Abstract
The AIDS-causing lentiviruses HIV and SIV effectively evade host immunity, and once established, infections with these viruses are only rarely controlled by immunologic mechanisms1-3. However, the initial establishment of infection in the first few days after mucosal exposure, prior to viral dissemination and massive replication, may be more vulnerable to immune control4. Here, we report that SIV vaccines that include rhesus cytomegalovirus (RhCMV) vectors5 establish indefinitely persistent, high frequency, SIV-specific effector-memory T cell (TEM) responses at potential sites of SIV replication in rhesus macaques (RM) and stringently control highly pathogenic SIVmac239 infection early after mucosal challenge. Thirteen of 24 RM receiving either RhCMV vectors alone or RhCMV vectors followed by adenovirus 5 (Ad5) vectors (vs. 0 of 9 DNA/Ad5-vaccinated RM) manifested early complete control of SIV (undetectable plasma virus), and in 12/13 of these RM, we observed long-term (≥1 year) protection characterized by: 1) occasional blips of plasma viremia that ultimately waned; 2) predominantly undetectable cell-associated viral load in blood and lymph node mononuclear cells; 3) no depletion of effector site CD4+ memory T cells; 4) no induction or boosting of SIVenv-specific antibodies (Abs); and 5) induction and then loss of T cell responses to an SIV protein (vif) not included in the RhCMV vectors. Protection correlated with the magnitude of the peak SIV-specific CD8+ T cell responses in the vaccine phase, and occurred without anamnestic T cell responses. Remarkably, long-term RhCMV vector-associated SIV control was insensitive to either CD8+ or CD4+ lymphocyte depletion, and at necropsy, cell-associated SIV was only occasionally measurable at the limit of detection with ultrasensitive assays, observations suggesting the possibility of eventual viral clearance. Thus, persistent vectors such as CMV and their associated TEM responses might significantly contribute to an efficacious HIV/AIDS vaccine.
Conventional prime-boost vaccine regimens with non-persistent vectors lead to lymphoid tissue-based memory T cell responses (“central memory” or TCM), which deliver peak effector responses only after TCM have undergone antigen-stimulated expansion, differentiation and trafficking6 -- too late to effectively control pathogens with the rapid replication and spread kinetics and highly developed immune evasion capabilities of the AIDS-causing lentiviruses2,4,5. As T cell effector responses are likely to be much more effective against the smaller, localized and less diverse viral populations present in the first hours and days of mucosally acquired HIV/SIV infection2,4,7,8, we hypothesized that a vaccine able to “pre-position” differentiated effector cells (TEM) at such early replication sites would demonstrate improved efficacy. Such TEM responses are the hallmark of persistent agents9,10, prompting our development of SIV vectors based on the persistent β-herpesvirus RhCMV. As recently reported5 and illustrated in Suppl. Fig. 1, RhCMV/SIV vectors can establish and indefinitely maintain high frequency SIV-specific, TEM-biased, CD4+ and CD8+ T cell responses in diverse tissue sites of RhCMV+ RM, and in a small efficacy study were associated with early control of intra-rectally administered SIVmac239. To evaluate potential differential effects of persistent vector/TEM-biased vs. non-persistent vector/TCM-biased, SIV-specific T cell responses on the outcome of mucosal SIVmac239 infection, we compared naturally RhCMV+ male RM vaccinated with: 1) RhCMV/SIV vectors alone (Group A); 2) RhCMV/SIV vectors followed by replication-defective Ad5 vectors (Group B); and 3) a standard DNA prime/Ad5 vector boost benchmark vaccine (Group C)11-13 vs. unvaccinated control RM (Group D; Fig. 1a). RhCMV/SIV vectors efficiently super-infected all Group A and B RM and elicited robust CD4+ and CD8+ T cell responses to all vector-encoded SIV proteins (Fig. 1b; Suppl. Figs. 2-4). The Ad5 vector boost of Group B RM, and the DNA/Ad5 regimen given to Group C RM were also strongly immunogenic (Fig. 1b; Suppl. Figs. 3-4). Although the pattern of development of the SIV-specific T cell responses differed between these vectors (Suppl. Fig. 3a), the magnitude of the total SIV-specific, CD4+ and CD8+ T cell responses at the end of the vaccine phase in Groups A, B, and C were similar (Fig. 1b, Suppl. Fig 4). Consistent with previous results5, RhCMV/SIV vector-elicited, SIV-specific CD8+ T cell responses exhibited different epitope targeting than the DNA- and/or Ad5 vector-elicited responses (Suppl. Fig. 3b), as well as maintained a markedly TEM-biased phenotype over the entire vaccine phase, in contrast to the development of a more TCM-biased response in the DNA/Ad5-vaccinated RM (Suppl. Fig. 5).
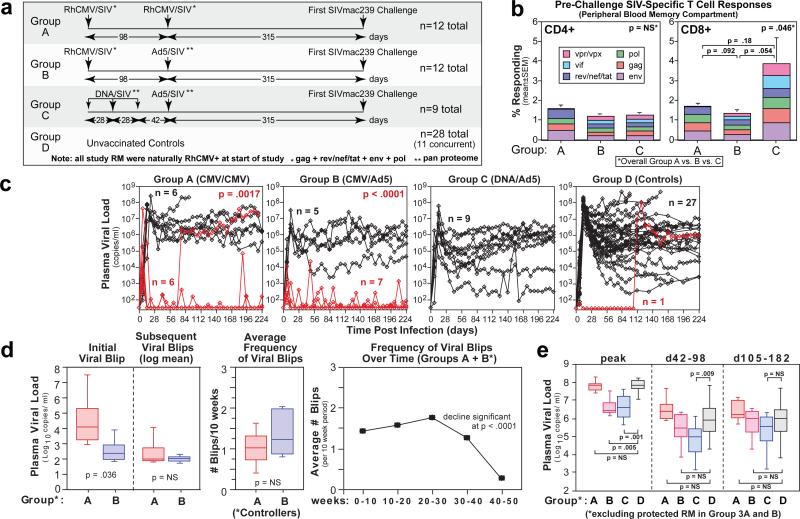
Figure 1. Immunogenicity and efficacy of RhCMV/SIV vectors.
