Abstract
DNA methyltransferase 1 (DNMT1) is crucial for maintenance of methylation, gene regulation and chromatin stability1-3. DNA mismatch repair, cell cycle regulation in post-mitotic neurons4,5 and neurogenesis6 are influenced by DNA methylation. Here we show mutations in DNMT1 cause both central and peripheral neurodegeneration in one form of hereditary sensory and autonomic neuropathy (HSAN1) with dementia and hearing loss7,8. Exome sequencing led to the identification of DNMT1 mutation c.A1484G (p.Tyr495Cys) in two American and one Japanese kindreds and a triple nucleotide change c.1470TCC-1472ATA (p.Asp490Glu-Pro491Tyr) in one European kindred. All mutations are within the targeting sequence (TS) domain of DNMT1. These mutations cause premature degradation of mutant proteins, reduced methyltransferase activity and impaired heterochromatin binding during the G2 cell cycle phase, leading to global hypomethylation and site specific hypermethylation. Our study demonstrates DNMT1 mutations cause aberrant methylation implicated in complex pathogenesis. The discovered DNMT1 mutations provide a new framework for the study of neurodegenerative diseases.
Neurodegenerative diseases are hallmarked by an increased rate of neuronal axonal loss. Neural development, neural survival and connectivity have been linked to DNA methylation and chromatin stability1. Here we studied one form of neurodegeneration with both peripheral and central involvement, namely hereditary sensory and autonomic neuropathy type 1 (HSAN1) with dementia and hearing loss. As a group, HSAN1 has autosomal dominant inheritance with neurodegeneration that clinically manifests with loss of sensation leading to painless extremity injuries, infections and resultant amputations9. Serine palmitoyltransferase (SPTLC1 and SPTLC2)10,11, atlastin-1 (ATL1)12 and Ras-related GTPase 7 (RAB7)13 have been linked to some HSAN1 kindreds, but the majority of causative genes remains unknown. The specific form of HSAN1 investigated here has early onset dementia and sensorineural hearing loss that tracks with sensory neuropathy7,8. Pathologic studies of affected persons' nerve7, brain14 and spinal cord (Fig. 1) show axonal loss without interstitial infiltrative inclusions. Affected persons typically have early mortality and often require total care due to dementia, hearing loss, and lose of ambulation from predominant sensory ataxia. Four kindreds (Fig. 2a-d) were investigated, two of them were previously reported in detail7,8. Affected persons had very similar phenotypes (Table 1) and were normal in youth, but developed worsening sensorineural deafness and sensory neuropathy by ages 20-35 years. Progressive cognitive and behavioral declines occurred by the fourth decade. Brain imaging of affected persons showed global atrophy and reduced weight of autopsied brains. Among the Japanese kindred PET and SPECT imaging suggested medial frontal and thalamic hypo metabolism8. Quantitative sensory testing, nerve conductions, and nerve biopsy were indicative of length dependent progressive sensory axonal loss.
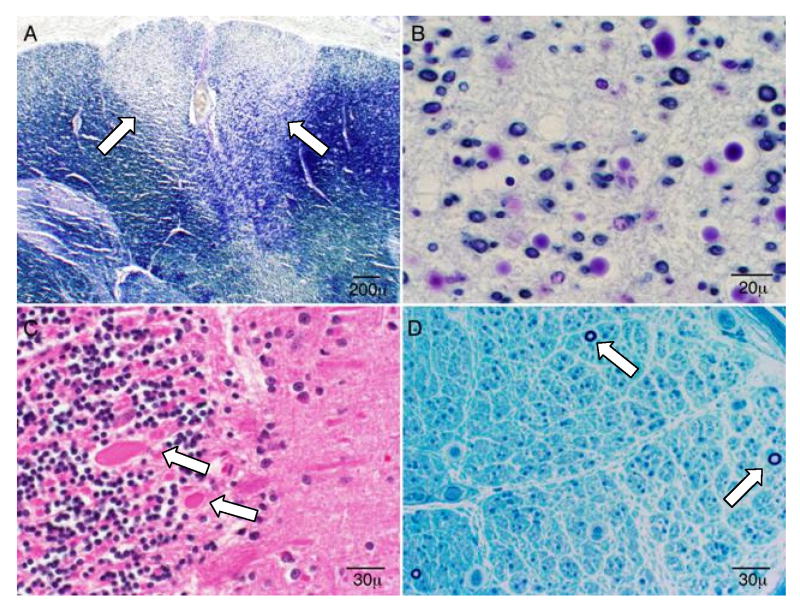
Figure 1.
Autopsy (a-c) and nerve biopsy (d) results of an affected person (VI-36, deceased at 48 y.o.) from Kindred 1. Similar results were also seen in two other affected and deceased persons from kindred 1. (a) Ascending spinal sensory tract degeneration with profound myelin and axonal loss (lighter staining, between arrowheads), involving the gracile fasciculus (medial posterior columns) at all spinal levels; (b). The neuronal loss described in panel a is shown at higher magnification; (c) Chronic cerebellar Purkinje cell swelling and axonal loss (arrowheads) with associated hyperplasia of the Bergman glia. There was also severe neuronal loss and gliosis of inferior olivary nucleus as well as generalized cerebral atrophy with brain weight of 1085 (normal 1300-1400) grams without distinct histopathologic alterations or inclusions which was determined by Bielschowsky silver and immunohistochemical stains for detection of beta-amyloid, tau, TDP-43, and alpha-synuclein; (d) Epoxy embedded sural nerve tissue showing severe loss of large and small myelinated fibers with only few fibers remaining (arrows) without distinctive interstitial infiltrative change.
