMEC. 48 hours after transfection, RNA was isolated and analyzed by RT-PCR in two knockdown HIMEC compared with controls. GAPDH expression served as a control.
Identification of genes regulated by NKX2-3
To analyze the effects of NKX2-3 knockdown on gene expression and identify genes with altered expression levels, cDNA microarray analysis was conducted with NKX2-3 knockdown and control cells from two HIMEC. Stringent criteria (fold change ≥1.5, up or down, p<0.0005) were used to filter the differentially expressed genes. The expression levels of 1746 genes were affected by NKX2-3 knockdown (935 down-regulated and 811 up-regulated) in the HIMEC 21B cell line as compared to control, and 1603 genes were affected by NKX2-3 knockdown (741 down-regulated and 862 up-regulated) in the HIMEC 432 cell line as compared to control. A total of 1000 shared genes were found to be affected by NKX2-3 knockdown in both HIMEC, including 996 (99.6%) genes in the same direction (432 genes down-regulated and 564 genes up-regulated by NKX2-3 knockdown in both HIMEC), and only 4 genes in the opposite direction. Taken together, the transcriptional profile of genes affected by NKX2-3 knockdown was highly consistent for both HIMEC. Table 1 shows the top 100 down-regulated and top 100 up-regulated genes by NKX2-3 knockdown with average fold changes in the two cell lines. In order to characterize the top genes affected by NKX2-3 knockdown, they were assigned to ontological functional groups based on IPA and references in the literature. These 200 genes grouped primarily within the following functional categories, which are listed as such in Table 1: immune and inflammatory response; cell growth and proliferation; metabolic process; cell adhesion; transcription regulation; transport and structure; and angiogenesis. Since the NKX2-3 transcription factor is associated with IBD and is up-regulated in CD patients [13], it is reasonable to assume that NKX2-3 could play a role in IBD pathogenesis by regulating inflammation-related genes. In fact, among the 200 genes regulated by NKX2-3, 70 genes were immune and inflammatory response genes, including IL8 (array ratio 0.4) and KLF2 (array ratio 3.92). Angiogenesis plays an important role in endothelial cell participation in inflammation [15], ANGPT2 is down-regulated (array ratio 0.18) and FGF2 is up-regulated (array ratio 4.0) by NKX2-3 knockdown.
Table 1. Top 200 genes regulated by NKX2-3 knockdown in HIMEC.
| Gene | Fold | p-value | Gene | Fold | p-value | Gene | Fold | p-value | Gene | Fold | p-value |
| Cell growth and proliferation | Transport and structure | Inflammation and immune response | Inflammation and immune response | ||||||||
| PIM3 | 0.3 | 2.31E-06 | SYT11 | 0.35 | 3.66E-07 | TNFRSF10D | 0.31 | 3.54E-07 | SAMD9 | 7.31 | 2.01E-10 |
| TFPI2 | 0.32 | 3.94E-06 | KRT81 | 0.4 | 3.31E-07 | TNFSF18 | 0.32 | 6.43E-05 | STAT1 | 7.72 | 1.98E-09 |
| NR2F1 | 0.37 | 4.64E-07 | RPS23 | 0.4 | 2.97E-07 | CERK | 0.33 | 1.39E-06 | TAP2 | 7.83 | 7.33E-10 |
| EIF4B | 0.38 | 2.68E-07 | EXOC6 | 0.47 | 2.09E-06 | FAM172A | 0.37 | 1.94E-07 | PSMB8 | 8.22 | 1.84E-10 |
| GHR | 0.39 | 3.69E-07 | RPL23 | 0.47 | 1.33E-06 | YPEL2 | 0.39 | 0.000952 | UBE2L6 | 8.33 | 9.08E-10 |
| HMMR | 0.39 | 3.79E-06 | RPS29 | 0.48 | 2.41E-06 | PAPSS2 | 0.39 | 8.84E-06 | PTGS2 | 8.63 | 2.77E-11 |
| REEP1 | 0.4 | 2.86E-07 | ADAP1 | 3.29 | 2.36E-08 | IL8 | 0.4 | 7.63E-06 | GBP1 | 9.24 | 2.23E-08 |
| ZMAT3 | 0.41 | 3.60E-07 | SLC25A28 | 3.71 | 1.99E-07 | UACA | 0.4 | 3.05E-05 | PARP9 | 9.46 | 6.09E-10 |
| MMP7 | 0.42 | 3.43E-06 | RTP4 | 6.47 | 0.000111 | GIMAP7 | 0.41 | 9.24E-07 | SP110 | 9.83 | 1.31E-10 |
| GDF15 | 0.42 | 1.22E-05 | GJD3 | 7.95 | 0.000154 | HES2 | 0.41 | 2.05E-07 | TAP1 | 10.62 | 7.18E-11 |
| MFNG | 0.42 | 2.32E-05 | SLC40A1 | 0.41 | 2.59E-07 | USP18 | 11.2 | 9.63E-11 | |||
| AGAP3 | 0.43 | 6.31E-07 | TNFAIP8L1 | 0.43 | 3.66E-07 | DDX58 | 11.39 | 3.09E-08 | |||
| TMEM158 | 0.44 | 1.25E-05 | Cell adhesion | F2RL1 | 0.43 | 0.000121 | HLA-B | 11.89 | 8.66E-12 | ||
| UNC5A | 0.44 | 7.44E-05 | CD36 | 0.43 | 5.14E-07 | PRIC285 | 12.35 | 1.67E-10 | |||
| SHMT2 | 0.44 | 1.55E-06 | POSTN | 0.22 | 6.28E-09 | PLD1 | 0.44 | 2.12E-07 | PSMB9 | 14.18 | 1.73E-10 |
| FAM198B | 0.45 | 3.44E-06 | MMRN1 | 0.35 | 3.33E-06 | CBS | 0.46 | 3.70E-06 | |||
| HNRNPA0 | 0.46 | 6.11E-05 | CXADR | 0.36 | 7.75E-07 | CEP55 | 0.47 | 3.69E-06 | |||
