Abstract
A prototype imaging surface plasmon resonance-based multiplex microimmunoassay for mycotoxins is described. A microarray of mycotoxin–protein conjugates was fabricated using a continuous flow microspotter device. A competitive inhibition immunoassay format was developed for the simultaneous detection of deoxynivalenol (DON) and zearalenone (ZEN), using a single sensor chip. Initial in-house validation showed limits of detection of 21 and 17 ng/mL for DON and 16 and 10 ng/mL for ZEN in extracts, which corresponds to 84 and 68 μg/kg for DON and 64 and 40 μg/kg for ZEN in maize and wheat samples, respectively. Finally, the results were critically compared with data obtained from liquid chromatography-mass spectrometry confirmatory analysis method and found to be in good agreement. The described multiplex immunoassay for the rapid screening of several mycotoxins meets European Union regulatory limits and represents a robust platform for mycotoxin analysis in food and feed samples.
Keywords: Imaging surface plasmon resonance, Multiplex immunoassay, Mycotoxins, Deoxynivalenol, Zearalenone
Introduction
Mycotoxins, secondary metabolites of fungi, are commonly found in food and feed commodities as well as in agricultural crops. Mycotoxin species were found as metabolic products of various groups of fungi, e.g., Aspergillus, Fusarium, Penicillium, etc. [1]. These secondary metabolites are poisonous for humans and animals. Mycotoxins play substantial roles in spoilage of food, beverages, and feed for livestock [2]. The occurrence of mycotoxins in food and feed products might have serious impact on economy. The regulations of maximum mycotoxin levels in food and animal feed have been established in many countries including European Union (EU) members [3].
Several analytical techniques have been reported and are widely used in order to detect the presence of mycotoxin contaminants [4–7]. The most common (conventional) confirmatory method for the detection of mycotoxins in food and feed samples is high-pressure liquid chromatography or gas chromatography in combination with UV-VIS spectroscopy or/and mass spectrometry [8–13]. These methods allow simultaneous analysis of several toxins; however, they require laboratory facilities, qualified personnel, as well as extensive cleanup procedures for sample preparation. Another group of detection techniques include rapid screening assays based on an immunoreaction between antibodies and antigens such as enzyme-linked immunosorbent assays (ELISAs) [14–18], lateral flow devices (LFDs) [19, 20], fluorescence polarization immunoassays (FPIA) [21–23], an optical waveguide biosensor [24], and a multiplex flow cytometric immunoassay (FCIA) [25]. Recently, two technically new approaches were found for mycotoxin detection utilizing an acoustic principle of sensing [26] and Raman spectroscopy [27]. However, this multitude of approaches has not led to a routinely and generally applicable approach. ELISA tests are well-known but have numerous disadvantages, such as a long assay development time and non-recoverable consumables. Simple LFD immunoassays are capable of providing mainly qualitative results without precise quantification of data. FPIA and FCIA assays are solution-based methods that require labeling in contrast with optical biosensor immunoassays. Finally, the developed array biosensor based on the optical waveguide technology requires a time-consuming fabrication procedure and is limited by the number of channels for toxin detection [24].
Eventually, surface plasmon resonance (SPR) has attracted attention as a fast label-free screening method for the detection of food contaminants, in particular, mycotoxins [28–34]. This technique has a number of advantages: SPR is a rapid label-free detection method that gives quantitative and qualitative information of the analyzed samples and provides the possibility to reuse the sensor chip for many analytical cycles [35]. Performing SPR in an imaging format (iSPR) allows multiplex screening of tens of different biointeractions using a microarray of sensing spots [36, 37]. So far, the maximum number of mycotoxins detected using a conventional SPR sensor chip is limited to four [30].
In this article, we report on the application of a multiplex microassay platform, and we developed a rapid screening of DON and zearalenone (ZEN) in feed commodities, which is also applicable to related mycotoxins. The detection principle is based on iSPR, which allows analysis for several toxins with a single multiplexed microassay sensor chip. The fabricated microassays were tested for cross-reactivity with other mycotoxins that often occur in food and feed products. The developed microassay was in-house validated for screening of mycotoxin-contaminated wheat and maize extracts. The obtained data were critically compared with a confirmatory liquid chromatography-mass spectrometry (LC-MS/MS) method.
