Abstract
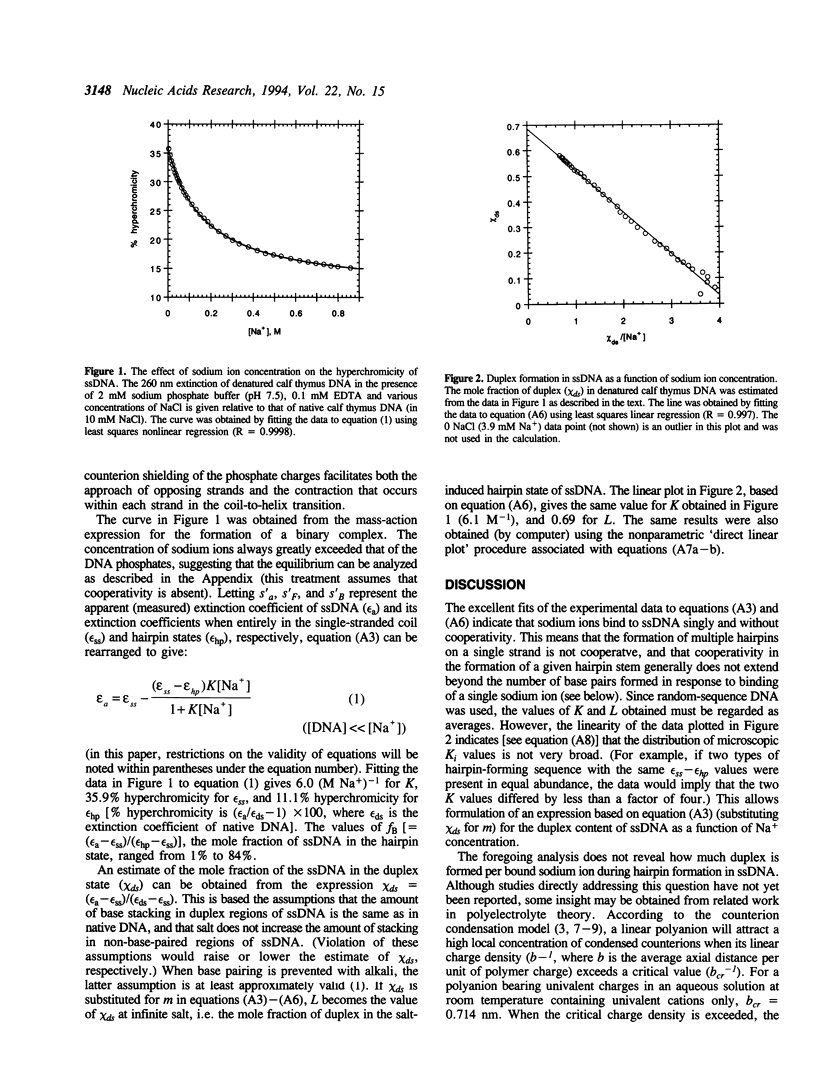
The salt-induced formation of duplex structure (primarily hairpin loops) in denatured calf thymus DNA was monitored by measuring the decrease in absorbance at 260 nm as a function of increasing sodium ion concentration. It was found that this process was noncooperative and could be accurately described by the mass-action expression for the reversible formation of a binary complex: single strand (coil) + free sodium ion <==> hairpin (with associated sodium ion). The equilibrium constant for the transition was found to be 6 (M Na+)-1. The extrapolated absorbance at infinite salt concentration represents 11% hyperchromicity, which is one third of the hyperchromicity of denatured DNA in the absence of salt (36%).