| PAWR | 0.46 | 4.71E-05 | SRPX | 0.37 | 7.57E-06 | GNG12 | 0.48 | 5.21E-05 | Others | ||
| DCLK1 | 0.47 | 7.04E-07 | FLRT2 | 0.46 | 8.02E-06 | TNFSF4 | 0.48 | 1.00E-05 | |||
| TTC3 | 0.47 | 5.53E-07 | AIF1L | 3.41 | 8.77E-08 | ARHGDIB | 0.49 | 2.55E-05 | ANKRD55 | 0.18 | 1.07E-07 |
| TFF3 | 0.47 | 1.73E-05 | CLEC1A | 0.49 | 1.96E-05 | C2CD4B | 0.22 | 2.48E-08 | |||
| PTPRE | 0.47 | 4.31E-06 | CXCL16 | 3.19 | 1.04E-05 | KIAA0114 | 0.27 | 2.03E-08 | |||
| UBE4B | 0.47 | 2.02E-06 | Transcription regulation | GCH1 | 3.32 | 5.31E-05 | NT5DC2 | 0.28 | 8.23E-09 | ||
| CDKN3 | 0.48 | 0.000101 | UNC93B1 | 3.46 | 1.36E-06 | EIF4BP7 | 0.29 | 3.73E-07 | |||
| CCNB2 | 0.49 | 2.92E-05 | SDPR | 0.44 | 1.95E-06 | CD68 | 3.51 | 3.49E-07 | FLJ41200 | 0.29 | 2.64E-08 |
| ANLN | 0.49 | 3.85E-05 | HOXD1 | 0.45 | 0.000147 | TRIM21 | 3.62 | 9.21E-06 | C13orf33 | 0.34 | 7.43E-06 |
| LGMN | 3.23 | 4.42E-07 | ZNFX1 | 3.52 | 2.22E-06 | APOBEC3G | 3.67 | 5.26E-06 | EIF4BP3 | 0.38 | 1.42E-07 |
| SEMA3A | 3.25 | 2.19E-06 | BATF2 | 4.11 | 1.75E-06 | TRIM5 | 3.82 | 4.78E-05 | FBN2 | 0.39 | 5.49E-06 |
| PNPT1 | 3.44 | 2.45E-05 | KLF2 | 3.92 | 7.66E-06 | C7orf41 | 0.4 | 5.68E-07 | |||
| MAP2 | 3.57 | 4.49E-09 | PLSCR1 | 4.05 | 2.58E-08 | SNHG9 | 0.41 | 0.000156 | |||
| PMAIP1 | 3.65 | 3.66E-08 | Metabolic process | MYD88 | 4.28 | 1.98E-06 | WRB | 0.42 | 1.18E-06 | ||
| EIF2AK2 | 3.73 | 9.30E-09 | CD38 | 4.28 | 6.55E-08 | BCYRN1 | 0.42 | 0.000157 | |||
| HEY1 | 3.85 | 2.33E-07 | ACO1 | 0.2 | 8.00E-07 | CX3CL1 | 4.33 | 0.000209 | ZNF704 | 0.44 | 5.90E-07 |
| CSRNP1 | 3.87 | 1.55E-06 | HSD17B2 | 0.23 | 2.48E-06 | STAT2 | 4.34 | 1.08E-05 | ANKRD37 | 0.46 | 5.41E-06 |
| C8orf4 | 3.95 | 5.53E-06 | PHGDH | 0.29 | 1.86E-08 | PLEKHA4 | 4.37 | 1.83E-05 | HNRPA1L-2 | 0.46 | 8.27E-06 |
| XAF1 | 4.17 | 2.88E-06 | PSAT1 | 0.3 | 1.24E-07 | IFNB1 | 4.37 | 3.68E-06 | TCTEX1D2 | 0.47 | 0.000153 |
| SERPINE2 | 4.33 | 2.29E-08 | ASNS | 0.31 | 1.09E-07 | FCN3 | 4.38 | 0.000198 | CENPW | 0.48 | 7.38E-05 |
| HEY2 | 4.6 | 4.55E-08 | GALNTL2 | 0.33 | 7.00E-06 | F2RL3 | 4.44 | 2.45E-07 | NETO2 | 0.48 | 1.93E-05 |
| PARP10 | 5.54 | 5.42E-08 | PKIA | 0.34 | 7.05E-05 | ZC3HAV1 | 4.6 | 5.95E-07 | C8orf45 | 0.48 | 8.75E-05 |
| CRYAB | 6.8 | 8.25E-06 | MYO5A | 0.38 | 5.34E-05 | HCG4 | 4.61 | 2.85E-09 | MAMDC2 | 3.19 | 6.34E-07 |
| KLF4 | 6.86 | 6.09E-10 | TXNDC12 | 0.4 | 1.43E-07 | HLA-C | 4.63 | 1.63E-09 | FBXO6 | 3.26 | 7.21E-08 |
| LY6E | 7.47 | 1.94E-10 | SNCAIP | 0.41 | 0.000257 | GBP2 | 4.71 | 4.44E-08 | C1orf74 | 3.48 | 0.000186 |
| TMEM100 | 15.53 | 3.36E-07 | CHST7 | 0.44 | 7.79E-07 | SMAD7 | 4.78 | 1.31E-06 | USP41 | 3.52 | 3.72E-06 |
| MCTP1 | 0.46 | 0.000231 | NT5C3 | 4.8 | 1.92E-07 | C19orf66 | 3.92 | 2.77E-08 | |||
| FAR2 | 0.48 | 0.000238 | HLA-H | 5.39 | 4.83E-10 | HIST2H2AC | 4.18 | 5.43E-08 | |||
| Angiogenesis | PRSS12 | 0.48 | 1.43E-06 | TRIM22 | 5.49 | 9.22E-08 | GCA | 5.14 | 5.15E-06 | ||
| BCHE | 0.49 | 3.81E-06 | DHX58 | 5.58 | 1.57E-06 | HIST2H2AA3 | 5.17 | 2.92E-09 | |||
| ANGPT2 | 0.18 | 1.42E-05 | ADAMTSL2 | 3.2 | 1.26E-06 | IL18BP | 5.66 | 3.92E-09 | HIST2H2AA4 | 5.28 | 2.11E-09 |
| SRPX2 | 0.33 | 0.000284 | TIPARP | 3.28 | 1.18E-07 | HLA-F | 5.79 | 2.68E-10 | RNF213 | 6.6 | 1.03E-06 |
| TRPC6 | 0.33 | 1.08E-07 | PLA1A | 4.75 | 3.24E-08 | CASP1 | 5.82 | 5.35E-10 | DDX60L | 6.75 | 1.14E-08 |
| BTG1 | 0.43 | 3.66E-05 | GMPR | 4.8 | 5.63E-08 | PARP12 | 6.28 | 1.97E-09 | LYPD1 | 6.75 | 3.01E-05 |
| BMP4 | 0.48 | 2.53E-05 | LAP3 | 6.07 | 9.20E-09 | IDO1 | 6.93 | 1.77E-08 | DDX60 | 7.87 | 2.71E-09 |
| ANGPTL4 | 3.37 | 1.01E-08 | UBA7 | 6.64 | 9.54E-08 | HCP5 | 6.99 | 3.95E-10 | FAM46A | 8.63 | 4.81E-06 |
| TYMP | 3.38 | 1.13E-08 | CMPK2 | 10.3 | 5.63E-08 | PARP14 | 7.01 | 1.24E-08 | MT1M | 8.77 | 6.76E-11 |
| FGF2 | 4 | 5.16E-05 | ALPL | 12.12 | 0.000474 | IL7R | 7.06 | 3.94E-05 | SAMD9L | 9.07 | 1.22E-08 |
Identification of pathways regulated by NKX2-3
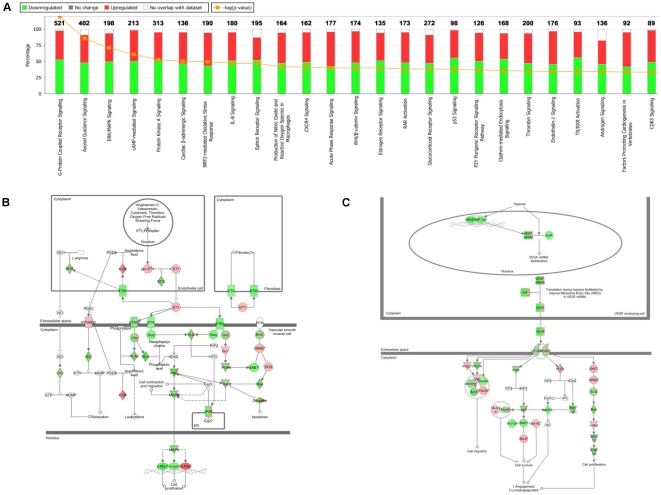
To assess the global effects of NKX2-3 on gene expression, IPA was used to systematically visualize the relationships among genes regulated by NKX2-3 knockdown. The 25 pathways most affected by NKX2-3 are shown in Fig. 2A. G-Protein Coupled Receptor Signaling (p = 1.71×10−14), Axonal Guidance Signaling (p = 5.80×10−11), and ERK/MAPK Signaling (p = 5.42×10−9) were the top three canonical pathways affected by NKX2-3 knockdown.
