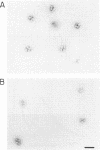
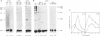Abstract
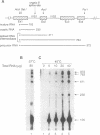
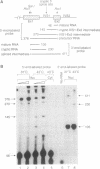
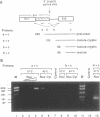
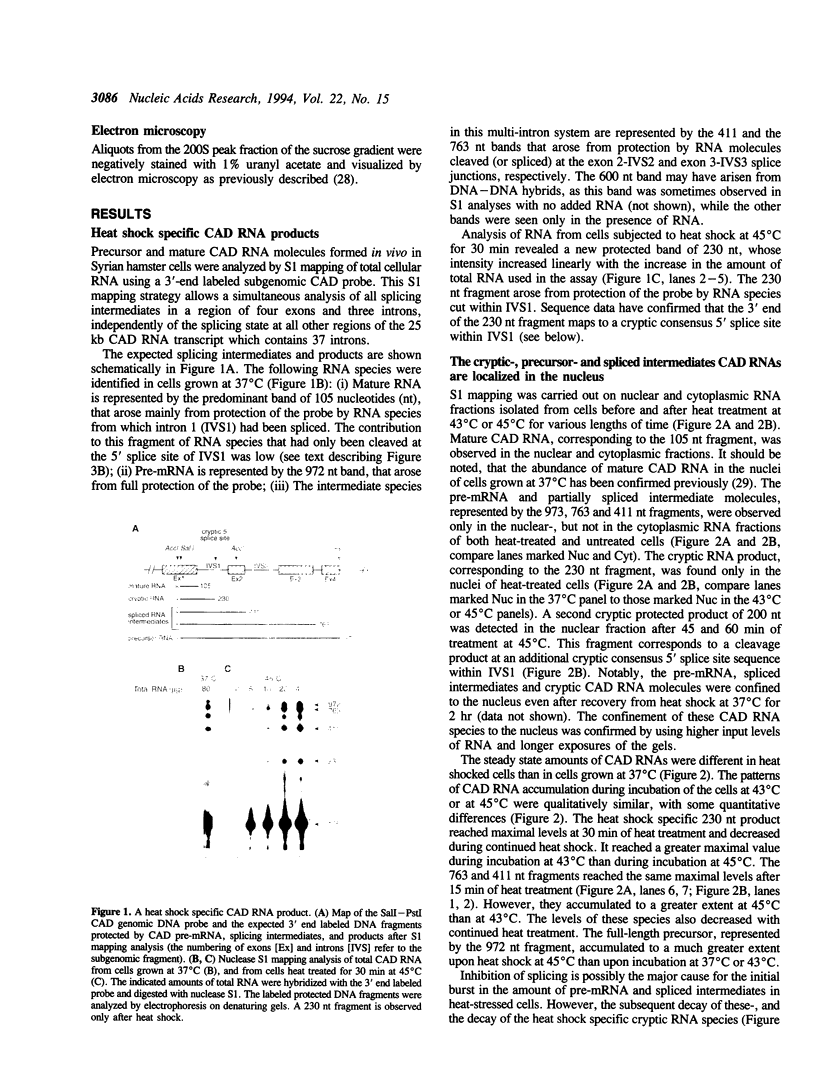
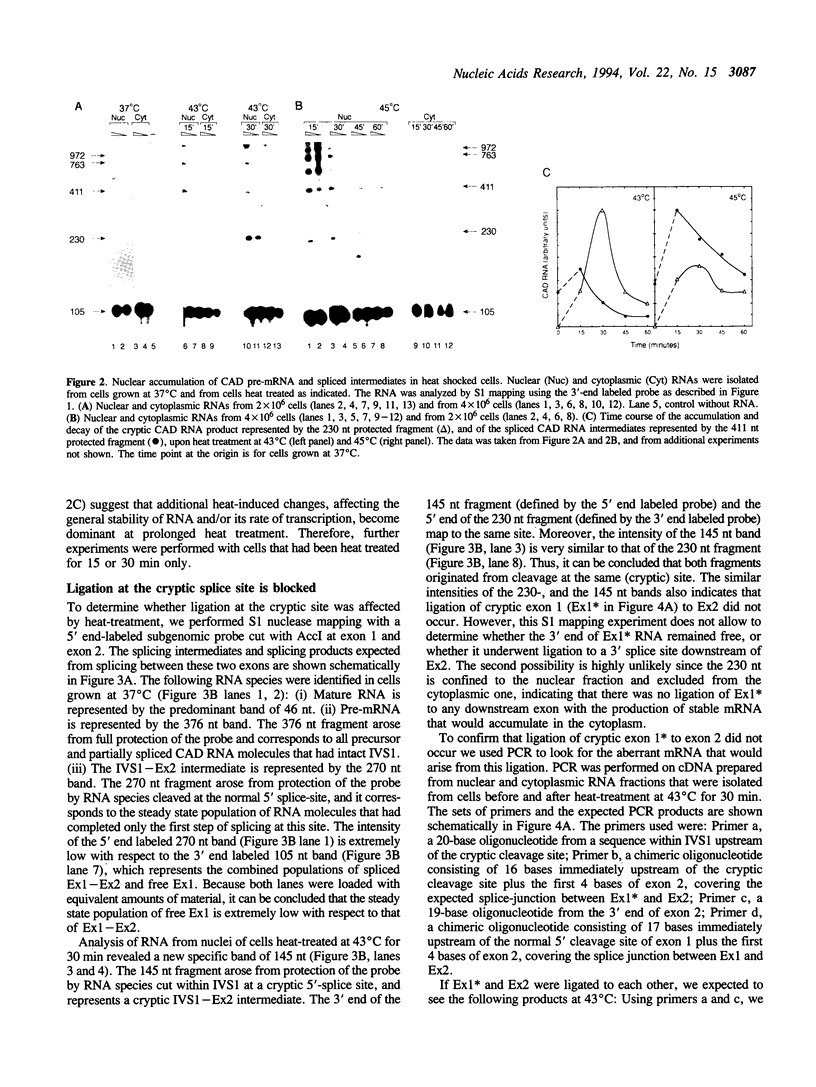
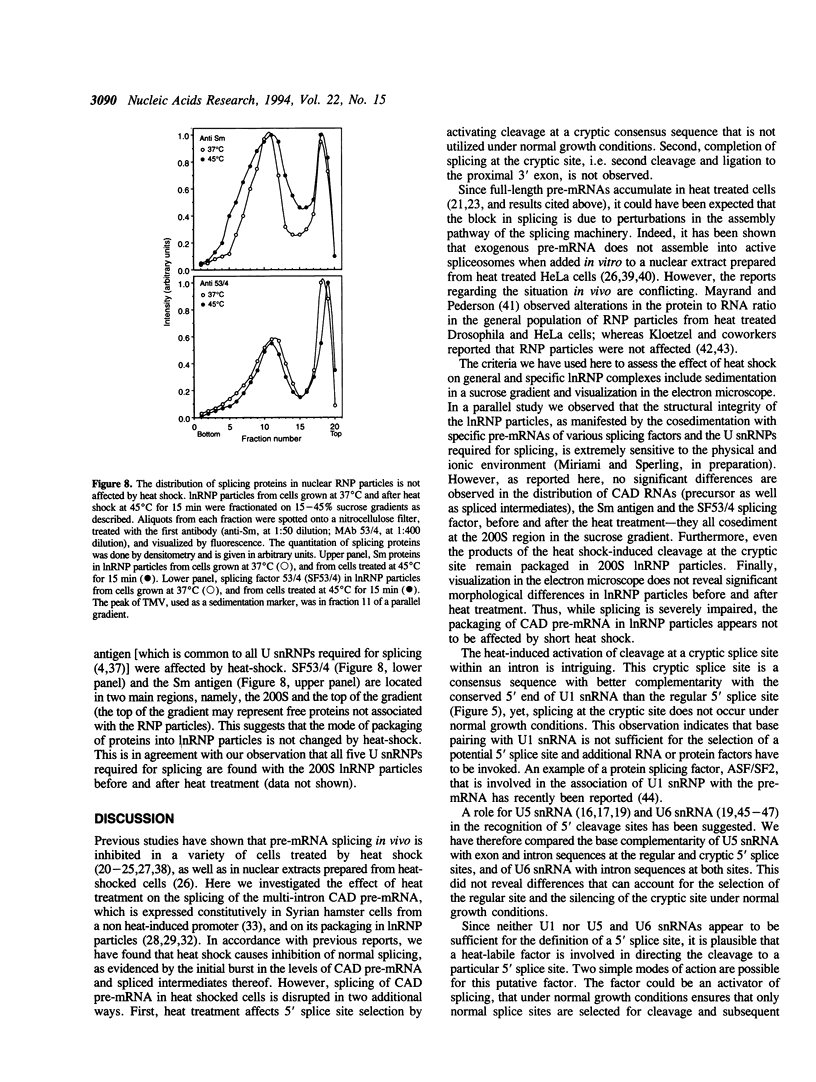
The effect of heat shock on the packaging and splicing of nuclear CAD pre-mRNA, a transcript expressed constitutively from a non heat-inducible promoter, was studied in vivo in Syrian hamster cells. While mild heat shock did not affect significantly the packaging of CAD RNA in 200S InRNP particles, it caused perturbation to splicing. First, the heat shock inhibited splicing of CAD pre-mRNA. Second, it affected 5' splice site selection by activating cleavage at a cryptic 5' splice site; yet ligation of the cryptic exon to the downstream proximal exon was not observed. Base complementarities of the cryptic site with U1, U5, or U6 snRNAs are comparable, or even better, than those with the neighboring normal site. Hence, the exclusion of the cryptic site under normal growth conditions cannot be attributed to weaker base pairing with these snRNAs. On the other hand, these results imply the involvement of a heat labile factor in the selection of the 5' cleavage site. The exclusion of the cryptic site at 37 degrees C and the aborted splicing at this site after heat shock may also be explained by a proposed nuclear checking mechanism that detects in-frame stop codons upstream of the 5' splice site, and aborts splicing at such sites to prevent the production of a defective message.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aebi M., Hornig H., Weissmann C. 5' cleavage site in eukaryotic pre-mRNA splicing is determined by the overall 5' splice region, not by the conserved 5' GU. Cell. 1987 Jul 17;50(2):237–246. doi: 10.1016/0092-8674(87)90219-4. [DOI] [PubMed] [Google Scholar]