Materials and methods
Chemical and reagents Mycotoxin standards, e.g., DON, deoxynivalenol-3-glucoside (DON3G), 3-acetyl-deoxynivalenol (3-AcDON), 15-acetyl-deoxynivalenol (15-AcDON), T-2 toxin, HT-2, nivalenol (NIV), ZEN, alfa-zearalenol (α-ZEL), beta-zearalenol (β-ZEL), aflatoxins B1, B2, G1, G2 (AFB1, AFB2, AFG1, AFG2), fumonisin B1, B2, B3, (FB1, FB2, FB3), and ochratoxin A (OTA) were purchased from Biopure Referenzsubstanzen GmbH (Tulln, Austria). The mycotoxin standard zearalenone sulfate (ZENS) was kindly provided by the Van ‘t Hoff Institute for Molecular Sciences of the University of Amsterdam (Amsterdam, The Netherlands). The iSPR rectangular sensor chips Easy2spot coated with pre-activated carboxylated dextran hydrogel were supplied by IBIS Technologies (Enschede, The Netherlands). The running HBS-EP buffer (containing 10 mM 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid pH 7.4, 150 mM sodium chloride, 3 mM EDTA, and 0.005% (v/v) surfactant polysorbate (P20)), 1 M ethanolamine hydrochloride pH 8.5, and 10 mM acetate buffers (pH 3.5 and 4.5) were received from GE Healthcare (Uppsala, Sweden). DON and ZEN ovalbumin conjugates (DON-OVA and ZEN-OVA) and monoclonal antibodies (Mabs) against deoxynivalenol (aDON) and zearalenone (aZEN) were purchased from Aokin AG (Berlin, Germany). Neutral liquid detergent RBS T230 was purchased from R. Borghgraef S.A. (Brussels, Belgium). Ovalbumin (OVA), sodium acetate, Tween 20, and anhydrous dimethyl sulfoxide were purchased from Sigma-Aldrich (Zwijndrecht, The Netherlands).
Microarray fabrication The toxin–protein conjugates DON-OVA and ZEN-OVA and unconjugated OVA were diluted in 10 mM sodium acetate buffer of different pH (3.5, 4, 4.5, and 5) until a final concentration of 50 μg/mL. The pH scouting for optimum covalent binding of the protein conjugates to the carboxylated surface of the sensor chip was performed in a Biacore 3000 (GE Healthcare, Uppsala, Sweden) SPR instrument using a pH scouting wizard. The optimum pH of 3.5 was used for the mycotoxin–OVA conjugates, and they were spotted on the pre-activated carboxylated polymeric hydrogel chip surface during 1 h using a continuous flow microspotter (CFM; Wasatch Microfluidics, USA). Ovalbumin was immobilized from 10 mM sodium acetate buffer at pH 4.5. Subsequently, the chip surface was blocked with ethanolamine for 10 min and rinsed with HBS-EP buffer and MilliQ water. In order to determine the optimal concentration, toxin–protein conjugates were immobilized at different concentrations (DON-OVA at 200, 100, 50, 25, 12.5 μg/mL; ZEN-OVA at 10, 5, 2.5, 1.25, 0.6253 μg/mL; OVA at 10, 5, 2.5, 1.25, 0.625 μg/mL).
iSPR measurements iSPR was carried out using the IBIS iSPR instrument (IBIS Technologies, Enschede, The Netherlands). The rectangular sensor chip containing the mycotoxins microarray was placed into the chip holder with the hemisphere prism, and a microfluidic cell was installed on top. Two syringe pumps were used either to deliver the sample solution to the chip surface or to flush/flow the analyte solution backward and forward at 20 μL/s over the sensor surface. The SPR angle shifts were measured for every microarray spot (region of interests (ROI)). For data analysis, Sprint software was used (IBIS Technologies). Sensorgrams were recorded during the injection of a mixture of 40 μL of the monoclonal antibodies (aDON 20 μg/mL and aZEN 5 μg/mL) and 40 μL of the multianalyte standard solution containing DON and ZEN toxins (0, 0.1, 1, 2.5, 5, 10, 25, 50, 100, 1,000 ng/mL). The injection volume into the microfluidic cell was 80 μL. All toxin and antibody solutions were diluted in HBS-EP buffer. To remove the bound Mabs and to prepare the sensor chip for the next analytical cycle, the chip surface was regenerated by the injection of 10 mM HCl for 30 s. The entire iSPR cycletime was approximately 14 min. The recorded sensorgrams were zeroed to the baseline of the buffer signal before the injection of the Mabs. For each mycotoxin spot, the maximum response was calculated from the data points recorded during the dissociation step. The sensorgrams were recorded at a constant temperature of 25 °C.