Figure 2. An interaction network generated using IPA analysis shows genes regulated by NKX2-3 in HIMEC.
(A) The top 25 signaling pathways affected by NKX2-3 knockdown. (B) The EDN1 pathway is regulated by NKX2-3 in 2 HIMEC. IPA analysis shows genes up-regulated (red) and down-regulated (green) by NKX2-3 knockdown. (C) The VEGF-PI3K/AKT-eNOS pathway is regulated by NKX2-3 in 2 HIMEC. IPA analysis shows genes up-regulated (red) and down-regulated (green) by NKX2-3 knockdown in the VEGF-PI3K/AKT-eNOS pathway.
We previously found that knockdown of NKX2-3 in human B cells [14] significantly affected the expression of genes, including EDN1 and NOS, known to have important roles in endothelial cell function. We sought to investigate whether NKX2-3 knockdown would affect important genes and signaling pathways in HIMEC.
EDN1 was found to be up-regulated by NKX2-3 knockdown in both HIMEC (array ratio 1.2/1.3, p<0.05). NKX2-3 knockdown significantly affected genes within the endothelin-1 pathway (p = 1.02×10−4), which was among the top 25 pathways affected by NKX2-3 (Fig. 2A). Fig. 2B illustrates the genes in the endothelin-1 pathway affected by NKX2-3 knockdown. NKX2-3 knockdown activated MAPK through EDN1-PLCβ-PLC pathway, thus regulated inflammation response and cell proliferation.
Nitric oxide (NO) which is generated by NO synthase (NOS) may play an important role in the pathogenesis of IBD [15]. Production of NO (in macrophages) is among the top 10 pathways affected by NKX2-3 (Fig. 2A). eNOS is down-regulated by NKX2-3 knockdown in both HIMEC (array ratio −1.3/−2.7, p<0.0001). IPA analysis showed that the regulation of eNOS can be through VEGF-PI3K/AKT pathway and that this pathway is significantly affected by NKX2-3 knockdown (p = 5.45×10−3) (Fig. 2C). Fold changes of key genes in this pathway regulated by NKX2-3 knockdown in HIMEC are: VEGF (−1.3/−1.3), PI3K (−1.5/−1.7), and AKT (−1.35/−1.35).
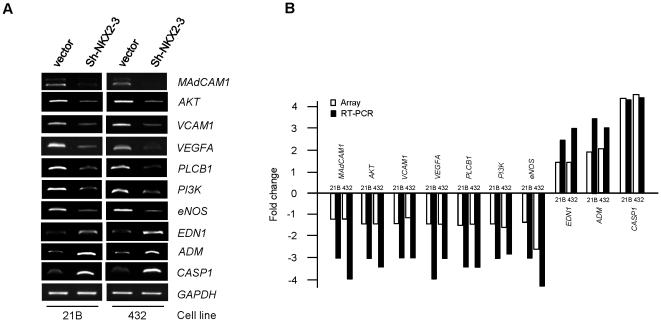
Validation of cDNA microarray by RT-PCR
RT-PCR on 10 genes (involved in or affected by EDN1 and VEGF pathways) was carried out as an independent verification method of the microarray results. Among the 10 genes, seven (MAdCAM1, AKT, VCAM1, VEGF, PLCB1, PI3K, and eNOS) were down-regulated by NKX2-3 knockdown in both HIMEC, and three (EDN1, ADM, and CASP1) were up-regulated by NKX2-3 knockdown in 2 HIMEC detected by cDNA microarray. All RT-PCR results were consistent with the microarray data. The fold changes in microarray between the 2 NKX2-3 knockdown cell lines and 2 control HIMEC lines were: MAdCAM1 (−1.2;−1.2), AKT (−1.35;−1.35), VCAM1 (−1.3;−1.14), VEGFA (−1.3;−1.3), PLCB1 (−1.6;−1.5), PI3K (−1.5;−1.7), eNOS (−1.3;−2.7), EDN1 (1.2;1.3), ADM (1.9;2.1), and CASP1 (4.6;5). The PCR ratios between the 2 NKX2-3 knockdown cell lines and 2 control HIMEC lines were: MAdCAM1 (−3;−4), AKT (−3;−3.5), VCAM1 (−3;−3), VEGFA (−4;−3), PLCB1 (−3.5;−3.5), PI3K (−3;−2.8), eNOS (−3;−4.5), EDN1 (2.5;3), ADM (3.5;3), and CASP1 (4.5;4.8) (Fig. 3).
Figure 3. Validation of microarray results by RT-PCR in two NKX2-3 knockdown HIMEC.
(A) RT-PCR analysis of mRNA expression levels of 10 genes in knockdown and control cells. EDN1, ADM, and CASP1 showed increased mRNA expression levels in NKX2-3 knockdown cells compared with control cells; MAdCAM1, AKT, VCAM1, VEGF, PLCB1, PI3K, and eNOS showed decreased mRNA expression levels in NKX2-3 knockdown cells compared with control cells. GAPDH expression served as a control. (B) Microarray results and corresponding RT-PCR for the 10 genes. The RT-PCR product bands on the photograph were scanned by densitometry. The relative mRNA expression level was expressed as gene expression levels in knockdown cells compared to gene expression levels in control cells.
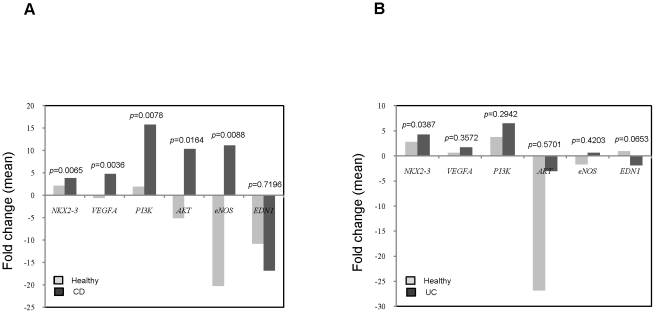
Expression of NKX2-3, EDN1, VEGFA, PI3K, AKT, and eNOS in intestinal tissues from IBD patients
NKX2-3 is reported to be up-regulated in intestinal tissues in CD patients [13] and VEGF showed markedly enhanced expression levels in both CD and UC tissues [16], while EDN1 showed both increased levels [17] and decreased levels in intestinal tissues from IBD patients [18]. Since NKX2-3 can affect the endothelin-1 and VEGF-PI3K/AKT-eNOS pathways (Fig. 2B and C) in HIMEC, we examined mRNA expression levels of the 6 genes in diseased and adjacent normal intestinal tissues from IBD patients to study clinical implication of NKX2-3 and its regulated genes. 31 CD and 32 UC patients are for this study (Table 2).