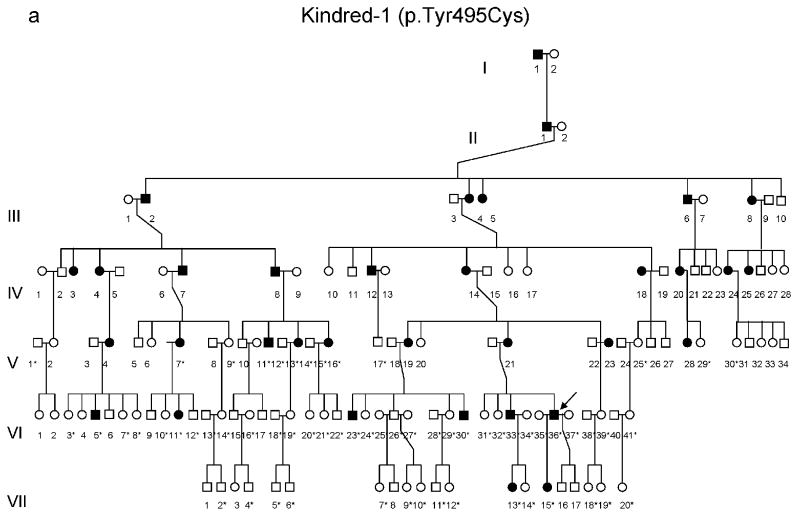
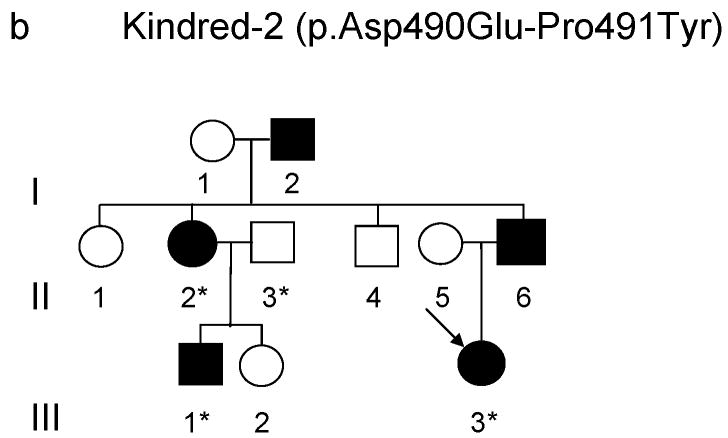
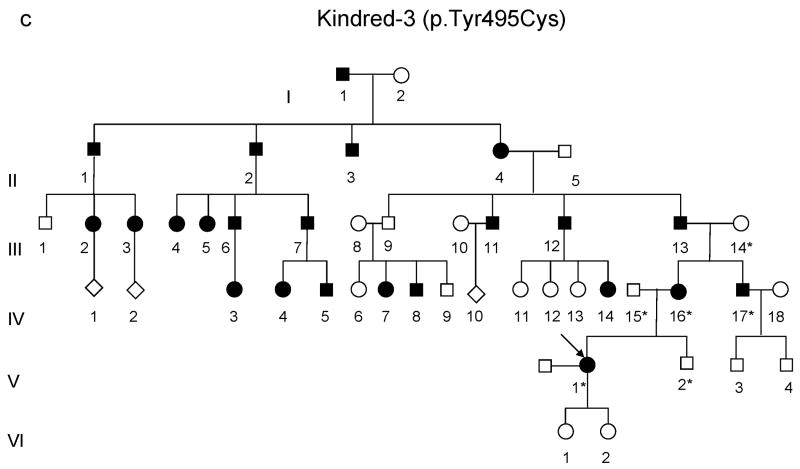
Figure 2.
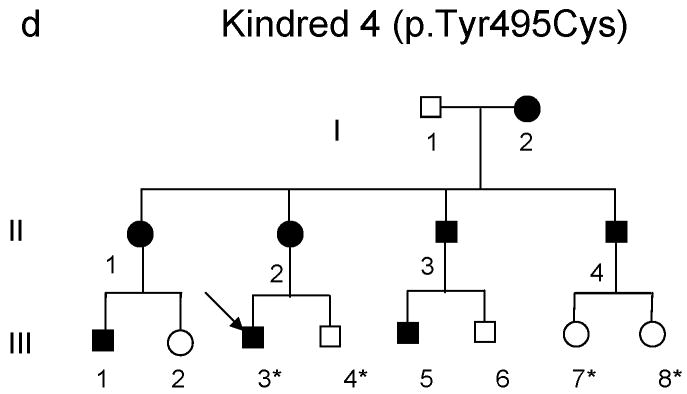
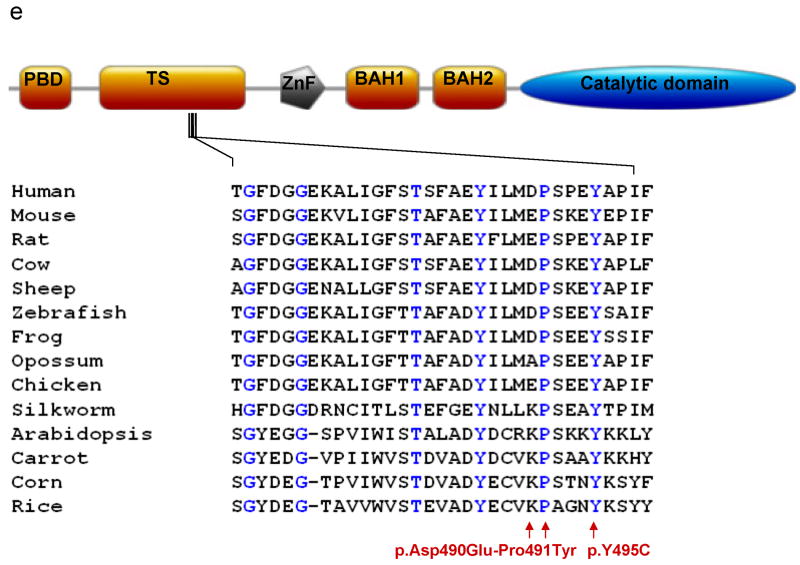
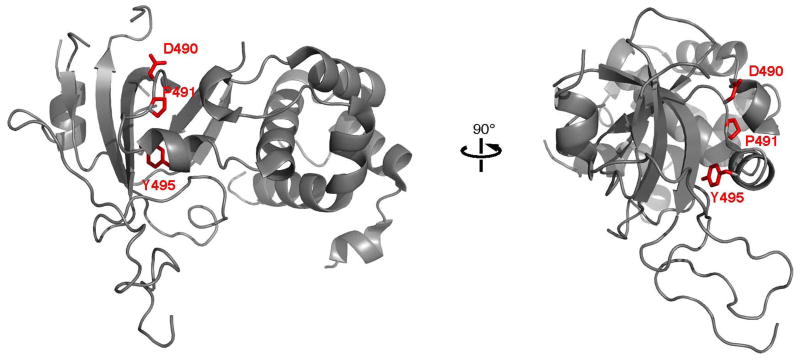
(a-d) Pedigrees of four kindreds are shown. A heterozygous mutation c.A1484G, resulting in p.Tyr495Cys, was identified in exon 20 of DNMT1 in the HSAN kindred 1(a), 3(c) and 4(d). Three consecutive heterozygous mutations c.1470TCC-1472ATA, resulting in p.Asp490Glu-Pro491Tyr substitution, also in exon 20 of DNMT1 was identified in the HSAN kindred 2(b). We sequenced all available samples (with asterisk) from these four kindreds to confirm the mutation is segregated with the disease. (e) Schematic overview of DNMT1 and its multiple domains in the N-terminal region (PBD, PCNA binding domain; TS, targeting sequence; ZnF, zinc finger; BAH1 & 2, bromo adjacent homology domains1 & 2). Below is the ClustalW alignment of the part of TS domain where mutations occur from multiple DNMT1 homologs. Comparison of human DNMT1 (P26358) and its orthologs in mouse (P13864), rat (Q9Z330), cow (Q24K09), sheep (Q865V5), zebrafish (Q8QGB8), frog (Q6GQH0), opossum (Q8MJ28), chicken (Q92072), silkworm (Q5W7N6), arebiopsis (Q9SEG3), carrot (O48867), corn (Q8LPU6) and rice (A2XMY1). Conserved amino acids are colored in blue. Red arrow points to the mutations. (f). Location of mutated residues in the TS domain of human DNMT1. Shown in red are the side chains of Asp490, Pro491 and Tyr495 in the crystal structure of the TS domain (Protein Data Bank accession number 3EPZ). The image was generated using PyMOL (http://www.pymol.org/).