Standard calibration curves and LOD The calibration curves were plotted from the calculated values of the relative binding (B/B0), where B is the response of the solution containing toxin plus antibodies and B0 is the response of the blank solution containing antibodies, versus concentration of the toxin in the standard solution or sample matrix extract, respectively. The calibration curves were fitted with a non-linear four-parameter model using GraphPad Prism software (GraphPad Software Inc., USA). From the calibration curves, the half maximal inhibitory concentration (IC50) was calculated. In addition, limits of detection (LODs) were estimated by analyzing blank samples six times and subtracting three times the standard deviation (3SD) from the average of the relative responses.
Specificity of the microassay The specificity of the microassay was tested against commonly occurring mycotoxins and masked toxins having similar chemical structures (DON3G, 3-AcDON, 15-AcDON, NIV, T2, HT2, α-ZEL, β-ZEL, AFB1, AFB2, AFG1, AFG2, FB1, FB2, FB3, OTA, and ZENS). The IC50 values were calculated from calibration curves, and the cross-reactivity was assessed with respect to DON and ZEN, respectively.
Preparation of wheat and maize extracts Samples of 2.5 g were milled and extracted with 10 mL of acetonitrile–water–formic acid (84:16:1). Of this extraction solvent, 100 μL was evaporated under a flow of nitrogen. In the case of spiked blank extracts, DON and ZEN were spiked at different concentrations (0.1–1,000 ng/mL). After evaporation, the residue in the vial was dissolved in 100 μL of HBS-EP buffer containing 10% of DMSO. The solution was centrifuged at 11,000 rpm for 10 min. Subsequently, 100 μL of the supernatant was collected for iSPR measurements. The entire preparation procedure for several samples in parallel can be performed within 1 h.
Liquid chromatography tandem mass spectrometry The LC-MS/MS system consisted of a Shimadzu (Kyoto, Japan) LC system equipped with an ABI Sciex (Foster City, USA) model QTRAP 5500 triple quadrupole mass spectrometer. For DON and 3- and 15-AcDON, the mass spectrometer was operated in the positive electrospray ionization mode at a capillary voltage of 5.0 kV, a desolvation temperature of 400 °C, and an entrance voltage of 10 V. Desolvation gas was nitrogen, and the CID gas was argon. ZEN, α-ZEL, and β-ZEL were ionized in the negative mode at −4.0 kV. Data were acquired in the multiple reaction monitoring mode using the following ion transitions: DON [M + H]+m/z 297 → 231 and m/z 297 → 249, AcDON [M + NH4]+m/z 356 → 137 and m/z 356 → 321; ZEN [M-H]-m/z 317 → 175 and m/z 317 → 131, ZEL [M-H]-m/z 319 → 160 and m/z 319 → 130. The analytical column was a 100 × 2.1 mm i.d. 3 μm Ultra Aqueous C18 (Restek, Bellefonte, PA, USA) column, kept in a column oven at 35 °C. The two mobile phases used consisted of (A) water/formic acid/1 M ammonium formate (100:1:0.1) and (B) methanol/water/formic acid/1 M ammonium formate (95:5:1:0.1), and the flow was 0.4 mL/min. Following a 1-min isocratic period at 0% B, a linear gradient was started towards 50% B at 2 min, isocratic until 3 min, then followed by a gradient towards 100% B at 8 min, and then kept at that composition until 10 min. The injection volume was 10 μL.The concentrations of the mycotoxins were calculated using the standard addition method following the addition of 500 μg/kg DON and 50 μg/kg ZEN to a second portion of the sample.