Table 2. Patient demographic data.
| Crohn's disease | Ulcerative colitis | |
| (n = 31) | (n = 32) | |
| Sex M/F | 14/17 | 16/16 |
| Family history for IBD (yes/no) | 8/23 | 5/27 |
| Average age | 39.9±15.1 | 48.4±16.3 |
| IBD location | ||
| terminal ileum | 16 | 1 |
| small bowel | 5 | 1 |
| colon | 8 | 26 |
| cecum | 2 | 3 |
| rectum | 0 | 1 |
As shown in Fig. 4A, the mRNA expression levels of NKX2-3, VEGFA, PI3K, AKT, and eNOS are significantly increased in diseased intestinal tissues compared with adjacent normal tissues in CD patients. The overall mean NKX2-3 expressions were: 3.88±2.42 (CD) vs. 2.11±1.4 (normal) (p = 0.0065); VEGFA expressions were: 4.8±15.57(CD) vs. −0.55±7.43 (normal) (p = 0.0036); PI3K expressions were: 15.78±35.76 (CD) vs. 1.91±7.61 (normal) (p = 0.0078); AKT expressions were: 10.32±30.75 (CD) vs. −5.05±24.58 (normal) (p = 0.0164); and eNOS expressions were: 11.13±48.31(CD) vs. −20.22±55.52 (normal) (p = 0.0088). Expression levels of EDN1 are decreased in diseased intestinal tissues compared with adjacent normal tissues in CD patients: −16.82±83.13 (CD) vs. −10.77±26.94 (normal) (p = 0.7196).
Figure 4. mRNA expression levels of 6 genes in intestinal tissues from CD and UC patients.
(A) Comparison of mRNA expression levels of 6 genes in diseased vs. adjacent normal intestinal tissues from CD patients: NKX2-3 (n = 30), VEGFA (n = 30), PI3K (n = 31), AKT (n = 28), eNOS (n = 24), and EDN1 (n = 30). RT-PCR was carried out on surgically excised diseased and adjacent normal intestinal tissues from CD patients. The RT-PCR product bands on the photograph were scanned by densitometry. RT-PCR results were normalized by GAPDH as fold change for each patient. Data are presented as the means. (B) Comparison of mRNA expression levels of 6 genes in diseased vs. adjacent normal intestinal tissues from UC patients: NKX2-3 (n = 30), VEGFA (n = 32), PI3K (n = 32), AKT (n = 21), eNOS (n = 23), and EDN1 (n = 27). RT-PCR was carried out on surgically excised diseased and adjacent normal intestinal tissues from UC patients. Data are presented as the means.
As shown in Fig. 4B, the mRNA expression levels of NKX2-3, VEGFA, PI3K, AKT, and eNOS are increased in diseased intestinal tissues compared with adjacent normal tissues in UC patients. The overall mean NKX2-3 expressions were: 4.26±3.39 (UC) vs. 2.78±1.68 (normal) (p = 0.0387); VEGFA expressions were: 1.7±1.53 (UC) vs. 0.65±3.13 (normal) (p = 0.3572); PI3K expressions were: 6.46±7.63 (UC) vs. 3.73±5.22 (normal) (p = 0.2942); AKT expressions were: −3±5.63 (UC) vs. −26.79±45.65 (normal) (p = 0.5701); and eNOS expressions were: 0.62±2.04 (UC) vs. −1.67±5.19 (normal) (p = 0.4203). Expression levels of EDN1 are decreased in diseased intestinal tissues compared with adjacent normal tissues in UC patients: −1.85±8.7 (UC) vs. 0.97±4.29 (normal) (p = 0.0653).
Taken together, expression levels of NKX2-3, VEGFA, PI3K, AKT, and eNOS are increased in intestinal tissues from IBD patients. On the other hand, expression levels of EDN1 are decreased in intestinal tissues from IBD patients. These results demonstrated the important roles of these genes in IBD pathogenesis.
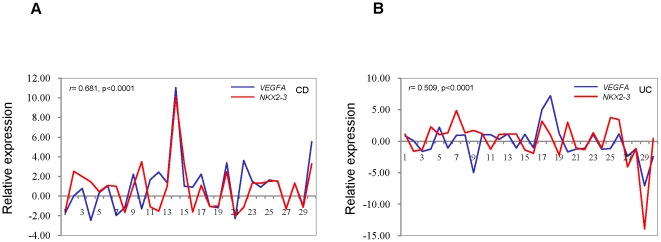
Positive correlation of gene expression of NKX2-3 with VEGFA in intestinal tissues from IBD patients
There are seven members of the vascular endothelial growth factors (VEGFs) family, ie, VEGF-A, -B, -C, -D, -E, -F, and placental growth factor. VEGFA is crucially involved in several chronic inflammatory disorders in which it not only promotes pathologic angiogenesis but directly fosters inflammation [19], [20]. VEGFA has been reported to over-express in humans with IBD [21], [22], [23]. VEGFA was down-regulated by NKX2-3 knockdown in 2 HIMEC (Fig. 2C and 3). Since both NKX2-3 and VEGFA are up-regulated in intestinal tissues from IBD patients (Fig. 4), next we examined whether NKX2-3 expression was correlated with VEGFA expression in IBD patients. We examined mRNA expression levels of NKX2-3 and VEGFA in diseased and adjacent normal intestinal tissues from 30 CD and 30 UC patients. Fold change results of normalized NKX2-3 or VEGFA were summarized as a ratio of medians (CD or UC: adjacent normal tissue) in every patient. Correlation analysis showed a positive correlation between expression of NKX2-3 and VEGFA (r = 0.681, p<0.0001) for CD (Fig. 5A) and for UC (r = 0.509, p<0.0001) (Fig. 5B).
Figure 5. The correlation between the expression levels of NKX2-3 and VEGFA in intestinal tissues from IBD patients.
(A) The correlation between the expression levels of NKX2-3 and VEGFA in diseased and adjacent normal intestinal tissues from 30 CD patients. RT-PCR was carried out on surgically excised intestinal tissues. The RT-PCR product bands on the photograph were scanned by densitometry. RT-PCR results were normalized by GAPDH for each sample. Fold change results of normalized NKX2-3 or VEGFA were summarized as a ratio of medians (CD: adjacent normal tissue) in every patient. Correlation coefficient (r) and p value are shown. (B) The correlation between the expression levels of NKX2-3 and VEGFA in diseased and adjacent normal intestinal tissues from 30 UC patients.