a, Schematic of the vaccination protocol used in this study. b, Comparison of the mean frequency (± SEM) of the overall SIV-specific CD4+ and CD8+ T cell responses and the contribution of the designated SIV proteins to these total responses in the blood memory compartments of Groups A-C RM at the end of the vaccine phase. The Kruskal-Wallis test was used to determine the significance of differences in total SIV-specific response frequencies among the 3 vaccine groups, with the Wilcoxon rank sum test used to perform pair-wise analysis for the CD8+ response. As these latter p values were > 0.05, we concluded that overall response frequencies of Groups A, B and C were not significantly different. c, Outcome of repeated, limiting dose, intra-rectal SIVmac239 challenge of Groups A-D. The significance of differences in the fraction of infected RM in each group that met controller criteria (see Methods) was determined by Fisher's exact test (closed symbols in Group D are concurrent controls; open, previous controls given the same challenge). d, Analysis of the magnitude and frequency of pvl “blips” in Group A and B controllers, with the significance of the differences in blip magnitude and frequency between Groups A and B determined by the Wilcoxon rank sum test, and the significance of the decline in blip frequency of Group A + B RM after 30 weeks pi determined by analysis of variance and linear trend tests. e, Comparison of pvl in Group A-D RM with progressive infection (excluding Group A and B controllers and Group D RM with protective MHC alleles not represented in Groups A-C) with the significance of differences between Groups A, B and C vs. Group D determined by the Wilcoxon rank sum test.
At week 59 post-initial vaccination, all RM were challenged via the intra-rectal route with highly pathogenic SIVmac239 using a repeated, limiting dose protocol5. The number of challenges required to achieve measureable infection – plasma viral load (pvl) > threshold (30 copies/ml) – was not significantly different between Groups A-D (Suppl. Fig. 6), but the subsequent course of infection in these groups was strikingly different (Fig. 1c). Of 28 unvaccinated controls (both concurrent and historical), 27 exhibited typical progressive SIVmac239 infection, and one exhibited an initially non-progressive infection (transient viremia) that spontaneously progressed 105 days later. Similarly, all DNA/Ad5-vaccinated RM (9 of 9) manifested progressive infection, albeit with reduced mean pvl compared to controls (see below). In contrast, 13 of the 24 RM that received RhCMV/SIV vectors (6/12 in Group A; 7/12 in Group B) presented with an initial burst of plasma SIV, ranging in magnitude from as few as 60 to as many as 4 × 107 SIV RNA copies/ml, which was followed by rapid control to undetectable levels (Fig. 1c/d). From 3-18 weeks post-infection (pi), all but one of these protected RM demonstrated one or more repeat episodes of transient viremia that were always controlled back down to below detection limits (Fig. 1c/d). These periodic viral blips were similar in magnitude in Group A and B controllers, and recurred on average about once every 7 weeks during the first 30 weeks pi (Fig. 1d). Notably, the frequency of these viral blips declined significantly after week 30 pi such that, by 52 weeks pi, viral blips were rarely observed (Fig. 1d). No SIV-mediated pathogenesis (loss of effector site CD4+ T cells) was noted in Group A and B controllers (Suppl. Fig. 7), and the vast majority of blood and lymph node mononuclear cell specimens from these RM were negative for cell-associated SIV RNA and DNA (Suppl. Fig. 8). Six of 12 Group A and 5 of 12 Group B RM were not protected in this novel manner, but rather, demonstrated a typical pattern of progressive infection with associated pathogenesis (Fig. 1c/e, Suppl. Fig. 7). The mean peak and plateau phase pvls of the Group A RM with progressive infection were not statistically different from Group D controls (Fig. 1e), indicating that once systemic, progressive infection was established, RhCMV/SIV vector-elicited responses were unable to control virus replication. The addition of Ad5/SIV vectors in the Group B vaccination regimen was associated with a significantly reduced peak viremia in Group B RM with progressive infection compared to Group D controls, but this difference was lost in plateau phase. Consistent with previous reports11-13, the benchmark DNA/Ad5-vaccinated RM (Group C) showed significantly reduced log mean peak and early plateau (6-14 weeks pi) pvls, but for most of these RM, this partial virologic control was short-lived, as log mean pvls in later plateau phase were also not different from Group D controls (Fig. 1c/e). Importantly, the stringent control of SIV infection in protected Group A and B RM was not associated with CD8+ T cell responses restricted by protective MHC alleles (Suppl. Fig. 3b) or with TRIM 5 polymorphisms associated with target cell susceptibility to SIV infection (Suppl. Fig. 9).
Taken together, these data suggest that RhCMV/SIV vector-elicited immune responses mediate a novel pattern of protection in which mucosally administered SIVmac239 is stringently controlled before the onset of progressive, systemic infection. As shown in Fig. 2a and Suppl. Fig. 10, the peak frequencies of SIV-specific CD8+ (but not CD4+) T cells during the vaccine phase (which occurred shortly after the boost), but not the frequencies immediately pre-challenge, significantly correlated with protection in both Groups A and B. These peak responses reflect the level of overall production of SIV-specific CD8+ T cells by the vaccine, and for a TEM-biased response would likely parallel the extent of TEM seeding at effector sites. SIVenv-specific antibody (Ab) responses are not generated by our RhCMV/SIV vectors5, and did not develop after SIV infection in Group A controllers (Fig. 2b). Although Ad5/SIVenv vector-vaccinated RM in Group B developed low titre SIVenv-specific (tissue culture-adapted SIVmac251-neutralizing) Ab responses prior to challenge, these titres did not predict control, and were not boosted by controlled infection. In contrast, with the exception of rapid progressors, SIVenv-specific Ab responses developed or were boosted in all RM with systemic, progressive SIV infection. These findings suggest that Ab responses are unlikely to significantly contribute to the protection observed in Group A and B RM, and further confirm the stringency of protection in RhCMV/SIV vector-vaccinated controllers, as SIV replication in these RM produced insufficient antigen to drive humoral immune responses.