Results and discussion
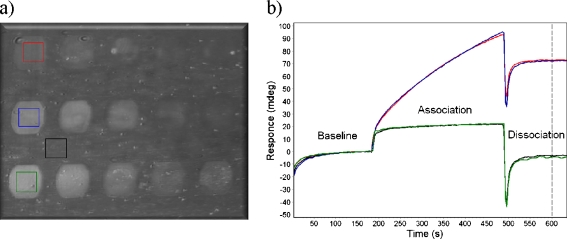
Development of the microarray sensor chip Different detection formats can be applied for designing an SPR immunoassay [35]. The competitive inhibition format is found to be the most suitable and robust format for the detection of small organic molecules like mycotoxins. Direct immobilization of mycotoxin–protein conjugates (DON ovalbumin and ZEN ovalbumin) on chip surface results in a robust and stable microassay in comparison with immobilized antibodies, which are susceptible to denaturation and proteolytic degradation [38]. In the inhibition format, during the injection over the sensor chip, the antibodies are mixed with the mycotoxins in solution. The higher the mycotoxin concentration in the solution, the lower is the binding of the antibodies to the mycotoxin–protein conjugates immobilized on the chip surface.A CFM was used in order to fabricate the microarray sensor chip. In contrast to contact spotting, this instrument yields a microarray with uniform morphology on the spot without crosstalk [39]. The commercially available DON-OVA and ZEN-OVA toxin–protein conjugates were covalently attached to the surface and coupled via the amine groups of the OVA to the gold substrate coated with pre-activated carboxylated polymeric hydrogel. Such a coupling is expected to provide a stable array with covalently attached toxin–protein conjugates on the surface [29, 30].Figure 1a shows a sensor chip image of different concentrations of the two mycotoxin–OVA conjugates and OVA successfully spotted on the surface of the sensor chip. OVA was immobilized as reference spots in order to control the mycotoxin-specific binding of the DON and ZEN Mabs. The microarray image demonstrates uniform morphology as well as precise separation between spots. The concentration needed for immobilization of ZEN-OVA conjugate is ten times lower than that for the DON-OVA conjugate, based on the response of the Mab binding. The toxin–protein conjugates were easy to spot from the buffer having a lower pH of 3.5. Next, SPR sensorgrams were measured from four different predefined ROIs during the injection of the mixture of the two antibodies, in order to test the performances of the binding and cross-interaction to the immobilized mycotoxins on the fabricated microarray (Fig. 1b). The sensorgrams begin with the baseline, followed by the association step, where the injection of the antibodies mixture results in rising of the responses due to the specific interactions of the antibodies in the solution to the mycotoxin pattern on the chip surface. Subsequently, buffer is injected for the dissociation step. The difference in responses of the defined ROIs of mycotoxin and reference spots (unmodified sensor chip surface) gives quantitative information of the specifically bound antibodies.In order to remove bound antibodies and to prepare the sensor chip for the next measurement cycle, an efficient regeneration step is vital. Incomplete regeneration or the loss of the capturing molecules from the chip surface will decrease the performance of the microarray. Depending on the capturing molecule and analyte, various regeneration solutions can be applied. The higher the multiplexity of the chip is the more complex will be the optimization of the regeneration phase. For the DON and ZEN multiplex microarray, the most suitable regeneration solution was a short pulse injection (30 s) of 10 mM HCl.
Fig. 1.
a Micropatterns of five different concentrations of ovalbumin (OVA)-conjugated mycotoxins on the chip surface (top row: DON-OVA (200 → 12.5 μg/mL), middle row: ZEN-OVA (10 → 0.63 μg/mL), and bottom row OVA (10 → 0.63 μg/mL)). The spot size is 400 × 600 μm. b SPR sensorgrams recorded on DON (red), ZEN (blue), OVA (green), and blank (black) ROIs during the injection of DON and ZEN antibodies mixture in the buffer solution. The dashed line indicates the response difference used for calculations
Cross-reactivity profile The fabricated microassay was tested with other mycotoxins which often occur in food and feed products. The DON and ZEN Mabs did not show interactions with the following mycotoxins: T-2, HT-2, NIV, AFB1, AFB2, AFG1, AFG2, FB1, FB2, FB3, OTA, and ZENS in a high concentration (1 μg/mL in buffer). However, the DON Mab demonstrated cross-reactivity with the conjugated (masked) forms of DON (Table 1). The ZEN Mab showed a high cross-reactivity towards α-ZEL (54%) and β-ZEL (137%; Table 1). The cross-reactivity of the Mabs might lead to some overestimation of mycotoxin concentration in the sample; however, for a rapid multiplex screening assay, this is not necessarily an obstacle. The presence of masked forms of parental DON and ZEN deserves attention during quantitative analysis using the immunoassay biosensors. The cross-reactivity of the DON and ZEN Mabs for the conjugated forms have been reported in several studies [14–16, 18, 33, 34, 40–42]. We have also observed high cross-reactivity of DON Mabs with acetylated forms of DON as well as ZEN Mabs with α- and β-ZEL masked mycotoxins. However, there are commercially available test kits demonstrating low cross-reactivity profile towards masked mycotoxins [40, 42]. Remarkably, ZEN antibodies did not show any cross-reactivity towards the sulfate conjugate (ZENS). The specificity and binding strengths of antibodies strongly depends on the method of antibody production [17, 33, 41], and detailed choice of antibodies will thus influence the performance (specificity and sensitivity) of an immunoassay.