Negative correlation of gene expression of NKX2-3 with EDN1 in intestinal tissues from IBD patients
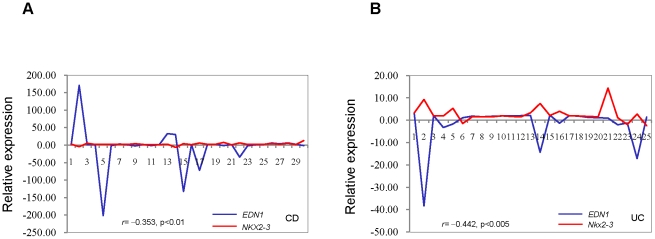
Endothelin-1 (EDN1) is a vasoactive peptide implicated in a number of pathological conditions, including human IBD [17]. EDN1 was up-regulated by NKX2-3 knockdown in 2 HIMEC by cDNA microarray analysis (Fig. 2B) and this observation was confirmed by RT-PCR (Fig. 3). Since NKX2-3 is up-regulated and EDN1 is down-regulated in intestinal tissues from IBD patients (Fig. 4), we examined whether NKX2-3 expression was correlated with EDN1 expression in IBD patients. We examined mRNA expression levels of NKX2-3 and EDN1 in diseased intestinal tissues from 30 CD and 25 UC patients. Correlation analysis showed a negative correlation between expression of NKX2-3 and EDN1 for CD (r = −0.353, p<0.01) (Fig. 6A) and for UC (r = −0.442, p<0.005) (Fig. 6B).
Figure 6. The correlation between the expression levels of NKX2-3 and EDN1 in intestinal tissues from IBD patients.
(A) The correlation between the expression levels of NKX2-3 and EDN1 in diseased intestinal tissues from 30 CD patients. RT-PCR was carried out on surgically excised intestinal tissues. The RT-PCR product bands on the photograph were scanned by densitometry. RT-PCR results were normalized by GAPDH in every patient. Correlation coefficient (r) and p value are shown. (B) The correlation between the expression levels of NKX2-3 and EDN1 in diseased intestinal tissues from 25 UC patients.
Discussion
Pathogenesis of IBD is not only restricted to those mediated by classic immune cells, such as T and B cells [24], but also involves nonimmune cells, including endothelial cells [15]. Endothelial cells play a key role in mucosal immune homeostasis by regulating the leukocytes migrating from the intravascular to the interstitial space, thus highlighting the endothelium as one of the pillars in inflammation pathogenesis [25]. The vascular response is a key component of inflammation. Microvascular endothelial cells could form capillary-like structures and display different functional sets of adhesion molecules, distinct chemokine secretory patterns, and activation of unique sets of genes in response to stress and inflammatory stimuli [26].
In this study, we performed genome-wide gene expression microarray analysis using two HIMEC with NKX2-3 knockdown, and identified 1746 genes in 21B cells and 1603 genes in 432 cells regulated by NKX2-3 knockdown. The regulation of these genes is highly consistent between the two HIMEC. Most of the NKX2-3 regulated genes are involved in immune and inflammatory response, cell proliferation and growth, metabolic process, and angiogenesis. Various aspects of immunity contribute to the development of an overall inflammatory immune response. Chemokines control leukocyte trafficking during homeostasis as well as inflammation. The migration of leukocytes into sites of inflammation is crucial in the pathogenesis of IBD. The CXC chemokine IL8/CXCL8 is down-regulated by NKX2-3 knockdown in HIMEC. IL8 is expressed in leucocytes and endothelial cells and plays an important role in inflammation and angiogenesis [27]. The expression level of IL8 is up-regulated in tissues after ischaemic injury [28]. One of the most novel aspects that directly implicates endothelial cell participation in inflammation is the process of angiogenesis. It is now well established that angiogenesis and microvascular remodeling are intrinsic components of the tissue remodeling in chronic inflammatory diseases [15]. ANGPT2 (angiopoietin 2) is down-regulated by NKX2-3 knockdown in HIMEC. ANGPT2 is responsible for the initiation of angiogenesis through recruitment and proliferation of endothelial cells [29]. Elevated serum ANGPT2 levels in IBD patients have been reported [30].
We used pathway analysis to systematically visualize the relationships of genes regulated by NKX2-3 knockdown. The top 25 pathways affected by NKX2-3 knockdown included those involved in inflammation and immune response, including the EDN1 and NO pathways. Generation of nitric oxide (NO) by NO synthase (NOS) is a central feature of chronic inflammatory diseases in the gastrointestinal tract [15]. eNOS is down-regulated by NKX2-3 knockdown in HIMEC. We identified that NKX2-3 regulated eNOS through the VEGF-PI3k/AKT pathway in HIMEC. VEGF, PI3K, and AKT are all down-regulated by NKX2-3 knockdown in HIMEC. A few reports have described overexpression of VEGFA in human intestinal tissues with IBD [21], [22], [23], but the functional significance of such up-regulation is not yet understood. In murine colonic-derived endothelial cells, VEGFA triggers an inflammatory phenotype by up-regulating CAMs and inducing adhesion of neutrophils and T cells, thus supporting an inflammatory role for this cytokine in the intestine [31]. One recent study showed that in vitro, VEGF-A induces both angiogenic activity and an inflammatory phenotype in human intestinal microvascular endothelial cells (HIMEC), whereas overexpression in vivo increases disease severity and blockade decreases disease severity in colitic mice [23]. Given that the activation of this VEGF–eNOS angiogenesis pathway results in maintenance production of NO, it is most likely to be implicated in the regulation of NO-controlled gene expression, as well as the NO-mediated angiogenic responses to VEGF. VEGF is known to be a potent activator of endothelial cells and a critical factor for the induction of angiogenesis during IBD development [23]. The importance of the VEGF–eNOS pathway for regulating angiogenesis and vascular remodeling makes these genes important for IBD pathogenesis. Our results showed that mRNA expression levels of NKX2-3, VEGFA, PI3K, AKT, and eNOS were increased in intestinal tissues from IBD patients. And the positive correlation between the expression levels of NKX2-3 and VEGFA in intestinal tissues from IBD patients (Fig. 5) suggests a regulatory role of NKX2-3 in VEGFA gene expression. The mechanism of regulation of VEGF by NKX2-3 is not clear. NKX2-3 could regulate expression of IL8, ANGPT2, KLF2, and KLF4 in HIMEC. IL8 Stimulates VEGF expression in endothelial cells [32] and ANGPT2 stimulates the synthesis of VEGF [33]. The KLF2 and KLF4 transcription factors have been shown to coordinate transcriptional programs important for the establishment of an anti-inflammatory, vasodilatory, and anti-thrombotic vascular endothelial phenotypes, thus acting as critical regulators of endothelial homeostasis [34]. KLF2 and KLF4 regulate expression of VEGF and eNOS in vascular endothelial cells [34]. It is reasonable to speculate that NKX2-3 regulates VEGF expression through IL8, ANGPT2, KLF2, and KLF4 or direct regulation of VEGF. Although the role of NKX2-3 in IBD is still unclear, its regulatory role in VEGF-eNOS strongly suggests that NKX2-3 could be involved in IBD pathogenesis by regulating the VEGF-PI3K/AKT-eNOS pathway.