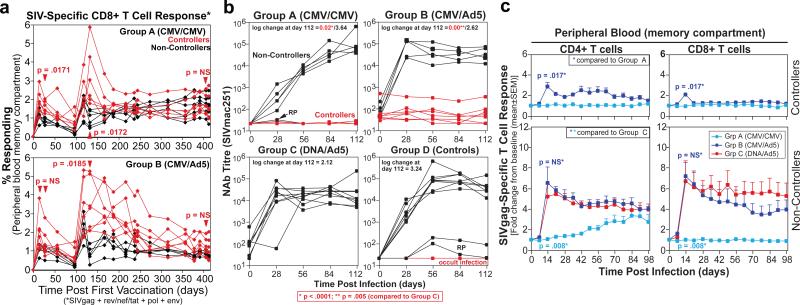
Figure 2. Immunologic correlates of RhCMV/SIV vector-associated control.
a, Analysis of total SIV-specific CD8+ T cell responses in the blood memory compartment during the vaccine phase of Group A and B RM with differences in the magnitude of these responses between controllers and non-controllers at the designated time points determined by the Wilcoxon rank sum test. b, Comparison of the anti-SIVenv antibody titres in plasma (as measured by neutralization of tissue culture-adapted SIVmac251) before and following infection of controller vs. non-controller RM among Group A-C and the concurrent Group D RM (RP = rapid progressor). The significance of the differences in log change in Ab titre from pre-infection to day 112 pi in Group A and B controllers vs. Group C RM was determined by the Wilcoxon rank sum test. c, Analysis of the change in the SIVgag-specific CD4+ and CD8+ T cell response frequency following controlled vs. progressive infection in Groups A, B and C with the significance of differences in peak response boosting between the designated groups determined by the Wilcoxon rank sum test.
We next investigated the effect of SIV infection on the magnitude of the vaccine-elicited T cell responses. Strikingly, Group A RM showed an almost complete lack of an anamnestic SIVgag-specific CD4+ or CD8+ T cell response to either progressive or controlled SIV infection (Fig. 2c; Suppl. Fig. 11). Group B RM demonstrated a modest anamnestic response in the setting of control, whereas in the setting of progressive infection these RM manifested a robust anamnestic response, similar to or only slightly less than that observed in Group C RM. Thus, despite the facts that Group B RM manifested circulating SIV-specific CD8+ T cells responses with the characteristic marked TEM-bias of RhCMV/SIV vector-elicited responses (Suppl. Fig. 5), and the early, abrupt RhCMV/SIV vector-associated pattern of protection (Fig. 1c), these RM appeared to maintain a distinct, Ad5 vector-elicited, SIV-specific TCM population capable of anamnestic expansion upon either controlled or progressive SIV infection. Importantly, Group A and B controllers robustly responded to infection with a de novo (Group A) or boosted (Group B) CD4+ and CD8+ T cell responses to SIVvif, an antigen not included in the RhCMV/SIV vectors used in this study (Suppl. Fig. 12), confirming both the presence of SIV infection in these RM, and the normal ability of their naïve T cell (Group A) and TCM (Group B) compartments to respond to the infection. These results indicate that not only does RhCMV/SIV vector-associated viral control occur in the absence of an overt anamnestic response, but that the SIV-specific TEM populations generated by RhCMV/SIV vectors alone appear unable to significantly expand after infection, regardless of whether antigen levels are limiting (controlled infection) or abundant (progressive infection). This lack of anamnestic expansion may account for the inability of Group A RM (in contrast to Group B RM) to manifest any suppression of viral replication once a systemic, progressive infection was established.
The decline in the frequency of SIV RNA blips in the plasma of RhCMV/SIV vector-vaccinated controllers over time suggests progressive loss of SIV-infected cells, either by immune clearance, virolysis or other attritive mechanisms. To explore the extent of residual infection in long-term RhCMV/SIV-vaccinated controllers, we used mAbs to deplete CD4+ or CD8+ lymphocytes from 2 Group A and 2 Group B controllers, in comparison to a Group C (DNA/Ad5) and a Group D (unvaccinated) RM with partial virologic control, for each treatment. Administration of the anti-CD4 huOKT4A mAb did not release viral replication in either Group C and D partial controllers or Group A and B complete controllers (Suppl. Fig. 13). In keeping with previous studies14,15, CD8+ lymphocyte depletion with mAb cM-T807 did result in a pronounced increase in pvl in a Group C and D RM with partial control, associated with a robust expansion of SIVvif-specific CD4+ T cells in effector sites (Fig. 3a). In contrast, CD8+ lymphocyte depletion failed to increase plasma viremia in RhCMV/SIV vector-vaccinated controllers, and the SIVvif-specific CD4+ T cell responses in these RM were unchanged following depletion, suggesting the absence of even a transient increase in viral replication not detectable by pvl measurements. These studies extend our previous data on the insensitivity of RhCMV/SIV vector-associated control to CD8+ lymphocyte depletion5 to RM that manifested a higher initial viremia as well as a period of subsequent, intermittent pvl blips.
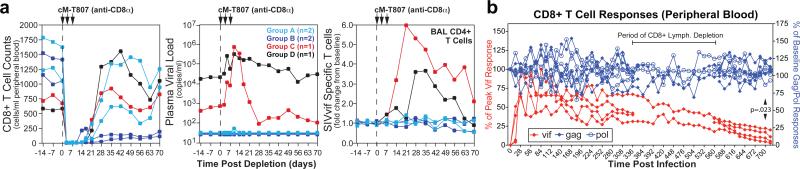
Figure 3. Immunologic characterization of long-term control associated with RhCMV/SIV vector vaccination.
a, Analysis of the effect of depletion of CD8+ lymphocytes with mAb cM-T807 on viral replication and boosting of SIVvif-specific T cell responses (in the non-depleted CD4+ subset) in 4 long-term RhCMV/SIV vector-vaccinated controllers (2 Group A and 2 Group B RM) vs. 2 conventional controllers (1 Group C, DNA/Ad5-vaccinated controller; 1 Group D spontaneous controller). b, Analysis of the frequencies of blood CD8+ T cells specific for SIV proteins that were (gag, pol) or were not (vif) included in the CMV/SIV vectors in the 4 Group A “controllers” for which long-term data is available. The response frequencies were normalized to the response frequencies immediately prior to SIV infection for the gag- and pol-specific responses, and to the peak frequencies following SIV infection for the vif-specific responses. The 4 RM used in this long-term response analysis include those subjected to transient CD4+ or CD8+ lymphocyte depletion (2 each). As Ag-specific CD8+ responses cannot be reliably determined during the period of overall CD8+ lymphocyte depletion, these periods are shown as gaps for 2 affected RM. The significance of differences in the maintenance of response frequencies of gag- and pol- vs. vif-specific CD8+ T cells in these RM was determined by Wilcoxon rank sum analysis.