Table 1.
Cross-reactivity (CR) of DON and ZEN antibodies with other mycotoxins calculated from IC50 values as a percentage relative to DON or ZEN, respectively
| Toxin | Cross-reactivity, % | |
|---|---|---|
| aDON | aZEN | |
| DON | 100 | 0 |
| 3-AcDON | 71 | 0 |
| 15-AcDON | 66 | 0 |
| DON3G | 36 | 0 |
| ZEN | 0 | 100 |
| α-ZEL | 0 | 59 |
| β-ZEL | 0 | 137 |
| ZENS | 0 | 0 |
| T-2 | 0 | 0 |
| HT-2 | 0 | 0 |
| NIV | 0 | 0 |
| AFB1 | 0 | 0 |
| AFB2 | 0 | 0 |
| AFG1 | 0 | 0 |
| AFG2 | 0 | 0 |
| FB1 | 0 | 0 |
| FB2 | 0 | 0 |
| FB3 | 0 | 0 |
| OTA | 0 | 0 |
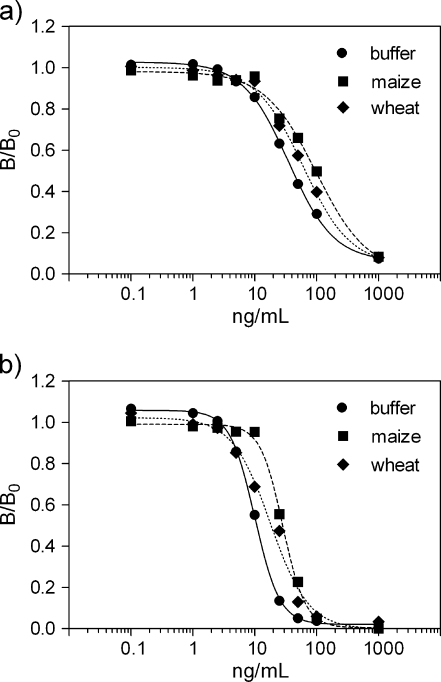
Initial in-house validation For critical evaluation of the sensitivity of the multiplex assay, calibration curves were constructed from sensorgrams recorded for the mixture of DON and ZEN standards in buffer with known concentrations (0–1000 ng/mL). From the calibration curves depicted in Fig. 2, the half-maximum inhibitory values (IC50) were calculated to be 35 and 10 ng/mL for DON and ZEN, respectively. The goodness and steepness of the curves show that the 4P model is the appropriate fitting model for the obtained data (Table 2). The LODs were calculated to be 9 ng/mL for DON and 4 ng/mL for ZEN. Comparatively to SPR immunoassays developed previously, the assay shows ∼2–3 times lower sensitivity [29, 30, 34]. Unfortunately, not all references provide comprehensive data allowing comparison of the developed assay [30]. The performance of the assay can be improved by selecting Mab clones having higher sensitivity as discussed above or by the dilution of extracts in order to decrease matrix effect [29, 30, 33]. Furthermore, a hardware upgrade will increase the sensitivity of the instrument. Next, the microassay was tested for the analysis of wheat and maize extracts spiked with DON and ZEN. Figure 2 shows that the calibration curves shift for matrix-derived products to the right, indicating a small decrease of the microassay sensitivity with respect to the results obtained in buffer. Decrease in the sensitivity is attributed to the influence of matrix components on the antibody binding in the solution and/or on the chip surface. The matrix effect on the performance of the immunoassay has been observed previously in sample extracts [33, 36]. Table 2 shows the 4P model fitting data for curves in sample extracts. The goodness of the curves demonstrates that 4P model also fits the maize- and wheat-derived data well. The steepness of curves did not change remarkably except the curve for ZEN-containing wheat extract. Subsequently, we have calculated LOD values for the mycotoxins in maize and wheat extracts. The results for DON and ZEN are 21 and 17 ng/mL in maize extracts and 16 and 10 ng/mL in wheat extracts, respectively. Evidently, due to matrix effects, these values show a decrease in sensitivity depending on the matrix and toxin. In practice, we recommend using a calibration sample prepared in a blank matrix. The LOD values calculated for the analysis of maize and wheat samples correspond to 84 and 68 μg/kg for DON and 64 and 40 μg/kg for ZEN. These are adequate with respect to the EU legislation limits established for DON in unprocessed durum wheat, maize, and oats (1750 μg/kg); unprocessed cereals (1250 μg/kg); breakfast cereal (500 μg/kg), and baby food (200 μg/kg). The EU legislation limits for ZEN are the following: in unprocessed maize, 200 μg/kg; unprocessed cereals other than maize, 100 μg/kg; cereals intended for direct human consumption, 75 μg/kg [3]. Note that we did not apply any extra cleanup procedure prior to injection in contrast to previously reported methods.Recently, various optical biosensors based on the SPR sensing principle were developed for the detection of mycotoxins. Nevertheless, most of them do not offer the possibility of multiplexing for the rapid screening of several toxins on one sensor chip [28–30, 33, 34]. The developed iSPR assay on the other hand can be upgraded for measurement up to 40 different toxins and does not require extensive sample preparation (incubation, dilution) or cleanup procedures [28, 29, 33]. The sensitivity range of the developed assay is in good agreement with the results reported previously for the SPR sensing of mycotoxins [29, 30, 33, 34].