- Ast G., Goldblatt D., Offen D., Sperling J., Sperling R. A novel splicing factor is an integral component of 200S large nuclear ribonucleoprotein (InRNP) particles. EMBO J. 1991 Feb;10(2):425–432. doi: 10.1002/j.1460-2075.1991.tb07964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baserga S. J., Benz E. J., Jr Nonsense mutations in the human beta-globin gene affect mRNA metabolism. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2056–2060. doi: 10.1073/pnas.85.7.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgrader P., Cheng J., Maquat L. E. Evidence to implicate translation by ribosomes in the mechanism by which nonsense codons reduce the nuclear level of human triosephosphate isomerase mRNA. Proc Natl Acad Sci U S A. 1993 Jan 15;90(2):482–486. doi: 10.1073/pnas.90.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond U. Heat shock but not other stress inducers leads to the disruption of a sub-set of snRNPs and inhibition of in vitro splicing in HeLa cells. EMBO J. 1988 Nov;7(11):3509–3518. doi: 10.1002/j.1460-2075.1988.tb03227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond U., Schlesinger M. J. The chicken ubiquitin gene contains a heat shock promoter and expresses an unstable mRNA in heat-shocked cells. Mol Cell Biol. 1986 Dec;6(12):4602–4610. doi: 10.1128/mcb.6.12.4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Maquat L. E. Nonsense codons can reduce the abundance of nuclear mRNA without affecting the abundance of pre-mRNA or the half-life of cytoplasmic mRNA. Mol Cell Biol. 1993 Mar;13(3):1892–1902. doi: 10.1128/mcb.13.3.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Daar I. O., Maquat L. E. Premature translation termination mediates triosephosphate isomerase mRNA degradation. Mol Cell Biol. 1988 Feb;8(2):802–813. doi: 10.1128/mcb.8.2.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eperon L. P., Estibeiro J. P., Eperon I. C. The role of nucleotide sequences in splice site selection in eukaryotic pre-messenger RNA. Nature. 1986 Nov 20;324(6094):280–282. doi: 10.1038/324280a0. [DOI] [PubMed] [Google Scholar]
- Green M. R. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu Rev Cell Biol. 1991;7:559–599. doi: 10.1146/annurev.cb.07.110191.003015. [DOI] [PubMed] [Google Scholar]
- Green M. R. Pre-mRNA splicing. Annu Rev Genet. 1986;20:671–708. doi: 10.1146/annurev.ge.20.120186.003323. [DOI] [PubMed] [Google Scholar]