As CD8+ T cell depletion with mAb cM-T807 is typically not complete in tissues (Suppl. Fig. 14), lack of viral rebound following such treatment of RhCMV/SIV vector-vaccinated controllers may simply reflect the potent anti-viral function of such residual SIV-specific CD8+ TEM cells, or possibly, the compensatory activity of anti-viral CD4+ TEM. On the other hand, these observations also raise the possibility that the frequency of SIV-infected and potentially infectious cells in long-term RhCMV/SIV vector-vaccinated controllers might have been reduced over time to levels that made detectable viral rebound unlikely. In this regard, we found that in Group A controllers, both CD8+ and CD4+ T cell responses to SIVvif, an Ag that was not included in the RhCMV/SIV vectors and therefore only available from SIV-infected cells, progressively waned over time to an average of <10% of their peak response immediately after (controlled) infection (Fig. 3b, Suppl. Fig. 15), suggesting that the numbers of productively infected cells present in these long-term controller RM are very few, below the threshold necessary to support the initially high frequency vif-specific responses. To further examine the extent of residual infection in long-term RhCMV/SIV-vaccinated controllers, we rigorously quantified SIV RNA and DNA at necropsy in 4 such RM (≥ week 52 post infection; lacking pvl blips for ≥10 weeks prior to necropsy) in comparison to an uninfected RM, two RM with SIV infections that were well-controlled by standard criteria, and an additional RM with poorly controlled, progressive SIV infection. As shown in Fig. 4, extensive analysis of lymphoid tissues and immune effector sites of RhCMV/SIV vector-vaccinated controllers with ultra-sensitive nested, quantitative PCR/RT-PCR assays (10 reactions per tissue specimen) demonstrated that cell-associated SIV RNA and DNA were undetectable (0/10 reactions +) in 72% and 80%, respectively, of specimens, and in those tissues where viral sequences were detected, the levels were extremely low (~ single copy per 107-108 cell equivalents). Notably, the majority of specimens with detectable SIV DNA or RNA (77% and 73%, respectively) were from outside the rectal mucosa. Cell-associated SIV RNA and DNA were not detected in any tissues from an SIV-negative RM, but were readily detected in all tissues of RM with conventionally controlled SIV infection. Overall, tissue levels in these conventional controllers averaged >3 logs higher than the measureable values of RhCMV/SIV vector-vaccinated controllers (p <.0001 by the Wilcoxon rank sum test). Levels of cell-associated SIV were higher still in an RM with poorly controlled infection. We also assayed lymphoid tissue cells from these RM for the presence of inducible, replication competent SIV by co-culture (Suppl. Table 1). All co-cultures (up to 20 replicates per specimen) from RhCMV/SIV vector-vaccinated controllers were negative for recoverable SIV, whereas replication-competent SIV was readily detected in co-cultures of tissue cells from the conventional controllers. The paucity of SIV nucleic acid and the lack of recoverable SIV in RhCMV/SIV vector-vaccinated controller RM are in sharp contrast to the levels of HIV or SIV found in either humans or RM receiving highly active anti-retroviral therapy or in elite controllers16-21, and suggest an unprecedented level of SIV control and even the possibility of progressive clearance of the SIV infection over time. Importantly, despite little or no SIV replication in the RhCMV/SIV vector-vaccinated controllers, peripheral blood T cells specific for SIV proteins included in the RhCMV/SIV vectors (e.g., gag and pol) were stably maintained at high frequency through 700 days pi (CD8+ response with 94 ± 0.5% TEM phenotype), in contrast to the SIV infection-elicited vif-specific responses; Fig. 3b, Suppl. Fig. 15). Thus, persistent RhCMV/SIV vectors provide for long-term maintenance of high frequency SIV-specific TEM responses, which would otherwise wane with stringent virologic control, thereby ensuring continuous, high-level surveillance for SIV-infected cells, even when only rare infected cells are present.
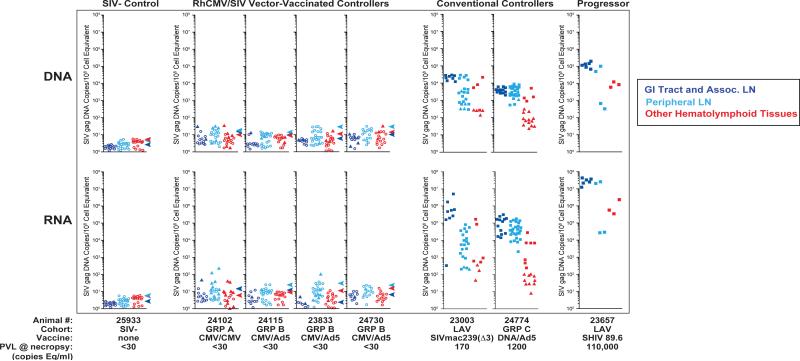
Figure 4. Measurement of SIV RNA and DNA in long-term RhCMV/SIV vector-vaccinated controllers.
Nested qPCR and qRT-PCR analysis of SIV DNA and RNA, respectively, on tissue obtained at necropsy from an uninfected RM, 4 long-term (>52 wk) RhCMV/SIV vector-vaccinated controller RM (1 Group A; 3 Group B), 2 conventional controller RM [a live attenuated SIV (LAV)-vaccinated RM that resisted wildtype SIVmac239 challenge 33 and 10 weeks prior to necropsy, and a Group C, DNA/Ad5 vaccine-protected RM at 55 weeks pi], and an RM with poorly controlled SIV infection [a LAV-vaccinated RM 24 weeks after wildtype SIVmac239 challenge]. Filled square plot symbols indicate DNA or RNA copy numbers based on directly measured values for samples giving 10/10 replicate reactions positive. Filled triangles indicate results for samples giving at least one, but <10, replicate reactions positive, with copy number imputed by Poisson distribution. Open circles indicate specimens that gave 0/10 replicates positive with the symbol's position in the plots at the threshold value corresponding to a Poisson distribution imputed copy number corresponding to 1/10 replicates positive. All values are normalized for nucleic acid input. Arrowheads indicate the highest threshold value for negative samples (0/10 replicates positive) for all of the tissues analyzed for that RM. “GI Tract and Associated LN” include colon/rectum, ileum, jejunum, superior/medial/inferior mesenteric and ileocecal LNs. “Peripheral LN” include axillary, submandibular, inguinal, iliosacral, and tracheobronchial LNs. “Other Hematolymphoid Tissues” include liver, spleen, bone marrow, tonsil, and thymus.