Table 1. Phenotypic Characteristics of HSAN 1 Subjects with Dementia and Sensorineural Hearing Loss.
| Patient | Symptom Onset (yrs) | Age (yr) First Examined | Gender | First Symptom(s) | Extremity Sensory Loss | Distal Limb Weakness | Cerebellar ataxia | Dementia Onset(yrs) | Death (yrs) |
|---|---|---|---|---|---|---|---|---|---|
| K1 V7 | 30 | 59 | F | Hearing loss | Yes* | No | No | 40s | 61 |
| K1 V-12 | 35 | 48 | M | Loss of sensation Hearing loss | Yes | No | No | 42 | Alive |
| K1 V-14 | 30 | 44 | F | Hearing loss | Yes | No | No | 41 | - |
| K1 V-16 | 32 | 38 | F | Loss of sensation Hearing loss | Yes | Yes | No | 30s | Alive |
| K1 VI-11 | 20s | 30 | F | Hearing loss | Yes | No | No | 30s | Alive |
| K1 VI-23 | 35 | 45 | M | Hearing loss | Yes* | No | No | 40 | 51 |
| K1 VI-30 | 20s | 30 | M | Loss of sensation | Yes | No | No | 30s | Alive |
| K1 V1-33 | 35 | 43 | M | Hearing loss | Yes* | Yes | No | 30s | 46 |
| K1 VI-36 | 35 | 42 | M | Hearing loss | Yes* | No | No | 40s | 48 |
| K1 VII 13 | 20s | 18 | F | Hearing loss | Yes | No | No | 30s | Alive |
| K1 VII 15 | 20s | 20 | F | Loss of sensation | Yes | No | No | 40s | Alive |
| K2 II-2 | 28 | 32 | F | Hearing loss | Yes* | No | No | 30s | 46 |
| K2 III-1 | 27 | 38 | M | Hearing loss | Yes* | No | Yes | 30s | Alive |
| K2 III-3 | 16 | 39 | F | Hearing loss | Yes* | No | Yes | 30s | Alive |
| K3 IV-16 | 35 | 47 | F | Hearing loss | Yes* | No | No | 40s | 55 |
| K3 IV-17 | 30s | 40 | M | Loss of sensation Hearing loss | Yes | No | Yes | 40s | 47 |
| K3 V-1 | 30s | 40 | F | Loss of sensation Hearing loss | Yes* | No | Yes | 40s | 44 |
| K4 III-3 | 30s | 40s | M | Loss of sensation Hearing loss | Yes | No | No | 30s | Alive |
Foot ulcers; -- Unavailable; K kindred
We performed genome-wide linkage analysis in Kindred-1 and mapped to19p13.2 with a maximal LOD score of 6.38. Fine mapping analysis with haplotype construction narrowed the region to 3.4 MB between D19S583 and D19S221. To increase the sequencing coverage and depth, we performed both Illumina GAII and Roche454 exome sequencing (Supplementary Table 1). All identified base alterations in the region were filtered against the dbSNP131 database. Bioinformatics analysis revealed a novel non-synonymous heterozygous mutation in DNMT1 at c.A1484G resulting in tyrosine495 changing to cysteine. Sequencing results confirmed this alteration segregated with disease status. We subsequently sequenced all 41 exons of DNMT1 and found kindred-3 (American kindred) and kindred-4 (Japanese kindred) have the same heterozygous mutation c.A1484G (p.Tyr495Cys) and kindred-2 (European Kindred) contain three consecutive nucleotides heterozygous change c.1470TCC-1472 ATA (p.Asp490Glu-Pro491Tyr) (Supplementary Fig. 1). All heterozygous mutations were absent in over 1500 unaffected controls and not reported in HGMD, the 1000 Genome and 10 exomes sequenced as part of the Environmental Genome Project.
DNMT1 has a large N-terminal regulatory region and a smaller C-terminal catalytic region (Fig. 2e). The highly conserved catalytic region of DNMT1 alone is not sufficient for enzymatic activity and an allosteric activation through direct interaction of the N-terminal regulatory and C-terminal catalytic regions is required for its enzymatic function15. The regulatory region contains many protein-protein interaction domains including PCNA binding, targeting sequence, zinc finger, bromo-adjacent homology domains. The identified mutations occurred in the TS domain. TS domain regulates DNMT1 binding to chromatin during late S phase and is responsible for persistent association during G2 and M phase. The proper folding of TS domain is crucial for DNMT1 function16. To probe the impact of the two mutations on the TS domain folding in vitro, we conducted the expression of wild type TS domain (aa 351-600) and corresponding p.Tyr495Cys and p.Asp490Glu-Pro491Tyr TS domain mutants in E. coli. While the three proteins are well produced in E. coli, only wt-TS is soluble (Supplementary Fig. 2). In contrast, the TS mutants form inclusion bodies, a strong indication that they are misfolded (Supplementary Fig. 2). Misfolding is not surprising for p.Tyr495Cys mutant, considering that Tyr495 is part of the protein hydrophobic core with an accessible solvent surface area (ASA) of only 0.55 Å2 in the crystal structure of wt-TS. Asp490 is solvent accessible (ASA is 75.3 Å2) and is therefore unlikely to contribute to the disease phenotype (Fig. 2f). In agreement, Asp490 is not a conserved residue and is replaced by a glutamate in other species (Fig. 2f). On the other hand, Pro491 has limited solvent accessibility with an ASA of 25.7 Å2 and together with Asp490 is part of the linker region between the β-sheet-based and α-helical subdomains of TS, suggesting that the constrained conformation of the trans-proline residue is needed for proper folding.