- Kandels-Lewis S., Séraphin B. Involvement of U6 snRNA in 5' splice site selection. Science. 1993 Dec 24;262(5142):2035–2039. doi: 10.1126/science.8266100. [DOI] [PubMed] [Google Scholar]
- Kay R. J., Russnak R. H., Jones D., Mathias C., Candido E. P. Expression of intron-containing C. elegans heat shock genes in mouse cells demonstrates divergence of 3' splice site recognition sequences between nematodes and vertebrates, and an inhibitory effect of heat shock on the mammalian splicing apparatus. Nucleic Acids Res. 1987 May 11;15(9):3723–3741. doi: 10.1093/nar/15.9.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempe T. D., Swyryd E. A., Bruist M., Stark G. R. Stable mutants of mammalian cells that overproduce the first three enzymes of pyrimidine nucleotide biosynthesis. Cell. 1976 Dec;9(4 Pt 1):541–550. doi: 10.1016/0092-8674(76)90036-2. [DOI] [PubMed] [Google Scholar]
- Kloetzel P. M., Bautz E. K. Heat-shock proteins are associated with hnRNA in Drosophila melanogaster tissue culture cells. EMBO J. 1983;2(5):705–710. doi: 10.1002/j.1460-2075.1983.tb01488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohtz J. D., Jamison S. F., Will C. L., Zuo P., Lührmann R., Garcia-Blanco M. A., Manley J. L. Protein-protein interactions and 5'-splice-site recognition in mammalian mRNA precursors. Nature. 1994 Mar 10;368(6467):119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- Krainer A. R., Maniatis T. Multiple factors including the small nuclear ribonucleoproteins U1 and U2 are necessary for pre-mRNA splicing in vitro. Cell. 1985 Oct;42(3):725–736. doi: 10.1016/0092-8674(85)90269-7. [DOI] [PubMed] [Google Scholar]
- Kretzner L., Rymond B. C., Rosbash M. S. cerevisiae U1 RNA is large and has limited primary sequence homology to metazoan U1 snRNA. Cell. 1987 Aug 14;50(4):593–602. doi: 10.1016/0092-8674(87)90032-8. [DOI] [PubMed] [Google Scholar]
- Lear A. L., Eperon L. P., Wheatley I. M., Eperon I. C. Hierarchy for 5' splice site preference determined in vivo. J Mol Biol. 1990 Jan 5;211(1):103–115. doi: 10.1016/0022-2836(90)90014-D. [DOI] [PubMed] [Google Scholar]
- Lesser C. F., Guthrie C. Mutations in U6 snRNA that alter splice site specificity: implications for the active site. Science. 1993 Dec 24;262(5142):1982–1988. doi: 10.1126/science.8266093. [DOI] [PubMed] [Google Scholar]