In summary, the 16 long-term RhCMV/SIV vector-vaccinated controllers described in this and our previous study5 unequivocally demonstrate a previously undescribed form of immune-mediated control of highly pathogenic SIV in which mucosally acquired infection is arrested prior to irreversible establishment of disseminated, progressive infection. Although stringently controlled, residual SIV infection is still present for weeks to months in most of these controllers, but wanes over time until eventually it is barely detectable by the most sensitive molecular virologic and immunologic criteria. The available data strongly suggest that this unique control is related to the high frequency CD8+, and possibly CD4+, TEM-biased, SIV-specific T cell responses that are elicited and indefinitely maintained by the persistent RhCMV/SIV vectors, are situated in both mucosal portals of entry and potential sites of distant viral spread, and can protect without anamnestic expansion (see Supplemental Discussion). The ability of RhCMV/SIV vectors to indefinitely maintain SIV-specific TEM responses in these sites, independent of the level of SIV replication, provides for continuous surveillance for SIV-infected cells, preventing relapse and, perhaps, ultimately clearing residual infection. Thus, CMV vectors provide a powerful new approach for HIV/AIDS vaccine development that could be used alone or in combination with complementary vaccine strategies that exploit different HIV immune vulnerabilities.
METHODS SUMMARY
Sixty-eight purpose-bred male RM (Macaca mulatta) of Indian genetic descent were used in this study. RhCMV/SIV vectors were given subcutaneously at a dose of 5 × 106 plaque-forming units per vector. DNA and Ad5 vectors were given intramuscularly at doses of 1.6 mg per vector and 2 × 1010 particle units per vector, respectively. RM were challenged intra-rectally with SIVmac239 using a repeated (weekly), limiting dose protocol5. After the onset of infection (pvl ≥30 copy eq/ml), RM were followed weekly until onset of AIDS or a minimum of 224 days for progressive infection and 365 days for controlled infection. SIV- and RhCMV-specific CD4+ and CD8+ T cell responses were measured in mononuclear cell preparations from blood and tissues by flow cytometric intracellular cytokine analysis5. SIVenv-specific Abs were determined by neutralization of tissue culture-adapted SIVmac251 using a luciferase reporter gene assay22. Levels of SIV RNA and DNA in plasma and from isolated cell preparations were quantified by standard quantitative real-time PCR and RT-PCR assays23,24. Tissue-associated SIV RNA and DNA at necropsy were quantified by an ultra-sensitive nested, quantitative real-time PCR and RT-PCR approach (see Full Methods). The presence of inducible, replication competent SIV in mononuclear cell preparations was detected by co-cultivation with CEMx174 cells, as previously described16.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institute of Allergy and Infectious Diseases (RO1 AI060392; contract #HHSN272200900037C); the International AIDS Vaccine Initiative (IAVI) and its donors, particularly the United States Agency for International Development (USAID); the Bill & Melinda Gates Foundation-supported Collaboration for AIDS Vaccine Discovery; the National Center for Research Resources (P51 RR00163; R24 RR016001); and the National Cancer Institute (contract HHSN261200800001E). We thank A. Sylwester, D. Seiss, R. Lum, H. Park and A. Okoye for specialized technical assistance; P. Barry, G. Pavlakis, G. Franchini, C. Miller, N. Wilson, and K. Reimann and Nonhuman Primate Reagent Resource for provision of crucial constructs or reagents; D. Watkins for MHC typing; D. Montefiori for nAb assays; N. Letvin and L. Shen for TRIM5a typing; S. Mongoue-Tchokote and M. Mori for statistical assistance; A. Townsend and T. Schroyer for figure preparation; and K. Früh, D. Watkins, B. Beresford, A. McDermott, R. King and W. Koff for helpful discussion and advice.
FULL METHODS
Rhesus macaques
A total of 68 purpose-bred juvenile and adult male rhesus macaques (RM) (Macaca mulatta) of Indian genetic descent were used in the experiments reported in this study, including 61 RhCMV+ RM in the vaccination/challenge experiment shown in Fig. 1a (Group A and B: n=12 each; Group C: n=9, and Group D: n=28; 11 concurrent and 17 historical controls), 4 long-term RhCMV/gag vector-vaccinated RM, 2 live attenuated SIV vector-vaccinated/wildtype SIVmac239-challenged RM (one controller and one non-controller), and one unvaccinated, uninfected RM. All RM were free of cercopithicine herpesvirus 1, D-type simian retrovirus, simian T-lymphotrophic virus type 1 and SIV infection at the start of the study. Group A-C and concurrent Group D included 4 RM each with the Mamu A*01 allele, but no RM with the B*08 and B*17 alleles associated with post-peak control of SIV replication4. Of the 18 historical Group D RM, 11 lacked A*01, B*08, and B*17 alleles, and 6 expressed either B*08 or B*17. The latter RM were excluded from analysis of viral load in progressive infection, as they expressed protective alleles that were not represented in the vaccine groups. RM were used with approval of the Oregon National Primate Research Center Animal Care and Use Committee under the standards of the US National Institutes of Health Guide for the Care and Use of Laboratory Animals. RhCMV/SIV vectors (RhCMV-gag, -rev/nef/tat, -env, -pol1 and -pol2, see below) were given subcutaneously at a dose of 5 × 106 plaque-forming units. Ad5-SIV vectors (Ad5-gag, -env, -pol, -nef and –VVVTR, see below) were given intramuscularly (IM) at a dose of 2 × 1010 particle units per vector, and DNA (IM at 1.6 mg per vector) was given at weeks 0, 4 and 8 for the DNA prime animals prior to Ad5-SIV boost at week 14. Plasmids expressing a fusion protein comprised of SIV vif, vpr, vpx, tat and rev, or individual gag, pol, env, and nef open reading frames (ORFs) were used for DNA priming. RM were challenged intra-rectally with highly pathogenic SIVmac239 using the repeated (weekly), limiting dose protocol described previously5. Plasma viral loads (pvl) were measured weekly, with challenge discontinued the week after detection of >30 copy equivalents per ml of SIV RNA (with the week prior to this first + pvl considered the day of infection). RM were considered controllers if pvl became undetectable within 2 weeks of the initial positive pvl and was then maintained below threshold for at least 4 of the subsequent 5 weeks. Challenged RM were followed until onset of AIDS or a minimum of 224 days for progressive infection and a minimum of 365 days for controlled infection. For CD8+ lymphocyte depletion, RM were administered 10, 5, 5, and 5 mg per kg body weight of the humanized monoclonal anti-CD8α antibody cM-T807, on days 0, 3, 7, and 10, respectively25. For CD4+ lymphocyte depletion, RM were administered one dose of the humanized monoclonal anti-CD4 antibody huOKT4A at 50 mg per kg body weight26. Mononuclear cell preparations were obtained from blood, bone marrow, broncho-alveolar lavage (BAL), lymph nodes, spleen, liver, tonsil, thymus and intestinal mucosa, as previously described27-29. For SIV quantification by nested qPCR/RT-PCR, whole tissue pieces obtained at necropsy were then flash frozen in liquid nitrogen and stored at -80°C prior to nucleic acid isolation.