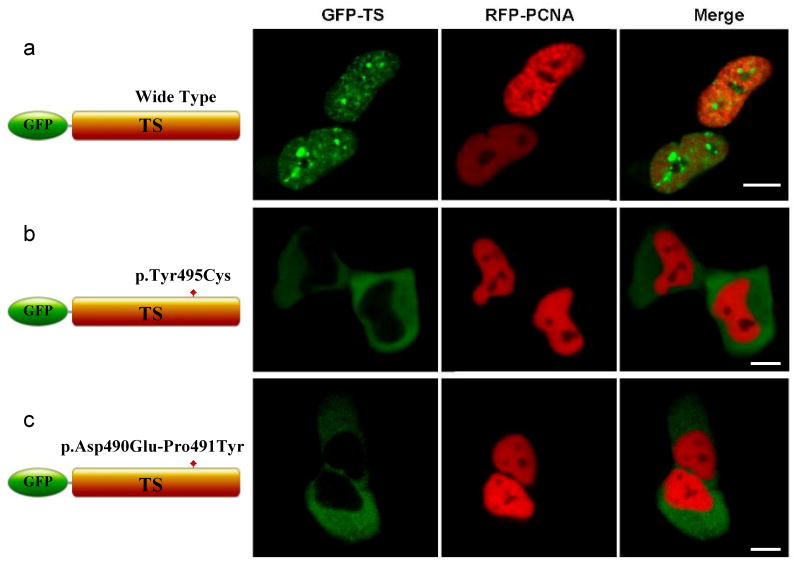
The TS domain has been expressed independently and shown as an important factor for the structure, function and localization of DNMT117,18,19, 16. Partial deletion of TS domain abolishes enzymatic activity of DNMT116. To investigate the functional implication of the mutant TS domain, we transfected GFP-tagged constructs encompassing wt-TS (aa 306-620) and the two TS mutants (p.Tyr495Cys-TS and p.Asp490Glu-Pro491Tyr-TS) into HeLa cells. While the wt-TS domain entered the nucleus and bound to the replication foci during S-phase and to heterochromatin during G2 phase, the two TS mutants localized to the cytoplasm and were not able to enter the nucleus (Fig. 3a-c). One of DNMT1's multiple nuclear localization signals is just upstream of the targeting-sequence domain (323KRRK326)19, the mutations likely disrupt the proper fold of dNMT1, preventing the mutant proteins from entering the nuclei. Mutations p.Tyr495Cys and p.Asp490Glu-Pro491Tyr also affect the stability and the enzymatic activity of full length DNMT1. We transfected GFP-tagged full length wild type and mutant DNMT1 into HeLa cells and treated cells with cycloheximide for different time lengths. Western blot analysis showed the mutant proteins were quickly degraded compared to wild type DNMT1 (Supplementary Fig. 3). The enzymatic activity assay showed that p.Tyr495Cys-DNMT1 and p.Asp490Glu-Pro491Tyr-DNMT1 have significantly decreased activity comparing to wt-DNMT1, (Supplementary Fig. 4 and Supplementary notes). These findings demonstrate the deleterious effects of these mutations in the TS domain on the structure, methyltransferase activity and stability of DNMT1.
Figure 3.
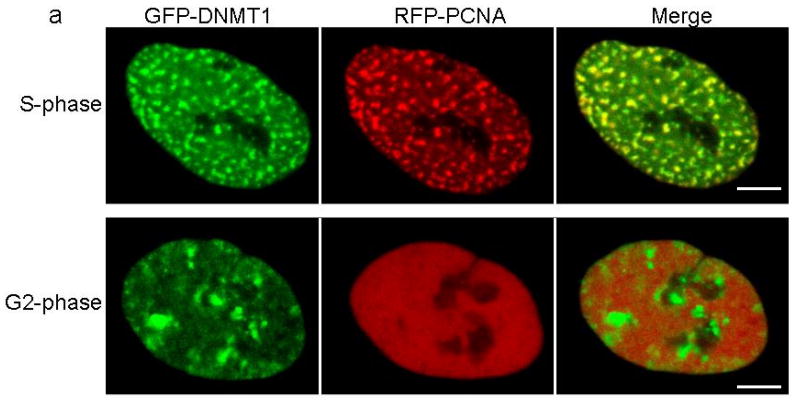
Confocal microscopy was performed using HeLa cells co-transfected with plasmids containing RFP-PCNA and GFP-DNMT1 wild type TS domain (panel a), p.Tyr495Cys-TS domain (panel b) or p.Asp490Glu-Pro491Tyr-TS domain (panel c). Wild type and mutant TS domains appear green in the right panels, PCNA appears red in the middle panels and merged images are shown in the left panels. Scale bar, 5um. In panel a, wild type TS domain enters nucleus during S phase when PCNA localizes to the toroidal structures of replication foci, wild type TS also binds to heterochromatin during G2 phase when PCNA showed diffused pattern and toroidal structures are no longer visible. Panel b and panel c showed mutant TS domains was unable to enter into the nucleus and remained in the cytoplasm.
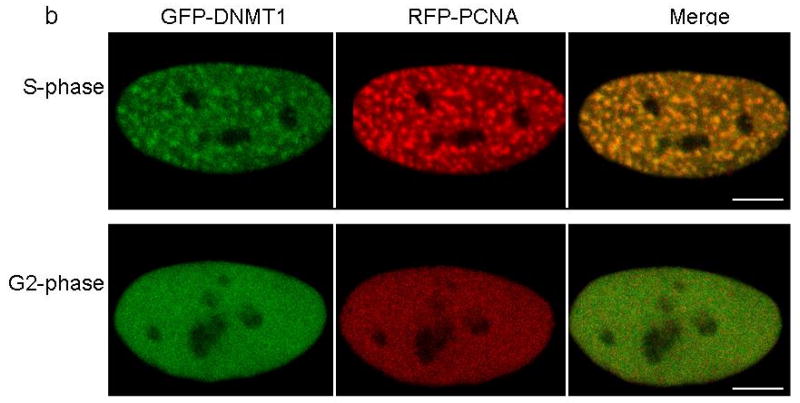
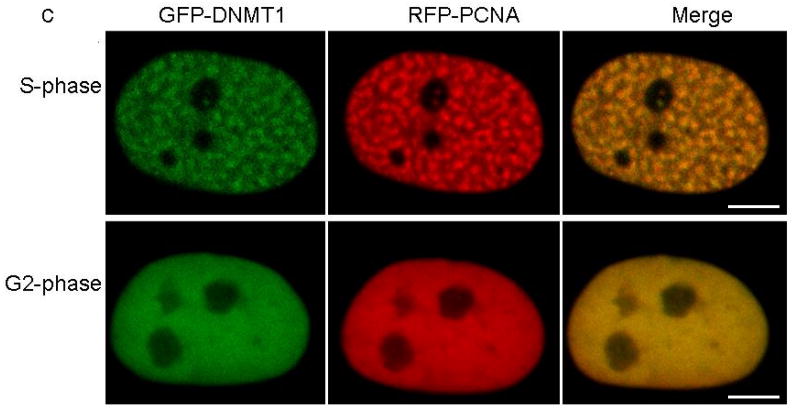
DNMT1 is one of the components of the DNA replication machinery during S phase of the cell cycle17. Studies have shown that maintenance of methylation could last past the replication stage with DNMT1 continuously loaded on heterochromatin after S phase. The TS domain was identified responsible for this persistent association20. Partial deletion of TS domain has been shown to abolish DNMT1 association with heterochromatin after S phase20. To evaluate the heterochromatin binding of mutant DNMT1, we co-transfected GFP-tagged wild type and p.Tyr495Cys-DNMT1 and p.Asp490Glu-Pro491Tyr-DNMT1 along with RFP-tagged proliferating cell nuclear antigen (PCNA) as cell cycle marker into HeLa cells. Our results show that the mutant DNMT1 only showed normal localization at replication foci during S phase as previously described20, but did not show binding of heterochromatin during G2 phase in contrast to wt-DNMT1 (Fig. 4a-c), indicating that the mutations abolished DNMT1 heterochromatin binding ability after S phase. Previous studies showed the replication machinery loading of DNMT1 is not required for the genome methylation maintenance21, suggesting DNMT1 binding to heterochromatin after S phase could be involved in the prolonged methylation maintenance, recruitment of histone deacetylases and chromatin stability preservation in the transcriptionally repressed genome region20. The impaired heterochromatin binding after S phase is likely to impair these important cellular functions.