- Maniak M., Nellen W. A developmentally regulated membrane protein gene in Dictyostelium discoideum is also induced by heat shock and cold shock. Mol Cell Biol. 1988 Jan;8(1):153–159. doi: 10.1128/mcb.8.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayrand S., Pederson T. Heat shock alters nuclear ribonucleoprotein assembly in Drosophila cells. Mol Cell Biol. 1983 Feb;3(2):161–171. doi: 10.1128/mcb.3.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhich M. L., Boothroyd J. C. Polycistronic transcripts in trypanosomes and their accumulation during heat shock: evidence for a precursor role in mRNA synthesis. Mol Cell Biol. 1988 Sep;8(9):3837–3846. doi: 10.1128/mcb.8.9.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeger L. K., Schoborg R. V., Zhao Q., Tullis G. E., Pintel D. J. Nonsense mutations inhibit splicing of MVM RNA in cis when they interrupt the reading frame of either exon of the final spliced product. Genes Dev. 1992 Jun;6(6):1107–1119. doi: 10.1101/gad.6.6.1107. [DOI] [PubMed] [Google Scholar]
- Nelson K. K., Green M. R. Splice site selection and ribonucleoprotein complex assembly during in vitro pre-mRNA splicing. Genes Dev. 1988 Mar;2(3):319–329. doi: 10.1101/gad.2.3.319. [DOI] [PubMed] [Google Scholar]
- Newman A. J., Norman C. U5 snRNA interacts with exon sequences at 5' and 3' splice sites. Cell. 1992 Feb 21;68(4):743–754. doi: 10.1016/0092-8674(92)90149-7. [DOI] [PubMed] [Google Scholar]
- Newman A., Norman C. Mutations in yeast U5 snRNA alter the specificity of 5' splice-site cleavage. Cell. 1991 Apr 5;65(1):115–123. doi: 10.1016/0092-8674(91)90413-s. [DOI] [PubMed] [Google Scholar]
- Ohshima Y., Gotoh Y. Signals for the selection of a splice site in pre-mRNA. Computer analysis of splice junction sequences and like sequences. J Mol Biol. 1987 May 20;195(2):247–259. doi: 10.1016/0022-2836(87)90647-4. [DOI] [PubMed] [Google Scholar]