RhCMV/SIV vectors
Construction and characterization of RhCMV-Gag, RhCMV-Retanef, and RhCMV-Env has been described5. Two additional RhCMV viruses expressing either the 5’ (protease/reverse transcriptase) (designated RhCMV-Pol1), or 3’ (RNAse H/integrase) (designated RhCMV-Pol2) of SIVmac239 Pol were constructed in an identical fashion by using E/T recombination and the RhCMV (68-1) bacterial artificial chromosome (BAC) (pRhCMV/BAC-Cre)30. Deletions were introduced into Pol to inactivate protease (Δ25-DTG-27), reverse transcriptase (Δ184-YMDD-187), RNaseH (ΔE478), and integrase (ΔD64, ΔD116, and ΔE152)31. Pol protein expression was placed under control of the EF1α promoter to achieve maximal expression. In all RhCMV/SIV vectors, the SIV antigen-expressing cassettes are inserted into the pRhCMV/BAC-Cre at nucleotide 207,630 within a non-coding region between rh213 and Rh214. RhCMV/SIV viruses were reconstituted by transfection of recombinant BAC DNA into RhCMV-permissive RM fibroblasts. Following virus reconstitution and BAC cassette ‘self-excision’, RhCMV/SIV vectors contain the entire wild-type (68-1) RhCMV genome30. Vector SIV antigen expression was confirmed by western blot analysis using antibodies to FLAG (Sigma-Aldrich; RhCMV-Gag), V5 (Invitrogen; RhCMV-Retanef), c-Myc/KK45 (Sigma-Aldrich; RhCMV-Env), and HA (Sigma-Aldrich; RhCMV-Pol1 and Pol2).
DNA vaccines
The plasmid vaccine immunogens used in this study covered the full SIVmac239 (NCBI M33261) genome. ORFs were sequence-optimized for expression in mammalian cells and cloned into a plasmid DNA expression vector. Expression of the intronless coding sequences was controlled by the human CMV (HCMV) promoter/enhancer and the bovine growth hormone polyadenylation signal32. Gag (12S), Pol (91S), Env (99S) and Nef (pCMV-Nef) were expressed as single polypeptides, while Vif, Vpr, Vpx, Tat and Rev were expressed as a fusion protein (pCMV-VVVTR). The 12S Gag plasmid expresses a myristoylated Gag protein spanning amino acids (aa) 1-508 and lacks 2 C-terminal aa, but is otherwise similar to a gag plasmid reported33. Plasmid 99S expresses a native form of gp160, as previously reported31,33. Sequence optimized Nef without a myristoylation signal was PCR-amplified from 179S plasmid31, and subsequently transferred into the pCMV vector. The Pol (91S) coding sequence contained deletion mutations to inactivate protease, reverse transcriptase, RNaseH, and integrase, as described31. Large-scale plasmid production for immunization was prepared by Aldevron, LLC. Expression of plasmids after transient transfection of HEK 293 cells was corroborated by western blot using polyclonal antibodies against SIVmac239 Gag, Env, Nef, and Rev proteins made in house, and anti-SIVmac251 serum from the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
Adenovirus vectors
Plasmid SIVmac239 sequences, again covering the full SIVmac239 genome, were cloned into an human adenovirus serotype 5 (Ad5), which lacks E1A and most of E1B and ΔE3 using the Adeasy Adenoviral vector system (Stratagene) to make the Ad5-SIVgag, Ad5-SIVenv, Ad5-SIVpol, AD5-SIVnef and Ad5-SIVVVVTR vectors. SIVmac239 genes were inserted into the E1 region of the Ad5 under the control of the HCMV immediate early promoter/enhancer and the SV40 polyadenylation signal. All vectors were rescued and propagated in HEK 293 cells and purified by double cesium chloride centrifugation34. Dosing was based on the physical number of particles (PU) of Ad5 as measured by spectrophotometry35. Expression of SIV proteins from Ad5 vectors after A549 infection was confirmed by western blot as described above.
SIVmac239 challenge virus
The pathogenic SIVmac239 challenge virus stock (generously provided by Dr. C. Miller) was generated by expanding the SIVmac239 clone in RM peripheral PBMC, and was quantified using the sMAGI cell assay and by quantitative RT-PCR for SIV genomic RNA5.
Immunologic assays
SIV- and RhCMV-specific CD4+ and CD8+ T cell responses were measured in mononuclear cell preparations from blood and tissues by flow cytometric intracellular cytokine analysis (FCICA), as previously described in detail5. Briefly, sequential 15-mer peptides (overlapping by 11 amino acids) comprising the SIVmac239 gag, rev/nef/tat, env, pol, vif and vpr/vpx proteins were used in the presence of co-stimulatory mAbs CD28 and CD49d (BD Biosciences). Cells were incubated with antigen and co-stimulatory molecules alone for 1 hr, followed by addition of Brefeldin A (Sigma-Aldrich) for an additional 8 hrs. Co-stimulation without antigen served as a background control. Cells were then stained with fluorochrome-conjugated mAbs, flow cytometric data collected on an LSR II (BD Biosciences) and data analyzed using the FlowJo software program (version 8.8.6; Tree Star, Inc.). Response frequencies (CD69+/TNF+ and/or CD69+/IFN-γ+) were first determined in the overall CD4+ and CD8+ population and then memory corrected (with memory T cell subset populations delineated on the basis of CD28 and CD95 expression), as previously described5,27. The data presented as “end of vaccine phase” response frequencies represent an average of values obtained from samples collected on days 379, 392, 401 and 413 after initial vaccination. Titres of SIVenv-specific Abs were determined by neutralization of tissue culture-adapted SIVmac251 using a luciferase reporter gene assay22.