Figure 4.
Confocal microscopy was performed using HeLa cells co-transfected with plasmids containing RFP-PCNA and full length (a) GFP-wild type DNMT1, (b) GFP-p.Tyr495Cys-DNMT1 or (c) GFP-p.Asp490Glu-Pro491Tyr-DNMT1. Wild type and mutant DNMT1 appear green in the right panels, PCNA appears red in the middle panels and merged images are shown in the left panels. Scale bar, 5um. Cell cycles are deciphered from the pattern of RFP-PCNA. In S phase, PCNA is present at the toroidal structures of the replication foci; in G2 phase, PCNA shows diffused pattern in the nucleus. In panel a, wild type DNMT1 co-localizes with PCNA at replication foci during both S phase and binds to heterochromatin during G2 phase. In panel b and c, p.Tyr495Cys-DNMT1 and p.Asp490GLu-Pro491Tyr-DNMT1 localize along with PCNA at the replication foci during S phase but did not show binding of heterochromatin during G2 phase.
Next, we examined the effect of mutant DNMT1 on the global methylation level using liquid chromatography-electrospray ionization tandem mass spectrometry (LC-ESI-MS/MS) to measure 5-methyl-2′-deoxycytidine (5-mdC) levels following enzymatic hydrolysis of genomic DNA22. Total 24 samples including 12 affected and 12 genetically related and unaffected samples from Kindred-1 were quantified. Our results showed 8% reduction in 5-mdC content in affected group comparing to the unaffected group (p<0.001) (Supplementary Fig. 5). This is predicted meaningful considering in DNMT1 knockout HCT116 cells only 10-20% of genomic DNA hypomethylation was observed, but the deleterious impact led to pronounced chromosomal defects and apoptosis23,24. Since methylation varies with age and sex, we specifically compared the content of m5dC between three pairs of same sex siblings and between two pairs of same sex cousins with similar age (<5 years difference). The results showed consistent 7-15% reduction of 5-mdC content in all affected sibling and cousins (Supplementary Table 2). Centromeric and pericentromeric regions have been shown to contribute to the majority of global methylation. We used Pyrosequencing assay of bisulfite treated DNA to investigate the CpG sites in the repetitive elements of Satellite-2, Line-1 and Satellite-α and Alu region. The results suggested reduced methylation at certain CpG sites in Satellite-2 region among affected group (Supplementary Table 3 and Supplementary notes). The Satellite-2 methylation reduction was further confirmed by Southern blot using genomic DNA digested by methylation sensitive enzyme BstB1 (Supplementary Fig. 6a-b). These results support global hypomethylation demonstrated by reduced 5-mdC content, suggesting the hypomethylation likely occurs at the Satellite-2 repetitive elements. The hypomethylation in repetitive and transposable elements has been linked to the loss of methyl-specific protein complexes that stabilize a high order of heterochromatin structure25. The reduction of total 5-mdC content and decreased methylation in Satellite-2 resulting from DNMT1 mutations may impair the heterochromatin stability.
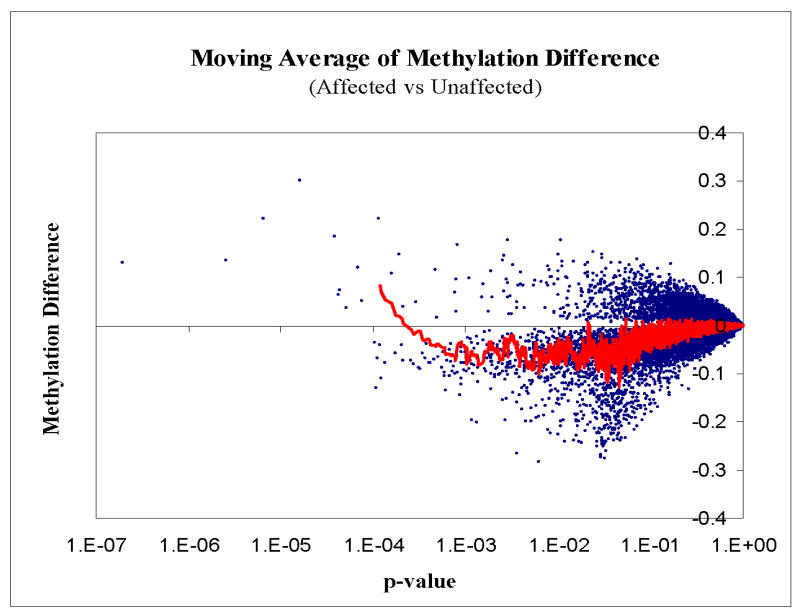
Since variation in methylation is usually linked to the transcriptional regulation of specific genes, we investigate the methylation of CpG sites in gene promoter regions. Illumina HumanMethylation27 BeadChip was used to survey genome-wide promoter methylation profiles. The 24 samples from kindred-1 were studied. Based on the moving average of methylation difference between affected and unaffected group, moderate reduction of methylation was observed in the affected samples (Fig. 5). Interestingly, the 10 promoter CpG sites with the most significant p value (p<0.00007) showed increased methylation (Fig. 5 and Supplementary Fig. 7). Global hypomethylation of cytosines and local hypermethylation of CpG islands are characteristics of the neoplastic epigenome26 but the investigated kindreds did not have any cancer histories. One of the characteristics of tumor cells is uncontrolled proliferation. In postmitotic neurons, DNA methylation undergoes turnover process in response to synaptic activity27 and failure of mitotic arrest may result in cell death4. Re-expression of various cell cycle proteins has been reported in patients with Alzheimer's dementia and other neurodegenerative disorders4, suggesting the initiation of cell cycle by abnormal expression of cell cycle proteins impairs neuronal health. It is intriguing to see that the methylation pattern in the affected persons mimics the methylation pattern of tumor cells, leading us to speculate that mutated DNMT1 could be linked to the loss of cell cycle arrest in postmitotic neuron. Defects of methylation have been shown to cause developmental and progressive human neurodegenerative diseases such as Rett (MeCP2)28 and ICF(DNMT3b)29 syndromes. Epigenetic dysregulation has also been suggested in Alzheimer's dementia through reduced histone acetylation30. Our chromosomally linked region at 19p13.2 also associates with late onset familial Alzheimer's (LOFAD)31, possible association of DNMT1 in LOFAD needs further investigation.
Figure 5.