Fig. 2.
Calibration curves for the multimycotoxin standard solution in buffer and in spiked extracts from maize and wheat samples calculated from the iSPR sensorgrams (DON (a), ZEN (b)). Solid, dashed, and dotted lines show 4P model fitting
Table 2.
Sensitivity of multiplex microassay and fitting parameters
| Toxin | Goodness of 4P fita, R2 | Curve steepness (mL/ng) | IC50 (ng/mL)b | LOD (ng/mL) |
|---|---|---|---|---|
| DON (buffer) | 0.9994 | −1.181 | 35 | 9 |
| DON (maize) | 0.9966 | −1.138 | 113 | 21 |
| DON (wheat) | 0.9922 | −1.048 | 62 | 17 |
| ZEN (buffer) | 0.9997 | −2.154 | 10 | 4 |
| ZEN (maize) | 0.9900 | −1.410 | 29 | 16 |
| ZEN (wheat) | 0.9979 | −2.392 | 19 | 10 |
aGoodness of the four-parameter model fit to the calibration curve
bIC50 value derived from the four-parameter model fit of the calibration curve
Application to real samples The developed multiplex sensor chip has been tested for the analysis of naturally contaminated samples. The values obtained from the recorded sensorgrams were fitted into calibration curves determined in maize and wheat extracts. The estimated concentrations for DON and ZEN mycotoxins are summarized in Table 3. The iSPR data thus obtained were essentially compared with the data from an accredited confirmatory LC-MS/MS method. The obtained data show that our method yields comparable results. However, the SPR assay demonstrated slightly lower levels of mycotoxins in extracts compared with the confirmatory LC-MS/MS method. To perform more accurate measurements, a protocol for the sample preparation should be developed and established depending on the matrix nature [33]. In combination with such protocol, the currently described approach thus offers the potential for rapid and accurate multianalyte analysis of mycotoxins in food.
Table 3.
Toxin concentrations in maize and wheat extracts measured by iSPR and LC-MS/MS
| Sample | iSPR | LC-MS/MS | ||
|---|---|---|---|---|
| DON (ng/mL) | ZEN (ng/mL) | DON (ng/mL) | ZEN (ng/mL) | |
| Maize | <LOD | – | 20 | – |
| Wheat | – | – | – | – |
| Maizea | 253 | <LOD | 270 | 12.5 |
| Wheata | 218 | – | 250 | 12.5 |
aSpiked maize/wheat extracts
Conclusions
The multiplex microarray immunosensor based on the iSPR sensing principle allows for the qualitative and quantitative simultaneous and label-free detection of several mycotoxins in multianalyte sample extracts with sensitivities of 84 and 68 μg/kg for DON and 64 and 40 μg/kg for ZEN in maize and wheat samples, respectively. The developed multiplex immunoassay is suitable for rapid screening of maize and wheat extract without a complex and time-consuming sample preparation procedure. Despite a small sample matrix effect, the sensitivity of the assay falls well into EU regulatory limits. The results obtained using a single sensor chip are in good agreement with LC-MS/MS data. The iSPR sensor chip platform has perspective for the development of a rapid multiplex screening method for up to 40 different mycotoxins.
Acknowledgments
The authors thank Mr. Ed Boers (RIKILT) for providing wheat and maize extracts and the LC-MS/MS data. Dr. Alex van der Kooi and Dr. Richard Schasfoort (IBIS Technologies) are acknowledged for fruitful discussions. The Nano4Vitality program of the provinces Gelderland and Overijssel is highly appreciated for financial support.