- Padgett R. A., Wahl G. M., Stark G. R. Structure of the gene for CAD, the multifunctional protein that initiates UMP synthesis in Syrian hamster cells. Mol Cell Biol. 1982 Mar;2(3):293–301. doi: 10.1128/mcb.2.3.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa H., Shimura Y. Association of U6 snRNA with the 5'-splice site region of pre-mRNA in the spliceosome. Genes Dev. 1992 Feb;6(2):244–254. doi: 10.1101/gad.6.2.244. [DOI] [PubMed] [Google Scholar]
- Shukla R. R., Dominski Z., Zwierzynski T., Kole R. Inactivation of splicing factors in HeLa cells subjected to heat shock. J Biol Chem. 1990 Nov 25;265(33):20377–20383. [PubMed] [Google Scholar]
- Siliciano P. G., Guthrie C. 5' splice site selection in yeast: genetic alterations in base-pairing with U1 reveal additional requirements. Genes Dev. 1988 Oct;2(10):1258–1267. doi: 10.1101/gad.2.10.1258. [DOI] [PubMed] [Google Scholar]
- Sontheimer E. J., Steitz J. A. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science. 1993 Dec 24;262(5142):1989–1996. doi: 10.1126/science.8266094. [DOI] [PubMed] [Google Scholar]
- Spann P., Feinerman M., Sperling J., Sperling R. Isolation and visualization of large compact ribonucleoprotein particles of specific nuclear RNAs. Proc Natl Acad Sci U S A. 1989 Jan;86(2):466–470. doi: 10.1073/pnas.86.2.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling R., Spann P., Offen D., Sperling J. U1, U2, and U6 small nuclear ribonucleoproteins (snRNPs) are associated with large nuclear RNP particles containing transcripts of an amplified gene in vivo. Proc Natl Acad Sci U S A. 1986 Sep;83(18):6721–6725. doi: 10.1073/pnas.83.18.6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling R., Sperling J., Levine A. D., Spann P., Stark G. R., Kornberg R. D. Abundant nuclear ribonucleoprotein form of CAD RNA. Mol Cell Biol. 1985 Mar;5(3):569–575. doi: 10.1128/mcb.5.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B., Kretzner L., Rosbash M. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5' cleavage site. EMBO J. 1988 Aug;7(8):2533–2538. doi: 10.1002/j.1460-2075.1988.tb03101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B., Rosbash M. Exon mutations uncouple 5' splice site selection from U1 snRNA pairing. Cell. 1990 Nov 2;63(3):619–629. doi: 10.1016/0092-8674(90)90457-p. [DOI] [PubMed] [Google Scholar]