Endothelin-1 (EDN1) is one of several vasoconstrictors that may play a role in the progression of IBD. Endothelin-1 is expressed by endothelial cells as a precursor peptide (proET-1) that is first cleaved to bigET-1 and then to the mature 21-amino acid peptide. EDN1 acts through two receptors, ET-A and ET-B. EDN1 binding to ET-B receptors in endothelial cells initiates a signaling cascade that leads to nitric oxide (NO) and endothelial-derived relaxing factor production. The signaling of NO production through ET-B receptors via the G-protein βγ subunit dimer has been linked to AKT phosphorylation of eNOS. The G-protein Galpha12 has been linked to increased levels of eNOS [35]. EDN1 increases leukocyte adhesion to intestinal microvasculature [36], resulting in oxidant stress and mucosal dysfunction [37]. Increased levels of EDN1 in intestinal tissues from IBD patients have been reported [17]. However, other reports have indicated that EDN1 levels were decreased in IBD [18], [38]. We showed that expression of EDN1 was down-regulated in intestinal tissues from IBD patients. Correlation study confirmed the negative correlation between the expression levels of NKX2-3 and EDN1 in intestinal tissues from IBD patients (Fig. 6), suggesting a regulatory role of NKX2-3 in EDN1 gene expression. NKX2-3 could negatively regulate EDN1 expression and is thus involved in IBD pathogenesis.
In summary, this work identified many inflammation-related genes and microvascular endothelial cell function related genes that are regulated by NKX2-3 in HIMEC. Our present results demonstrate that a decrease in NKX2-3 gene expression level can profoundly affect the signaling pathways relevant to the pathogenesis and progression of IBD such as the EDN1 and VEGF signaling and PI3K/AKT-eNOS pathways. Further functional studies of the genes and pathways affected by NKX2-3 using HIMEC are underway to confirm and expand upon the current work.
Materials and Methods
Cell lines
Two HIMEC (432 and 21B) were isolated from normal ileal intestinal tissue from patients undergoing surgery at the Cleveland Clinic Lerner Research Institute as approved by the Institutional Review Board of University Hospitals of Cleveland. Cell lines were cultured in MCDB131 medium (Invitrogen, USA) supplemented with 20% fetal bovine serum (FBS), heparin, endothelial cell growth factor and antibiotics. HIMEC were incubated at 37°C in an atmosphere of 5% CO2.
Transfection
A small hairpin RNA (shRNA) vector targeting human NKX2-3 was generated using the pSUPER vector system as described previously [14], [39]. The 19-nucleotide sequence within NKX2-3 targeted by the shRNA oligonucleotide pair was 5′-AGGAACATGAAGAGGAGCC-3′. Forward and reverse primers were synthesized containing this sequence in sense and antisense orientations with an intervening linker. Forward and reverse primers were annealed and ligated into the pSUPER.retro.puro vector according to the manufacturer's instructions (OligoEngine, WA, USA).
pSUPER.retro.puro.shRNA-NKX2-3 and empty vector were transfected into two HIMEC lines with TransPass™ HUVEC transfection reagent (New England BioLabs, USA) according to the manufacturer's instructions. Forty-eight hours posttransfection, to confirm the suppression of NKX2-3 expression, mRNA from was isolated from the transfected HIMEC lines for further examination.
Microarray and data analysis
Illumina Human HT12 v 3 Expression BeadChips (Illumina, CA) were used in this study. This BeadChip targets >25,000 genes with >48,000 probes derived from the RefSeq (Build 36.2, rel22) and UniGene (Build 99) databases. 300 ng of total RNA collected from two independent cultures of control and shNKX2-3-expressing HIMEC cells was reverse transcribed into cRNA and biotin-UTP labeled using the Illumina Total Prep RNA Amplification Kit (Ambion, Austin, TX). Microarray hybridization, data collection, and analysis were performed at the Genomics Core of the Cleveland Clinic Lerner Research Institute.
Raw data filtering and quantile normalization were performed using the Bioconductor package lumi, a Beadarray specific software package for Illumina microarray data. Due to the small sample size, the moderated t-statistic implemented in the Bioconductor LIMMA package was used to detect differentially expressed genes. This statistic has the same interpretation as the standard t-statistic; however, standard errors were calculated to shrink toward a common value by empirical Bayes model to borrow information across all genes [40]. The p-values from moderated t-tests were adjusted by Benjamini and Hochberg's method to control false discovery rate. All data is MIAME (Minimum Information About a Microarray Experiment) compliant and that the raw data has been deposited in a MIAME compliant database (GEO at NCBI, accession number GSE28656).
Ingenuity pathway analysis (IPA)
IPA was used to identify gene networks according to biological functions and/or diseases in the Ingenuity Pathways Knowledge Base (Ingenuity Systems, Redwood City, CA). Known genes' expression levels served as input to the Ingenuity Pathways Analysis (IPA) Knowledge Base v4.0. Lists of top pathways associated with genes with changes in expression relative to controls were generated with corresponding Benjamini and Hochberg's p values. Expression data was overlaid upon canonical pathways associated with altered gene expression.
Patients and intestinal tissue samples
Intestinal tissues were obtained from IBD patients undergoing surgery at the Penn State Hershey Medical Center. All human tissues were approved by the Human Subjects Protection Offices of The Pennsylvania State University College of Medicine, and were undertaken with the understanding and written consent of each subject. Macroscopically normal areas of intestine and areas of intestine with obvious disease were classified by a pathologist. The intestinal tissues were immediately submerged in RNAlater Solution (Ambion, CA, USA) and stored at 4°C overnight. Tissues were stored frozen at −70°C until total RNA extraction.
RNA isolation and reverse transcription PCR
Total RNA was extracted from HIMEC and intestinal tissues using the RNeasy mini kit (QIAGEN Sciences, MD, USA) according to the manufacturer's instructions. cDNA was synthesized from 1.0 µg of total RNA using a Superscript III 1st Strand Synthesis Kit (Invitrogen, CA, USA).
Primer sequences are listed in table 3. Primers were designed using Primer3 software. PCR amplifications were performed at 94°C for 30 seconds, 60°C for 45 seconds, and 72°C for 45 seconds for 35 cycles (all genes except GAPDH) or 26 cycles (GAPDH), followed by a final extension at 72°C for 5 min. PCR products were visualized on 2% agarose gels stained with ethidium bromide. RT-PCR product bands were scanned by densitometry and results were normalized by GAPDH for each cell line.