Moving average of methylation difference of ∼25000 CpG sites between affected and unaffected groups from kindred-1. Kindred 1 was chosen to optimize same number of gender and the first degree relation between affected and unaffected group. The methylation profile of affected and unaffected groups was compared. Y-axis represents the methylation difference between the two groups. X-axis represents p-value. Red colored line represents the moving average of methylation difference between affected vs. unaffected group. Each blue dot represents methylation difference for a CpG site. Blue dots below the 0 line represents reduced methylation in the affected group while blue dots above 0 line represents increased methylation in the affected group. The red moving average line suggests local hypermethylation and moderate global hypomethylation in the affected group, consistent with the 5-mdC content measurement by LC-ESI-MS/MS (supplementary fig. 5).
In this report, we identified mutations in DNMT1 that cause HSAN1 with dementia and hearing loss. These mutations affect DNMT1's proper folding, decrease enzymatic activity, impair heterochromatin binding ability after S phase and lead to the occurrence of global hypomethylation with local hypermethylation. Our results provide a direct link between DNMT1 alteration and a neurodegenerative disorder of both central and peripheral nervous system. DNMT1 is highly expressed in postmitotic neurons and adult central nervous system32, it interacts with a series of important cell cycle regulating proteins33 and is involved in neuronal differentiation, migration and central neural connection1,34,35, suggesting DNMT1 participates in a precise mechanism of dynamic regulation of the nervous system. The affected individuals in our kindreds had apparent normal intelligence and nerve function until early adulthood, then the typical symptoms began and worsened with age, implicating DNMT1's influence is beyond embryogenesis and suggesting a progressive course with gradual accumulating effect. Our study emphasizes that even moderate global hypomethylation in association with local hypermethylation can have a severe impact on neuronal survival and function. The pathogenic mechanism of mutant DNMT1 is potentially complex and provides a new direction for the study of neurodegeneration.
Methods
Subjects and Samples
This study was approved by the Mayo Institutional Review Boards. Blood and other samples were taken after obtaining written informed consent. This study included total 4 HSAN1 kindreds with sensorineural deafness and dementia - two American families, one of German origin and the other of Northern European descent described previously 7, one Japanese family with no known European ancestry also described previously8 and one Australian family of English descent.
Linkage Analysis
We performed a 8-cM genome wide linkage scan on 63 individuals (12 affected, 51 unaffected including 14 unaffected spouses) from kindred-1 using 500 deCODE microsatellite markers spaced at approximately 8-cM intervals. To allow for potential ambiguities of the relationship between the underlying genotype and affection status, penetrance was assumed to be 0.1 for noncarriers and 0.9 for carriers. Two-point logarithm of the odds (LOD) scores was calculated using the MLINK program of Linkage, assuming a rare susceptibility allele (frequency 0.001) and an autosomal dominant mode of inheritance. Multipoint analysis and haplotypes were performed by SIMWALK2.17. To confirm and refine the localized region at chromosome 19q, we identified additional ten microsatellite markers for haplotype analysis between D19S884 and D19S226, spanning ∼6.5-cM distance with an average 0.8-cM density.
Exome capture and Next-Generation sequencing
a. Illumina GAII Paired End Exome Sequencing
Exome capture is carried out using Agilent's SureSelect Human All Exon kit-v1. The flow cells are sequenced as 76×2 paired end reads on an Illumina GAIIx using SBS sequencing kit-v4 and SCS-v2.5 software. The image processing and base calling was performed using the Illumina Pipeline Software. The reads with phred-like consensus quality ≥ 40 were aligned to human genome-36 (hg18) and the single nucletoide variants (SNVs) and Indels (insertion-deletions) were called using MAQ (http://maq.sourceforge.net/) and BWA (http://bio-bwa.sourceforge.net/bwa.shtml). SNVs and Indels were then filtered using Agilent's ‘on-target bed file’ and annotated using SIFT and SeattleSEQ annotation servers. The run statistics is summarized in Supplementary Table 1.
b. Roche 454 GS-FLX sequencing with NimbleGen 2.1 M Human Exome capture
The sample was prepared using the GS-FLX Titanium optimized sequence capture protocol using the Roche-NimbleGen 2.1 M Human Exome array. Exome-captured proband DNA sample was sequenced on 454 Genome Sequencer FLX instrument with GS-FLX Titanium Kits. Two full 454 GS-FLX (Titanium) runs were performed. High quality data were aligned to the hg18 human genome reference. Variants were identified using the GSMapper software 2.0.01 and were included in AllDiff (all differences) and HCDiff (high confidence differences) reports. The average read length is 382bp with mode of 490bp. We screened novel mutations in both AllDiff and HCDiff files within the identified 3.4 MB linked region. The run statistics is summarized in Supplementary Table 1.
Sanger Sequencing Analysis
Genomic DNA was extracted from whole peripheral blood, transformed lymphoblastoid cell lines or available tissues using standard methods (Qiagen). Resequencing the region of the interest was carried out using dye termination chemistry (Big Dye Terminator with the model 3730xl sequencer; Applied Biosystems, CA). Primer sets for PCR were designed using the web-based design tool Primer3 (Supplementary Table 4). PCR were carried out using the standard protocol. After PCR reactions, the amplicons were treated with the ExoSAP-IT (USB) to degrade unincorporated PCR primers and deoxynucleotide triphosphates. The cleaned products were mixed with 10 picomoles of the forward or reverse PCR primers for sequencing. DNA sequence variants were identified using Mutation Surveyor Analysis Software (SoftGenetics).
Expression Vectors for TS domain, Full length DNMT1 and PCNA
The nucleotide sequence encompassing wild type DNMT1 TS domain (aa 306-620) for HeLa cell transfection was custom synthesized (Origene) and this cDNA was cloned into the pCMV6-N-GFP vector. The plasmid pEX-N-His-TS containing wild type TS domain (aa 351-600) for bacterial expression and purification was also custom synthesized (Origene). The expression constructs pEGFP-human DNMT1 and pmRFP-human PCNA were kindly provided by Dr. H. Leonhardt (Ludwig Maximilians University Munich, Germany). The pEGFP-hDNMT1 contains a CMV promoter and enhanced GFP fused to N-terminus of full length human DNMT1 (NM_001379). Similarly pmRFP-human PCNA contains a CMV promoter and mRFP cDNA fused to the N-terminus of human PCNA. Mutants were generated using QuikChange mutagenesis kit (Stratagene). For primer sequences, see Supplementary Table 5 online. All constructs were confirmed by direct sequencing.
DNMT1 TS Domain Expression and Purification in E. coli
Plasmids of wild type, p.Tyr495Cys and p.Asp490Glu-Pro491Tyr TS domain of DNMT1 were transformed in BL21(DE3) E. coli cells and grown in LB broth supplemented with 100 mg/mL ampicillin at 37°C up to an OD600 of ∼0.8, after which the temperature was decreased to 15°C for about 1h and then 1mM IPTG (final concentration) was added. After 16 h, the cells were harvested by centrifugation, resuspended in 50mM sodium phosphate, pH 7.5, 300 mM NaCl (buffer A) and 20 mM PMSF and then lyzed with an Emulsiflex C-5 high-pressure homogenizer (Avestin). After centrifugation, the soluble fraction was purified by passing through a Ni-NTA column pre-equilibrated with buffer A; washing the column with buffer A containing 5 mM imidazole (buffer B); and eluting the protein with buffer A containing 500 mM imidazole (buffer C). Fractions containing the TS domain were pooled and passed through a Superdex 200 FPLC column (GE Healthcare) using buffer A as the running buffer. The cell debris were resuspended in buffer A containing 8 M urea, and centrifuged. The resulting supernate (referred to as insoluble fraction) was purified similarly as above but using buffers B and C with 6 M urea. At various stages during the purification, sample aliquots were obtained and later assessed by SDS-PAGE. Protein purification was carried out at 4°C.