- Urlaub G., Mitchell P. J., Ciudad C. J., Chasin L. A. Nonsense mutations in the dihydrofolate reductase gene affect RNA processing. Mol Cell Biol. 1989 Jul;9(7):2868–2880. doi: 10.1128/mcb.9.7.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utans U., Behrens S. E., Lührmann R., Kole R., Krämer A. A splicing factor that is inactivated during in vivo heat shock is functionally equivalent to the [U4/U6.U5] triple snRNP-specific proteins. Genes Dev. 1992 Apr;6(4):631–641. doi: 10.1101/gad.6.4.631. [DOI] [PubMed] [Google Scholar]
- Wassarman D. A., Steitz J. A. Interactions of small nuclear RNA's with precursor messenger RNA during in vitro splicing. Science. 1992 Sep 25;257(5078):1918–1925. doi: 10.1126/science.1411506. [DOI] [PubMed] [Google Scholar]
- Weber S., Aebi M. In vitro splicing of mRNA precursors: 5' cleavage site can be predicted from the interaction between the 5' splice region and the 5' terminus of U1 snRNA. Nucleic Acids Res. 1988 Jan 25;16(2):471–486. doi: 10.1093/nar/16.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yost H. J., Lindquist S. Heat shock proteins affect RNA processing during the heat shock response of Saccharomyces cerevisiae. Mol Cell Biol. 1991 Feb;11(2):1062–1068. doi: 10.1128/mcb.11.2.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yost H. J., Lindquist S. RNA splicing is interrupted by heat shock and is rescued by heat shock protein synthesis. Cell. 1986 Apr 25;45(2):185–193. doi: 10.1016/0092-8674(86)90382-x. [DOI] [PubMed] [Google Scholar]
- Yost H. J., Lindquist S. Translation of unspliced transcripts after heat shock. Science. 1988 Dec 16;242(4885):1544–1548. doi: 10.1126/science.3201243. [DOI] [PubMed] [Google Scholar]
- Yost H. J., Petersen R. B., Lindquist S. RNA metabolism: strategies for regulation in the heat shock response. Trends Genet. 1990 Jul;6(7):223–227. doi: 10.1016/0168-9525(90)90183-7. [DOI] [PubMed] [Google Scholar]
- Zieg J., Clayton C. E., Ardeshir F., Giulotto E., Swyryd E. A., Stark G. R. Properties of single-step mutants of Syrian hamster cell lines resistant to N-(phosphonacetyl)-L-aspartate. Mol Cell Biol. 1983 Nov;3(11):2089–2098. doi: 10.1128/mcb.3.11.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]