References
- 1.Betina V (1989) Mycotoxins chemical, biological and environmental aspects Elsevier Amsterdam-Oxford-New York-Tokyo 19–39 192–237
- 2.Bennett JW, Klich M. Clin Microbiol Rev. 2003;16:497–516. doi: 10.1128/CMR.16.3.497-516.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Commission Regulation EC No 1881 Off J Eur Union L. 2006;364:5–24. [Google Scholar]
- 4.Krska R, Molinelli A. Anal Bioanal Chem. 2007;387:145–148. doi: 10.1007/s00216-006-0797-3. [DOI] [PubMed] [Google Scholar]
- 5.Krska R, Welzig E, Berthiller F, Molinelli A, Mizaikoff B. Food Addit Contam. 2005;22:345–353. doi: 10.1080/02652030500070192. [DOI] [PubMed] [Google Scholar]
- 6.Krska R, Schubert-Ullrich P, Molinelli A, Sulyok M, McDonald S, Crews C. Food Addit Contam. 2008;25:152–163. doi: 10.1080/02652030701765723. [DOI] [PubMed] [Google Scholar]
- 7.Cigic IK, Procen H. Int J Mol Sci. 2009;10:65–115. [Google Scholar]
- 8.Cahill LM, Kruger SC, McAlice BT, Ramsey CS, Prioli R, Kohn B. J Chromatogr A. 1999;859:23–28. doi: 10.1016/S0021-9673(99)00846-8. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka T, Yoneda A, Inoue S, Sugiura Y, Ueno Y. J Chromatogr A. 2000;882:23–28. doi: 10.1016/S0021-9673(00)00063-7. [DOI] [PubMed] [Google Scholar]
- 10.Schothorst RC, Jeker AA. Food Chem. 2001;73:111–117. doi: 10.1016/S0308-8146(00)00321-6. [DOI] [Google Scholar]
- 11.Berthiller F, Schuhmacher R, Buttinger G, Krska R. J Chromatogr A. 2005;1062:209–216. doi: 10.1016/j.chroma.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 12.Pussemier L, Pierard JY, Anselme M, Tangni EK, Motte JC, Larondelle Y. Food Addit Contam. 2006;23:1208–1218. doi: 10.1080/02652030600699312. [DOI] [PubMed] [Google Scholar]
- 13.Vendl O, Berthiller F, Crews C, Krska R. Anal Bioanal Chem. 2009;395:1347–1354. doi: 10.1007/s00216-009-2873-y. [DOI] [PubMed] [Google Scholar]
- 14.Sinha RC, Savard ME, Lau R. J Agric Food Chem. 1995;43:1740–1744. doi: 10.1021/jf00054a061. [DOI] [Google Scholar]
- 15.Casale WL, Pestka J, Hart LP. J Agric Food Chem. 1998;36:663–668. doi: 10.1021/jf00081a064. [DOI] [Google Scholar]
- 16.Maragos CM, McCormick SP. Food Agric Immunol. 2000;12:181–192. doi: 10.1080/09540100050140722. [DOI] [Google Scholar]
- 17.Schneider L, Pichler H, Krska RJ. Anal Chem. 2000;367:98–100. doi: 10.1007/s002160051609. [DOI] [PubMed] [Google Scholar]
- 18.Yoshizava T, Kohno H, Ikeda K, Shinoda T, Yokohama H, Morita K, Kusada O, Kobayashi Y. Biosci Biotechnol Biochem. 2004;68:2076–2085. doi: 10.1271/bbb.68.2076. [DOI] [PubMed] [Google Scholar]
- 19.Kolosova AY, Sibanda L, Dumolin F, Lewis J, Duveiller E, Peteghem C, Saeger S. Anal Chim Acta. 2008;616:235–244. doi: 10.1016/j.aca.2008.04.029. [DOI] [PubMed] [Google Scholar]
- 20.Goryacheva IY, Rusanova TY, Burmistrova NA, Saeger S. J Anal Chem. 2009;10:768–785. doi: 10.1134/S1061934809080024. [DOI] [Google Scholar]
- 21.Maragon CM, Plattner RD. J Agric Food Chem. 2002;50:1827–1832. doi: 10.1021/jf011487d. [DOI] [PubMed] [Google Scholar]
- 22.Lippolis V, Pascale M, Visconti A. J Food Prot. 2006;11:2712–2719. doi: 10.4315/0362-028x-69.11.2712. [DOI] [PubMed] [Google Scholar]