Table 3. Primers for RT-PCR.
| Gene | Primer sequences | Sizes | Accession |
| EDN1 | F: 5′- ctttgagggacctgaagctg-3′ | 392 bp | NM_001955 |
| R: 5′- ctgttgcctttgtgggaagt-3′ | |||
| ADM | F: 5′- acttcggagttttgccattg-3′ | 230 bp | NM_001124 |
| R: 5′- ctcttcccacgactcagagc-3′ | |||
| PI3K | F: 5′- tgctttgggacaaccataca-3′ | 394 bp | NM_006218 |
| R: 5′- cggttgcctactggttcaat-3′ | |||
| AKT | F: 5′- ggtgatcctggtgaaggaga-3′ | 379 bp | NM_005163 |
| R: 5′- cttaatgtgcccgtccttgt-3′ | |||
| eNOS | F: 5′- tgctggcatacaggactaag-3′ | 385 bp | NM_000603 |
| R: 5′- taggtcttggggttgtcagg-3′ | |||
| PLCB1 | F: 5′- cgtggctttccaagaagaag-3′ | 305 bp | NM_015192 |
| R: 5′- ggcaaaggttgttgaggaaa-3′ | |||
| CASP1 | F: 5′- acctctgacagcacgttcct-3′ | 329 bp | NM_033292 |
| R: 5′- ggtgtggaagagcagaaagc-3′ | |||
| VCAM1 | F: 5′- attgacttgcagcaccacag-3′ | 391 bp | NM_001078 |
| R: 5′- ttccagggacttcctgtctg-3′ | |||
| VEGFA | F: 5′- tcctcacaccattgaaacca-3 | 379 bp | NM_001171623 |
| R: 5′- caccgatcagggagagagag-3′ | |||
| NKX2-3 | F: 5′-ccacccctttctcagtcaaa-3′ | 210 bp | NM_145285 |
| R: 5′-ctgcggctagtgagttcaaa-3′ | |||
| GAPDH | F: 5′-tgatgacatcaagaaggtggtgaag-3′ | 236 bp | NM_002046 |
| R: 5′-tccttggaggccatgtgggccat-3′ |
Statistical analysis
The paired data (RT-PCR results) were analyzed with a one sample permutation t-test with 500,000 permutations. The analysis was performed using R (version 2.12.0) and the onet.permutation function in the R library DAAG (version 1.03).
Acknowledgments
We thank Dr. Claudio Fiocchi (Department of Pathobiology, the Cleveland Clinic Foundation) for kindly providing two HIMEC lines. Gene expression profiling microarray experiments were performed at the Genomics Core of the Cleveland Clinic Lerner Research Institute (contact P.W. Faber, Ph.D., faberp@ccf.org). We thank Dr. Faber for his assistance with the cDNA microarray experiments.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by a grant from the Philadelphia Health Care Trust (WAK), the Carlino fund for IBD research at the Milton S. Hershey Medical Center (WAK), Penn State College of Medicine, and the Milton S. Hershey Penn State College of Medicine Surgery Research Feasibility Grant (WY). Budget: 942-82HY, Fund: 1940. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Cho JH. The genetics and immunopathogenesis of inflammatory bowel disease. Nat Rev Immunol. 2008;8:458–466. doi: 10.1038/nri2340. [DOI] [PubMed] [Google Scholar]
- 2.Wellcome Trust Case Control Consortium (WTCCC) Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franke A, Balschun T, Karlsen TH, Hedderich J, May S, et al. Replication of signals from recent studies of Crohn's disease identifies previously unknown disease loci for ulcerative colitis. Nat Genet. 2008;40:713–715. doi: 10.1038/ng.148. [DOI] [PubMed] [Google Scholar]
- 4.Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 5.Lawrence PA, Morata G. Homeobox genes: their function in Drosophila segmentation and pattern formation. Cell. 1994;78:181–189. doi: 10.1016/0092-8674(94)90289-5. [DOI] [PubMed] [Google Scholar]
- 6.Buchberger A, Pabst O, Brand T, Seidl K, Arnold HH. Chick NKx-2.3 represents a novel family member of vertebrate homologues to the Drosophila homeobox gene tinman: differential expression of cNKx-2.3 and cNKx-2.5 during heart and gut development. Mech Dev. 1996;56:151–163. doi: 10.1016/0925-4773(96)00521-7. [DOI] [PubMed] [Google Scholar]
- 7.Fu Y, Yan W, Mohun TJ, Evans SM. Vertebrate tinman homologues XNkx2-3 and XNkx2-5 are required for heart formation in a functionally redundant manner. Development. 1998;125:4439–4449. doi: 10.1242/dev.125.22.4439. [DOI] [PubMed] [Google Scholar]
- 8.Pabst O, Schneider A, Brand T, Arnold HH. The mouse Nkx2-3 homeodomain gene is expressed in gut mesenchyme during pre- and postnatal mouse development. Dev Dyn. 1997;209:29–35. doi: 10.1002/(SICI)1097-0177(199705)209:1<29::AID-AJA3>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 9.Wang CC, Biben C, Robb L, Nassir F, Barnett L, et al. Homeodomain factor Nkx2-3 controls regional expression of leukocyte homing coreceptor MAdCAM-1 in specialized endothelial cells of the viscera. Dev Biol. 2000;224:152–167. doi: 10.1006/dbio.2000.9749. [DOI] [PubMed] [Google Scholar]
- 10.Tarlinton D, Light A, Metcalf D, Harvey RP, Robb L. Architectural defects in the spleens of Nkx2-3-deficient mice are intrinsic and associated with defects in both B cell maturation and T cell-dependent immune responses. J Immunol. 2003;170:4002–4010. doi: 10.4049/jimmunol.170.8.4002. [DOI] [PubMed] [Google Scholar]
- 11.Pabst O, Zweigerdt R, Arnold HH. Targeted disruption of the homeobox transcription factor Nkx2-3 in mice results in postnatal lethality and abnormal development of small intestine and spleen. Development. 1999;126:2215–2225. doi: 10.1242/dev.126.10.2215. [DOI] [PubMed] [Google Scholar]
- 12.Hatoum OA, Binion DG. The vasculature and inflammatory bowel disease: contribution to pathogenesis and clinical pathology. Inflamm Bowel Dis. 2005;11:304–313. doi: 10.1097/01.mib.0000160772.78951.61. [DOI] [PubMed] [Google Scholar]
- 13.Yu W, Lin Z, Kelly AA, Hegarty JP, Poritz LS, Wang Y, Li T, Schreiber S, Koltun WA. Association of a Nkx2-3 polymorphism with Crohn's disease and expression of Nkx2-3 is up-regulated in B cell lines and intestinal tissues with Crohn's disease. Journal of Crohn's and Colitis. 2009;3:189–195. doi: 10.1016/j.crohns.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 14.Yu W, Lin Z, Hegarty JP, John G, Chen X, et al. Genes regulated by Nkx2-3 in siRNA-mediated knockdown B cells: implication of endothelin-1 in inflammatory bowel disease. Mol Genet Metab. 2010;100:88–95. doi: 10.1016/j.ymgme.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Deban L, Correale C, Vetrano S, Malesci A, Danese S. Multiple pathogenic roles of microvasculature in inflammatory bowel disease: a Jack of all trades. Am J Pathol. 2008;172:1457–1466. doi: 10.2353/ajpath.2008.070593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsiolakidou G, Koutroubakis IE, Tzardi M, Kouroumalis EA. Increased expression of VEGF and CD146 in patients with inflammatory bowel disease. Dig Liver Dis. 2008;40:673–679. doi: 10.1016/j.dld.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 17.Murch SH, Braegger CP, Sessa WC, MacDonald TT. High endothelin-1 immunoreactivity in Crohn's disease and ulcerative colitis. Lancet. 1992;339:381–385. doi: 10.1016/0140-6736(92)90077-g. [DOI] [PubMed] [Google Scholar]