Viral detection assays
Quantitative real-time RT-PCR and PCR assays targeting a highly conserved sequence in Gag were used for standard measurements of plasma SIV RNA and cell-associated SIV RNA and DNA within peripheral blood and lymph node mononuclear cells, as previously described23,24. For plasma testing of a sample to score as positive, duplicate amplification reactions yielding ≥ 30 copy Eq/mL were required. Isolated viral blips were also repeated in a duplicate sample in almost all instances, and in RM where infection was manifest only by isolated viral blips (Group A and B controllers), infection was confirmed by the development (Group A) or boosting (Group B) of T cell responses specific for SIVvif, an SIV Ag not included in the RhCMV/SIV vectors (Suppl. Fig. 12), as described5. To further address the possibility of false positive amplification reactions, we analyzed 136 known SIV-negative (pre-challenge) samples from the present study, and from other studies from the same facility and investigators and others. Samples were run in the same laboratory, over approximately the same period of time, using the same procedures, with negative specimens interspersed with positive samples in assay runs. Zero of 136 known SIV-negative samples scored positive by the above criteria, which was significantly different from 84 positive samples (viral “blips”) of 678 total samples (12.4%) obtained from RhCMV/SIV vector-vaccinated controllers in vaccine Groups A and B (p < .0001, Fischer's exact test). To more precisely characterize levels of SIV DNA and RNA from tissues of RhCMV/SIV vector-vaccinated controllers (which were below the sensitivity threshold of the standard assays), we utilized a new ultra-sensitive, nested, quantitative real-time PCR/RT-PCR method. Tissue pieces were collected directly into extraction tubes and immediately frozen using liquid nitrogen. Samples were stored at -80°C and handled on dry ice until stabilized in extraction solution. Specimens of approximately 100mg or less were homogenized in 1 ml of TriReagent (Molecular Research Center, Inc.) in 2 ml extraction tubes of Lysing Matrix D using FastPrep instrumentation (MP Biomedicals) according to the manufacturer's recommendations. Total RNA and DNA were prepared from the homogenates following manufacturer's recommendations, but specifically following the alternative, back-extraction method for DNA extraction. Recovered RNA and DNA were dissolved in minimal volumes of 10 mM TrisCl, pH 8.0 and 10 mM TrisCl, pH 9.0, respectively, as appropriate for replicate testing in qRT-PCR and qPCR protocols. Tissue specimens greater than 100 mg in mass were initially pulverized to powder under cryogenic conditions prior to extraction of RNA and DNA. Pulverization was accomplished in 15 ml polycarbonate extraction tubes with stainless steel grinding balls using rapid vertical shaking, all being maintained at appropriate temperatures in aluminum blocks pre-chilled in liquid N2 (GenoGrinder, SPEX SamplePrep). Pulverized tissue powder was then suspended in 3-10 ml of TriReagent, depending on the starting amount of tissue. Total RNA and DNA were then prepared from 1 ml of TriReagent suspension as described above; residual suspension was archived at -80° C for additional analysis, as necessary. To maximize sensitivity, “nested” quantitative real-time PCR/RT-PCR protocols were designed to accommodate higher amounts of input nucleic acid, and potential inhibitors, than are typically tolerated in standard assays. Reaction conditions and thermal profiles followed those referenced above for the plasma and isolated cell assays23,24 with two exceptions: 1) in the qRT-PCR assay, the “nested” reverse primer, as opposed to random hexamers, was used to prime cDNA synthesis specifically for SIV sequence, thereby avoiding generation of non-specific targets and further enhancing the direct sensitivity of detection of SIV RNA and 2) 2.5 units of PlatinumTaq (Invitrogen), rather than 1.25 units of TaqGold (Applied Biosystems), were employed in the amplification steps. A “nested” or “pre-amplification” of cDNA or DNA was performed for 12 cycles with the application of primers, SIVnestF01 (GATTTGGATTAGCAG AAAGCCTGTTG) and SIVnestR01 (GTTGGTCTACTTGTTTTTGGCATAGTTTC), flanking the SIV Gag target region. Five microliters of this first amplification were then transferred to 50 microliters of cocktail for amplification of SIV DNA gag in duplex, or not for RNA samples, with amplification of a single copy rhesus CCR5 target sequence for normalization, as referenced above23,24, and real-time PCR was then performed. For both RNA and DNA determinations, 12 replicate reactions were tested per sample including a spike of 10 copies of RNA or DNA internal control sequence standard in two of the 12 reactions to assess overall amplification efficiency and assess potential inhibition of the PCR or RT-PCR. Samples showing greater than a 5 cycle shift in amplification of the spiked standard, compared to amplification in the absence of specimen nucleic acid, corresponding to less than 74% overall amplification efficiency, were diluted and re-assayed. Quantitative determinations for samples showing amplification in all replicates were derived directly with reference to a standard curve. Quantitative determinations for samples showing less than 10 positive amplifications in replicates were derived from the frequency of positive amplifications, corresponding to the presence of at least one target copy in a reaction, according to a Poisson distribution of a given median copy number per reaction. It should be noted that this assay yielded no positive reactions out of a total of 1,100 total reactions (RNA and DNA) from tissues derived from an SIV-uninfected RM, which was significantly different from 178 positive reactions of 4,310 total reactions (4.1%) from tissues derived from the 4 RhCMV/SIV vector-vaccinated controllers studied at necropsy (P < .0001; Fischer's exact test). The presence of inducible, replication competent SIV in mononuclear cell preparations derived from different tissue sites at necropsy was detected by co-cultivation with CEMx174 cells, as previously described16,36. To detect shedding of RhCMV/SIV vectors in the urine of vaccinated RM, virus was concentrated from cleared pan-collected urine and co-cultured with RM fibroblasts. Cell lysates were collected after development of cytopathic effect or after 28 days, and assessed for vector replication based on expression of SIV antigen-specific epitope tags by western blot analysis5.
Statistical analysis
We performed statistical analysis with SAS version 9.1 (Statistical Analysis System). Individual tests are described in the Figure Legends for all analyses. Briefly, Fisher's exact test was used to determine significance of categorical data such as the fraction of controllers vs. non-controllers in the different vaccine groups. The Wilcoxon rank sum and Kruskal-Wallis tests were used to compare populations of continuous data for groups of 2 and ≥3, respectively. Analysis of variance and a test for linear trend were used to determine the significance of the reduction in viral blip frequency over time in RhCMV vector-vaccinated controllers. In all analyses, we used a two-sided significance level (alpha) of 0.05, with correction made for multiple comparisons using the Bonferroni method.