Cell Culture and Western Blot Analysis
HeLa Cells were cultured in RPMI supplemented with 10% fetal Bovine serum. Cells were transfected using FugeneHD (Roche). For Cycloheximide treatment, fresh media contain 0.05 mg/mL l of cycloheximide (Sigma) were added after 24 h of transfection. Cell lysate were collected using NP-40 lysis buffer (Invitrogen) at different time points for western blotting analysis. BCA assay was used to ensure the equal loading. Cell lysate were separated on 4-12% gradient SDS–PAGE and transferred to a PVDF-membrane (Bio-Rad). The membrane was blocked with 1×Casein (Vector) in TBST and incubated overnight at 4°C with an anti-GFP mouse monoclonal antibody (Allele Biotechnology). After washing, the blots were incubated with the appropriate secondary antibody conjugated with horseradish peroxidase at room temperature for 1 h. Immunoreactive bands were washed and visualized with SuperSignal Pico Western Blot Detection Kit (Thermo Scientific).
Confocal Microscopy
HeLa cells were seeded on Lab Tek II chambered cover glass (Nunc) 24 h before transfection. Cells were transfected using Fugene HD (Roche). Twenty-four hours after transfection, live cell microscopy was performed with a LSM510 (Carl Zeiss) confocal setup mounted on an Axiovert 200 M inverted microscope using a 100× phase contrast Plan-Neofluar oil, NA 1.3 heated to 37°C. Chambered cover glass was maintained at 37°C using a live-cell chamber and temperature controller (PeCon GmbH, Erbach, Germany). Enhanced GFP and mRFP were excited sequentially at 488nm, 543nm to minimize the crosstalk.
Global Genomic DNA Methylation Analyses
Genomic DNA methylation was assessed by direct measurement of the level of 5-methyl-2′-deoxycytidine (5-mdC) using liquid chromatography-electrospray ionization mass spectrometry (LC-ESI-MS/MS), as described previously22. Briefly, 1 μg genomic DNA samples were enzymatically hydrolyzed with Nuclease P1 or Nuclease S1 into component nucleosides, and loaded, along with standards, onto an LC-ESI/MS/MS system. All samples were analyzed in duplicate and proper controls were tested before and after sample measurement to ensure the accuracy.
Illumina Infinium HumanMethylation27 BeadChip Analysis
One microgram DNA from each selected sample was bisulfite-treated using Zymo Methylation kit (Zymo Research). After bisulfite conversion, 250 ng of each sample undergo a whole-genome amplification (WGA) followed by hybridization. Allele-specific primer annealing is followed by single-base extension with labeled nucleotides. After extension, the array is scanned and the intensities of the unmethylated and methylated bead types are measured. The methylation level at each CpG site was determined by measuring the methylation fraction (beta), defined as the fraction of methylated signal over the total signal (unmethylated + methylated). The beta values are continuous variables between 0 (unmethylated) and 1 (completely methylated). The Infinium Assay includes redundant, built-in, bisulfite conversion quality controls that measure the conversion rate of non-CpG cytosines and background signal. Using within-assay controls evaluated with Illumina GenomeStudio software, we assessed background signal and bisulfite conversion rate and ensured that non-methylated cytosines in all 24 samples were efficiently bisulfite converted. ∼8% CpG sites that did not reveal reliable signals, as determined by their detection p-value, were excluded from further analysis. Differential methylation analysis and construction of a volcano plot was performed using the Partek Statistical Package (Partek). We employed two-way analysis of variance (ANOVA) to account for the potential batch effects. The type I error was controlled by the use of Benjamini and Hochberg false discovery rate (FDR) controlling procedure.
Supplementary Material
Acknowledgments
This study was supported by the National Institute of Neurologic Disorders and Strokes (NINDS) K08 (NS065007), NINDS R01 (NS36797) and a previous grant from the Muscular Dystrophy Association. The authors wish to thank Mayo Clinic Bioinformatics Core and Sequencing Core for their excellent assistance. We would also like to thank the support from the Clinical Core of the Mayo Clinic Center for Cell Signaling in Gastroenterology (P30DK084567). DCW and MS would like to acknowledge the outstanding clinical contributions of Ms. Julia Platt. Their portion of this work was supported by NIH grants NS21328 and NS070298.
Footnotes
Authors Contributions: C.J.K., D.I.S. and P.J.D. directed the study. C.J.K. wrote the paper. C.J.K., P.J.D., G.A.N., S.H., K.H., H.Y., D.C.W., M.S., C.L., L.A.B., G.E.S., W.J.L., evaluated or collated patient data. E.J.A. did the linkage and haplotype analysis. S.M. and B.B. did next generation sequencing analysis. C.J.K., C.J.W. and Y.W. did the cell culture and protein expression studies, gene sequencing and southern blot analysis. J.M.C. and A.R.K. did the methylation analysis. M.V.B. and G.M. did the mutagenesis, bacterial protein expression and structural analysis. J.E.P. provided pathologic analysis of autopsy material.
Competing Financial Interests: The authors declare no competing financial interests.
Accession number:
Protein Data Bank accession number for DNMT1 targeting sequence domain, 3EPZ.
Exome sequencing data is available upon request through FTP site (NCBI Sequence Read Archive repositories has been discontinued for next generation sequencing data).
URLs:
Human Genome Mutation Database (HGMD), http://www.hgmd.cf.ac.uk/.
The 1000 Genome Browser, http://browser.1000genomes.org/index.html.
Environmental Genome Project, http://www.niehs.nih.gov/research/supported/programs/egp/.