- 18.Rachmilewitz D, Eliakim R, Ackerman Z, Karmeli F. Colonic endothelin-1 immunoreactivity in active ulcerative colitis. Lancet. 1992;339:1062. doi: 10.1016/0140-6736(92)90589-u. [DOI] [PubMed] [Google Scholar]
- 19.Takahashi H, Shibuya M. The vascular endothelial growth factor (VEGF)/VEGF receptor system and its role under physiological and pathological conditions. Clin Sci (Lond) 2005;109:227–241. doi: 10.1042/CS20040370. [DOI] [PubMed] [Google Scholar]
- 20.Lee YC. The involvement of VEGF in endothelial permeability: a target for anti-inflammatory therapy. Curr Opin Investig Drugs. 2005;6:1124–1130. [PubMed] [Google Scholar]
- 21.Danese S. Inflammation and the mucosal microcirculation in inflammatory bowel disease: the ebb and flow. Curr Opin Gastroenterol. 2007;23:384–389. doi: 10.1097/MOG.0b013e32810c8de3. [DOI] [PubMed] [Google Scholar]
- 22.Koutroubakis IE, Tsiolakidou G, Karmiris K, Kouroumalis EA. Role of angiogenesis in inflammatory bowel disease. Inflamm Bowel Dis. 2006;12:515–523. doi: 10.1097/00054725-200606000-00012. [DOI] [PubMed] [Google Scholar]
- 23.Scaldaferri F, Vetrano S, Sans M, Arena V, Straface G, et al. VEGF-A links angiogenesis and inflammation in inflammatory bowel disease pathogenesis. Gastroenterology. 2009;136:585–595 e585. doi: 10.1053/j.gastro.2008.09.064. [DOI] [PubMed] [Google Scholar]
- 24.Fiocchi C. Intestinal inflammation: a complex interplay of immune and nonimmune cell interactions. Am J Physiol. 1997;273:G769–775. doi: 10.1152/ajpgi.1997.273.4.G769. [DOI] [PubMed] [Google Scholar]
- 25.Danese S, Dejana E, Fiocchi C. Immune regulation by microvascular endothelial cells: directing innate and adaptive immunity, coagulation, and inflammation. J Immunol. 2007;178:6017–6022. doi: 10.4049/jimmunol.178.10.6017. [DOI] [PubMed] [Google Scholar]
- 26.Binion DG, West GA, Ina K, Ziats NP, Emancipator SN, et al. Enhanced leukocyte binding by intestinal microvascular endothelial cells in inflammatory bowel disease. Gastroenterology. 1997;112:1895–1907. doi: 10.1053/gast.1997.v112.pm9178682. [DOI] [PubMed] [Google Scholar]
- 27.Koch AE, Polverini PJ, Kunkel SL, Harlow LA, DiPietro LA, et al. Interleukin-8 as a macrophage-derived mediator of angiogenesis. Science. 1992;258:1798–1801. doi: 10.1126/science.1281554. [DOI] [PubMed] [Google Scholar]
- 28.Kocher AA, Schuster MD, Bonaros N, Lietz K, Xiang G, et al. Myocardial homing and neovascularization by human bone marrow angioblasts is regulated by IL-8/Gro CXC chemokines. J Mol Cell Cardiol. 2006;40:455–464. doi: 10.1016/j.yjmcc.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 29.Ramsauer M, D'Amore PA. Getting Tie(2)d up in angiogenesis. J Clin Invest. 2002;110:1615–1617. doi: 10.1172/JCI17326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oikonomou KA, Kapsoritakis AN, Kapsoritaki AI, Manolakis AC, Tiaka EK, et al. Angiogenin, angiopoietin-1, angiopoietin-2, and endostatin serum levels in inflammatory bowel disease. Inflamm Bowel Dis. doi: 10.1002/ibd.21410. [DOI] [PubMed] [Google Scholar]
- 31.Goebel S, Huang M, Davis WC, Jennings M, Siahaan TJ, et al. VEGF-A stimulation of leukocyte adhesion to colonic microvascular endothelium: implications for inflammatory bowel disease. Am J Physiol Gastrointest Liver Physiol. 2006;290:G648–654. doi: 10.1152/ajpgi.00466.2005. [DOI] [PubMed] [Google Scholar]
- 32.Martin D, Galisteo R, Gutkind JS. CXCL8/IL8 stimulates vascular endothelial growth factor (VEGF) expression and the autocrine activation of VEGFR2 in endothelial cells by activating NFkappaB through the CBM (Carma3/Bcl10/Malt1) complex. J Biol Chem. 2009;284:6038–6042. doi: 10.1074/jbc.C800207200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang YS, Park YG, Kim BK, Han SY, Jee YH, et al. Angiotensin II stimulates the synthesis of vascular endothelial growth factor through the p38 mitogen activated protein kinase pathway in cultured mouse podocytes. J Mol Endocrinol. 2006;36:377–388. doi: 10.1677/jme.1.02033. [DOI] [PubMed] [Google Scholar]
- 34.Villarreal G, Jr, Zhang Y, Larman HB, Gracia-Sancho J, Koo A, et al. Defining the regulation of KLF4 expression and its downstream transcriptional targets in vascular endothelial cells. Biochem Biophys Res Commun. 391:984–989. doi: 10.1016/j.bbrc.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andreeva AV, Vaiskunaite R, Kutuzov MA, Profirovic J, Skidgel RA, et al. Novel mechanisms of G protein-dependent regulation of endothelial nitric-oxide synthase. Mol Pharmacol. 2006;69:975–982. doi: 10.1124/mol.105.018846. [DOI] [PubMed] [Google Scholar]
- 36.Boros M, Massberg S, Baranyi L, Okada H, Messmer K. Endothelin 1 induces leukocyte adhesion in submucosal venules of the rat small intestine. Gastroenterology. 1998;114:103–114. doi: 10.1016/s0016-5085(98)70638-9. [DOI] [PubMed] [Google Scholar]
- 37.Oktar BK, Coskun T, Bozkurt A, Yegen BC, Yuksel M, et al. Endothelin-1-induced PMN infiltration and mucosal dysfunction in the rat small intestine. Am J Physiol Gastrointest Liver Physiol. 2000;279:G483–491. doi: 10.1152/ajpgi.2000.279.3.G483. [DOI] [PubMed] [Google Scholar]
- 38.McCartney SA, Ballinger AB, Vojnovic I, Farthing MJ, Warner TD. Endothelin in human inflammatory bowel disease: comparison to rat trinitrobenzenesulphonic acid-induced colitis. Life Sci. 2002;71:1893–1904. doi: 10.1016/s0024-3205(02)01923-9. [DOI] [PubMed] [Google Scholar]
- 39.Yu W, Lin Z, Pastor DM, Hegarty JP, Chen X, et al. Genes Regulated by Nkx2-3 in Sporadic and Inflammatory Bowel Disease-Associated Colorectal Cancer Cell Lines. Dig Dis Sci. 2010;55:3171–3180. doi: 10.1007/s10620-010-1138-0. [DOI] [PubMed] [Google Scholar]
- 40.Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21:2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]