- 23.Lattanzio VMT, Pascale M, Visconti A. Trends Anal Chem. 2009;28:758–768. doi: 10.1016/j.trac.2009.04.012. [DOI] [Google Scholar]
- 24.Ngundi MM, Qadri SA, Wallace EV, Moore MH, Lassman ME, Shriver-Lake LC, Ligler FS, Taitt CR. Environ Sci Technol. 2006;40:2352–2356. doi: 10.1021/es052396q. [DOI] [PubMed] [Google Scholar]
- 25.Peters J, Bienenmann-Ploum M, de Rijk T, Haasnoot W (2011) Mycotoxin Research doi:10.1007/s12550-010-0077-0 [DOI] [PMC free article] [PubMed]
- 26.Juodeikine G, Basinskiene L, Vidmantiene D, Bartkeine E, Kunigelis V, Koe WJ. World Mycotoxins J. 2008;1:267–274. doi: 10.3920/WMJ2008.1018. [DOI] [Google Scholar]
- 27.Liu Y, Delwiche SR, Dong Y. Food Addit Contam. 2009;26:1396–1401. doi: 10.1080/02652030903013310. [DOI] [PubMed] [Google Scholar]
- 28.Schnerr H, Vogel RF, Niessen L. Food Agric Immunol. 2002;14:113–321. doi: 10.1080/0954010021000096346. [DOI] [Google Scholar]
- 29.Tudos AJ, Lucas ER, Stigter ECA. J Agric Food Chem. 2003;51:5843–5848. doi: 10.1021/jf030244d. [DOI] [PubMed] [Google Scholar]
- 30.Graag B, Spath S, Dietrich H, Stigter ECA. Food Control. 2003;51:251–254. [Google Scholar]
- 31.Hodnik V, Anderluh G. Sensors. 2009;9:1339–1354. doi: 10.3390/s9031339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meneely JP, Sulyok M, Baumgartner S, Krska R, Elliott CT. Talanta. 2010;81:630–636. doi: 10.1016/j.talanta.2009.12.055. [DOI] [PubMed] [Google Scholar]
- 33.Meneely JP, Fodey T, Armstrong L, Sulyok M, Krska R, Elliott CT. J Agric Food Chem. 2010;58:8936–8941. doi: 10.1021/jf101517s. [DOI] [PubMed] [Google Scholar]
- 34.Kadota T, Takezawa Y, Hirano S, Tajima O, Maragos CM, Nakajima T, Tanaka T, Kamata Y, Sugita-Konishi Y. Anal Chim Acta. 2010;673:173–178. doi: 10.1016/j.aca.2010.05.028. [DOI] [PubMed] [Google Scholar]
- 35.Homola J. Chem Rev. 2008;108:462–493. doi: 10.1021/cr068107d. [DOI] [PubMed] [Google Scholar]
- 36.Rebe Raz S, Bremer MGEG, Giesbers M, Norde W. Biosens Bioelectron. 2008;24:552–557. doi: 10.1016/j.bios.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 37.Rebe Raz S, Bremer MGEG, Haasnoot W, Norde W. Anal Chem. 2009;81:7743–7749. doi: 10.1021/ac901230v. [DOI] [PubMed] [Google Scholar]
- 38.Maragos CM. Anal Bioanal Chem. 2009;395:1205–1213. doi: 10.1007/s00216-009-2728-6. [DOI] [PubMed] [Google Scholar]
- 39.Natarajan S, Katsamba PS, Miles A, Eckman J, Papalia GA, Rich RL, Gale BK, Myszka DG. Anal Biochem. 2008;373:141–146. doi: 10.1016/j.ab.2007.07.035. [DOI] [PubMed] [Google Scholar]
- 40.Zachariasova M, Hajslova J, Kostelanska M, Poustka J, Krplova A, Cuhra P, Hochel I. Anal Chim Acta. 2008;625:77–86. doi: 10.1016/j.aca.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 41.Burmistrova NA, Goryacheva IY, Basova EY, Franki A-S, Elewaut D, Beneden K, Deforce D, Peteghem C, Saeger S. Anal Bioanal Chem. 2009;395:1301–1307. doi: 10.1007/s00216-009-2913-7. [DOI] [PubMed] [Google Scholar]
- 42.Tangni EK, Motte JC, Callebaut A, Pussemier L. J Agric Food Chem. 2010;58:12625–12633. doi: 10.1021/jf103025e. [DOI] [PubMed] [Google Scholar]