Footnotes
AUTHOR CONTRIBUTIONS:
SGH planned and performed experiments and analyzed data, assisted by JCF, MSL, ABV, CMH, LC-J and NW. TS, AWL and MKA managed the animal protocols. MAJ designed, constructed and characterized the RhCMV vectors. MP, Jr. and JDL planned and performed SIV quantification studies, assisted by KO and RS. CLP and MJC designed and constructed the DNA and Ad5 vectors used in this study. JAN was involved in conception of the RhCMV vector strategy. LJP conceived the RhCMV vector strategy, supervised experiments, analyzed data, and wrote the paper, assisted by SGH, MAJ and JDL.
REFERENCES
- 1.Grovit-Ferbas K, Pappas T, O'Brien WA. In: Persistent Viral Infections. Ahmed R, Chen AI, editors. John Wiley & Sons; 1999. pp. 3–45. [Google Scholar]
- 2.Haase AT. Perils at mucosal front lines for HIV and SIV and their hosts. Nat. Rev. Immunol. 2005;5:783–792. doi: 10.1038/nri1706. [DOI] [PubMed] [Google Scholar]
- 3.Goulder PJ, Watkins DI. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat. Rev. Immunol. 2008;8:619–630. doi: 10.1038/nri2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haase AT. Targeting early infection to prevent HIV-1 mucosal transmission. Nature. 2010;464:217–223. doi: 10.1038/nature08757. [DOI] [PubMed] [Google Scholar]
- 5.Hansen SG, et al. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat. Med. 2009;15:293–299. doi: 10.1038/nm.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robinson HL, Amara RR. T cell vaccines for microbial infections. Nat. Med. 2005;11:S25–32. doi: 10.1038/nm1212. [DOI] [PubMed] [Google Scholar]
- 7.Keele BF, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. USA. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chun TW, et al. Early establishment of a pool of latently infected, resting CD4(+) T cells during primary HIV-1 infection. Proc. Natl. Acad. Sci. USA. 1998;95:8869–8873. doi: 10.1073/pnas.95.15.8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bachmann MF, Kundig TM, Hengartner H, Zinkernagel RM. Protection against immunopathological consequences of a viral infection by activated but not resting cytotoxic T cells: T cell memory without “memory T cells”? Proc. Natl. Acad. Sci. U S A. 1997;94:640–645. doi: 10.1073/pnas.94.2.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ochsenbein AF, et al. A comparison of T cell memory against the same antigen induced by virus versus intracellular bacteria. Proc. Natl. Acad. Sci. U S A. 1999;96:9293–9298. doi: 10.1073/pnas.96.16.9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casimiro DR, et al. Attenuation of simian immunodeficiency virus SIVmac239 infection by prophylactic immunization with dna and recombinant adenoviral vaccine vectors expressing Gag. J. Virol. 2005;79:15547–15555. doi: 10.1128/JVI.79.24.15547-15555.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Letvin NL, et al. Preserved CD4+ central memory T cells and survival in vaccinated SIV-challenged monkeys. Science. 2006;312:1530–1533. doi: 10.1126/science.1124226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson NA, et al. Vaccine-induced cellular immune responses reduce plasma viral concentrations after repeated low-dose challenge with pathogenic simian immunodeficiency virus SIVmac239. J. Virol. 2006;80:5875–5885. doi: 10.1128/JVI.00171-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedrich TC, et al. Subdominant CD8+ T-cell responses are involved in durable control of AIDS virus replication. J. Virol. 2007;81:3465–3476. doi: 10.1128/JVI.02392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reynolds MR, et al. Macaques vaccinated with live-attenuated SIV control replication of heterologous virus. J. Exp. Med. 2008;205:2537–2550. doi: 10.1084/jem.20081524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen A, et al. Resting CD4+ T lymphocytes but not thymocytes provide a latent viral reservoir in a simian immunodeficiency virus-Macaca nemestrina model of human immunodeficiency virus type 1-infected patients on highly active antiretroviral therapy. J. Virol. 2003;77:4938–4949. doi: 10.1128/JVI.77.8.4938-4949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dinoso JB, et al. A simian immunodeficiency virus-infected macaque model to study viral reservoirs that persist during highly active antiretroviral therapy. J. Virol. 2009;83:9247–9257. doi: 10.1128/JVI.00840-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.North TW, et al. Viral sanctuaries during highly active antiretroviral therapy in a nonhuman primate model for AIDS. J. Virol. 2010;84:2913–2922. doi: 10.1128/JVI.02356-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruiz L, et al. Protease inhibitor-containing regimens compared with nucleoside analogues alone in the suppression of persistent HIV-1 replication in lymphoid tissue. AIDS. 1999;13:F1–8. doi: 10.1097/00002030-199901140-00001. [DOI] [PubMed] [Google Scholar]
- 20.Blankson JN, et al. Isolation and characterization of replication-competent human immuno-deficiency virus type 1 from a subset of elite suppressors. J. Virol. 2007;81:2508–2518. doi: 10.1128/JVI.02165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anton PA, et al. Multiple measures of HIV burden in blood and tissue are correlated with each other but not with clinical parameters in aviremic subjects. AIDS. 2003;17:53–63. doi: 10.1097/00002030-200301030-00008. [DOI] [PubMed] [Google Scholar]
- 22.Montefiori DC. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr. Protoc. Immunol. Chapter. 2005;12 doi: 10.1002/0471142735.im1211s64. Unit 12.11. [DOI] [PubMed] [Google Scholar]
- 23.Cline AN, Bess JW, Piatak M, Jr., Lifson JD. Highly sensitive SIV plasma viral load assay: practical considerations, realistic performance expectations, and application to reverse engineering of vaccines for AIDS. J. Med. Primatol. 2005;34:303–312. doi: 10.1111/j.1600-0684.2005.00128.x. [DOI] [PubMed] [Google Scholar]
- 24.Venneti S, et al. Longitudinal in vivo positron emission tomography imaging of infected and activated brain macrophages in a macaque model of human immunodeficiency virus encephalitis correlates with central and peripheral markers of encephalitis and areas of synaptic degeneration. Am. J. Pathol. 2008;172:1603–1616. doi: 10.2353/ajpath.2008.070967. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.