References
- 1.Feng J, Fan G. The role of DNA methylation in the central nervous system and neuropsychiatric disorders. Int Rev Neurobiol. 2009;89:67–84. doi: 10.1016/S0074-7742(09)89004-1. [DOI] [PubMed] [Google Scholar]
- 2.Chen WG, et al. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–9. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- 3.Tohgi H, et al. Reduction with age in methylcytosine in the promoter region -224 approximately -101 of the amyloid precursor protein gene in autopsy human cortex. Brain Res Mol Brain Res. 1999;70:288–92. doi: 10.1016/s0169-328x(99)00163-1. [DOI] [PubMed] [Google Scholar]
- 4.Herrup K, Yang Y. Cell cycle regulation in the postmitotic neuron: oxymoron or new biology? Nat Rev Neurosci. 2007;8:368–78. doi: 10.1038/nrn2124. [DOI] [PubMed] [Google Scholar]
- 5.Brooks PJ, Marietta C, Goldman D. DNA mismatch repair and DNA methylation in adult brain neurons. J Neurosci. 1996;16:939–45. doi: 10.1523/JNEUROSCI.16-03-00939.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan G, et al. DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals. J Neurosci. 2001;21:788–97. doi: 10.1523/JNEUROSCI.21-03-00788.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wright A, Dyck PJ. Hereditary sensory neuropathy with sensorineural deafness and early- onset dementia. Neurology. 1995;45:560–2. doi: 10.1212/wnl.45.3.560. [DOI] [PubMed] [Google Scholar]
- 8.Hojo K, et al. Hereditary sensory neuropathy with deafness and dementia: a clinical and neuroimaging study. Eur J Neurol. 1999;6:357–61. doi: 10.1046/j.1468-1331.1999.630357.x. [DOI] [PubMed] [Google Scholar]
- 9.Klein CJ, Dyck P. Hereditary Sensory and Autonomic Neuropathies (HSAN): Clinical Features Pathologic Classification, and Molecular Genetics. In: Dyck PJ, T P, editors. Peripheral Neuropathy. Fourth. Vol. 2. Elsevier; 2005. pp. 1809–1845. [Google Scholar]
- 10.Dawkins JL, Hulme DJ, Brahmbhatt SB, Auer-Grumbach M, Nicholson GA. Mutations in SPTLC1, encoding serine palmitoyltransferase, long chain base subunit-1, cause hereditary sensory neuropathy type I. Nat Genet. 2001;27:309–12. doi: 10.1038/85879. [DOI] [PubMed] [Google Scholar]
- 11.Rotthier A, et al. Mutations in the SPTLC2 subunit of serine palmitoyltransferase cause hereditary sensory and autonomic neuropathy type I. Am J Hum Genet. 2010;87:513–22. doi: 10.1016/j.ajhg.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guelly C, et al. Targeted high-throughput sequencing identifies mutations in atlastin-1 as a cause of hereditary sensory neuropathy type I. Am J Hum Genet. 2011;88:99–105. doi: 10.1016/j.ajhg.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verhoeven K, et al. Mutations in the small GTP-ase late endosomal protein RAB7 cause Charcot-Marie-Tooth type 2B neuropathy. American Journal of Human Genetics. 2003;72:722–7. doi: 10.1086/367847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hojo K, Kawamata T, Tanaka C, Maeda K. Inflammatory glial activation in the brain of a patient with hereditary sensory neuropathy type 1 with deafness and dementia. Neurosci Lett. 2004;367:340–3. doi: 10.1016/j.neulet.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 15.Fatemi M, Hermann A, Pradhan S, Jeltsch A. The activity of the murine DNA methyltransferase Dnmt1 is controlled by interaction of the catalytic domain with the N-terminal part of the enzyme leading to an allosteric activation of the enzyme after binding to methylated DNA. J Mol Biol. 2001;309:1189–99. doi: 10.1006/jmbi.2001.4709. [DOI] [PubMed] [Google Scholar]
- 16.Margot JB, et al. Structure and function of the mouse DNA methyltransferase gene: Dnmt1 shows a tripartite structure. J Mol Biol. 2000;297:293–300. doi: 10.1006/jmbi.2000.3588. [DOI] [PubMed] [Google Scholar]
- 17.Leonhardt H, Page AW, Weier HU, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–73. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- 18.Fellinger K, Rothbauer U, Felle M, Langst G, Leonhardt H. Dimerization of DNA methyltransferase 1 is mediated by its regulatory domain. J Cell Biochem. 2009;106:521–8. doi: 10.1002/jcb.22071. [DOI] [PubMed] [Google Scholar]
- 19.Cardoso MC, Leonhardt H. DNA methyltransferase is actively retained in the cytoplasm during early development. J Cell Biol. 1999;147:25–32. doi: 10.1083/jcb.147.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Easwaran HP, Schermelleh L, Leonhardt H, Cardoso MC. Replication-independent chromatin loading of Dnmt1 during G2 and M phases. EMBO Rep. 2004;5:1181–6. doi: 10.1038/sj.embor.7400295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spada F, et al. DNMT1 but not its interaction with the replication machinery is required for maintenance of DNA methylation in human cells. J Cell Biol. 2007;176:565–71. doi: 10.1083/jcb.200610062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song L, James SR, Kazim L, Karpf AR. Specific method for the determination of genomic DNA methylation by liquid chromatography-electrospray ionization tandem mass spectrometry. Anal Chem. 2005;77:504–10. doi: 10.1021/ac0489420. [DOI] [PubMed] [Google Scholar]
- 23.Chen T, et al. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat Genet. 2007;39:391–6. doi: 10.1038/ng1982. [DOI] [PubMed] [Google Scholar]
- 24.Brown KD, Robertson KD. DNMT1 knockout delivers a strong blow to genome stability and cell viability. Nat Genet. 2007;39:289–90. doi: 10.1038/ng0307-289. [DOI] [PubMed] [Google Scholar]
- 25.Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- 26.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–28. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 27.Nelson ED, Kavalali ET, Monteggia LM. Activity-dependent suppression of miniature neurotransmission through the regulation of DNA methylation. J Neurosci. 2008;28:395–406. doi: 10.1523/JNEUROSCI.3796-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amir RE, et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–8. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 29.Xu GL, et al. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402:187–91. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
- 30.Cao X, Sudhof TC. A transcriptionally [correction of transcriptively] active complex of APP with Fe65 and histone acetyltransferase Tip60. Science. 2001;293:115–20. doi: 10.1126/science.1058783. [DOI] [PubMed] [Google Scholar]
- 31.Wijsman EM, et al. Evidence for a novel late-onset Alzheimer disease locus on chromosome 19p13.2. Am J Hum Genet. 2004;75:398–409. doi: 10.1086/423393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tawa R, Ono T, Kurishita A, Okada S, Hirose S. Changes of DNA methylation level during pre- and postnatal periods in mice. Differentiation. 1990;45:44–8. doi: 10.1111/j.1432-0436.1990.tb00455.x. [DOI] [PubMed] [Google Scholar]
- 33.Spada F, Rothbauer U, Zolghadr K, Schermelleh L, Leonhardt H. Regulation of DNA methyltransferase 1. Adv Enzyme Regul. 2006;46:224–34. doi: 10.1016/j.advenzreg.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 34.Golshani P, Hutnick L, Schweizer F, Fan G. Conditional Dnmt1 deletion in dorsal forebrain disrupts development of somatosensory barrel cortex and thalamocortical long-term potentiation. Thalamus Relat Syst. 2005;3:227–233. doi: 10.1017/S1472928807000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rampon C, et al. Enrichment induces structural changes and recovery from nonspatial memory deficits in CA1 NMDAR1-knockout mice. Nat Neurosci. 2000;3:238–44. doi: 10.1038/72945. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.