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson C. F., Record M. T., Jr The relationship between the poisson-boltzmann model and the condensation hypothesis: an analysis based on the low salt form of the Donnan coefficient. Biophys Chem. 1980 Jun;11(3-4):353–360. doi: 10.1016/0301-4622(80)87008-6. [DOI] [PubMed] [Google Scholar]
- Bujalowski W., Lohman T. M. A general method of analysis of ligand-macromolecule equilibria using a spectroscopic signal from the ligand to monitor binding. Application to Escherichia coli single-strand binding protein-nucleic acid interactions. Biochemistry. 1987 Jun 2;26(11):3099–3106. doi: 10.1021/bi00385a023. [DOI] [PubMed] [Google Scholar]
- Cornish-Bowden A., Eisenthal R. Estimation of Michaelis constant and maximum velocity from the direct linear plot. Biochim Biophys Acta. 1978 Mar 14;523(1):268–272. doi: 10.1016/0005-2744(78)90030-x. [DOI] [PubMed] [Google Scholar]
- Cornish-Bowden A., Eisenthal R. Statistical considerations in the estimation of enzyme kinetic parameters by the direct linear plot andother methods. Biochem J. 1974 Jun;139(3):721–730. doi: 10.1042/bj1390721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOWD J. E., RIGGS D. S. A COMPARISON OF ESTIMATES OF MICHAELIS-MENTEN KINETIC CONSTANTS FROM VARIOUS LINEAR TRANSFORMATIONS. J Biol Chem. 1965 Feb;240:863–869. [PubMed] [Google Scholar]
- Eisenthal R., Cornish-Bowden A. The direct linear plot. A new graphical procedure for estimating enzyme kinetic parameters. Biochem J. 1974 Jun;139(3):715–720. doi: 10.1042/bj1390715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G. S. The molecular theory of polyelectrolyte solutions with applications to the electrostatic properties of polynucleotides. Q Rev Biophys. 1978 May;11(2):179–246. doi: 10.1017/s0033583500002031. [DOI] [PubMed] [Google Scholar]
- Mascotti D. P., Lohman T. M. Thermodynamic extent of counterion release upon binding oligolysines to single-stranded nucleic acids. Proc Natl Acad Sci U S A. 1990 Apr;87(8):3142–3146. doi: 10.1073/pnas.87.8.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmsted M. C., Anderson C. F., Record M. T., Jr Importance of oligoelectrolyte end effects for the thermodynamics of conformational transitions of nucleic acid oligomers: a grand canonical Monte Carlo analysis. Biopolymers. 1991 Nov;31(13):1593–1604. doi: 10.1002/bip.360311314. [DOI] [PubMed] [Google Scholar]
- Record M. T., Jr, Anderson C. F., Lohman T. M. Thermodynamic analysis of ion effects on the binding and conformational equilibria of proteins and nucleic acids: the roles of ion association or release, screening, and ion effects on water activity. Q Rev Biophys. 1978 May;11(2):103–178. doi: 10.1017/s003358350000202x. [DOI] [PubMed] [Google Scholar]
- Record M. T., Jr, Lohman M. L., De Haseth P. Ion effects on ligand-nucleic acid interactions. J Mol Biol. 1976 Oct 25;107(2):145–158. doi: 10.1016/s0022-2836(76)80023-x. [DOI] [PubMed] [Google Scholar]
- Record M. T., Jr, Woodbury C. P., Lohman T. M. Na+ effects on transition of DNA and polynucleotides of variable linear charge density. Biopolymers. 1976 May;15(5):893–915. doi: 10.1002/bip.1976.360150507. [DOI] [PubMed] [Google Scholar]
- Studier F. W. Conformational changes of single-stranded DNA. J Mol Biol. 1969 Apr;41(2):189–197. doi: 10.1016/0022-2836(69)90384-2. [DOI] [PubMed] [Google Scholar]
- Walz F. G., Jr Rapid kinetic studies on the conformation of single-stranded DNA. Biopolymers. 1972;11(11):2365–2379. doi: 10.1002/bip.1972.360111115. [DOI] [PubMed] [Google Scholar]
- Wilson W. D., Lopp I. G. Analysis of cooperativity and ion effects in the interaction of quinacrine with DNA. Biopolymers. 1979 Dec;18(12):3025–3041. doi: 10.1002/bip.1979.360181210. [DOI] [PubMed] [Google Scholar]
- Wolfe A., Shimer G. H., Jr, Meehan T. Polycyclic aromatic hydrocarbons physically intercalate into duplex regions of denatured DNA. Biochemistry. 1987 Oct 6;26(20):6392–6396. doi: 10.1021/bi00394a013. [DOI] [PubMed] [Google Scholar]


