Abstract
The kinetics of PaeR7 endonuclease-catalysed cleavage reactions of fluorophor-labeled oligonucleotide substrates have been examined using fluorescence resonance energy transfer (FRET). A series of duplex substrates were synthesized with an internal CTCGAG PaeR7 recognition site and donor (fluorescein) and acceptor (rhodamine) dyes conjugated to the opposing 5' termini. The time-dependent increase in donor fluorescence resulting from restriction cleavage of these substrates was continuously monitored and the initial rate data was fitted to the Michaelis-Menten equation. The steady state kinetic parameters for these substrates were in agreement with the rate constants obtained from a gel electrophoresis-based fixed time point assay using radiolabeled substrates. The FRET method provides a rapid continuous assay as well as high sensitivity and reproducibility. These features should make the technique useful for the study of DNA-cleaving enzymes.
Full text
PDF

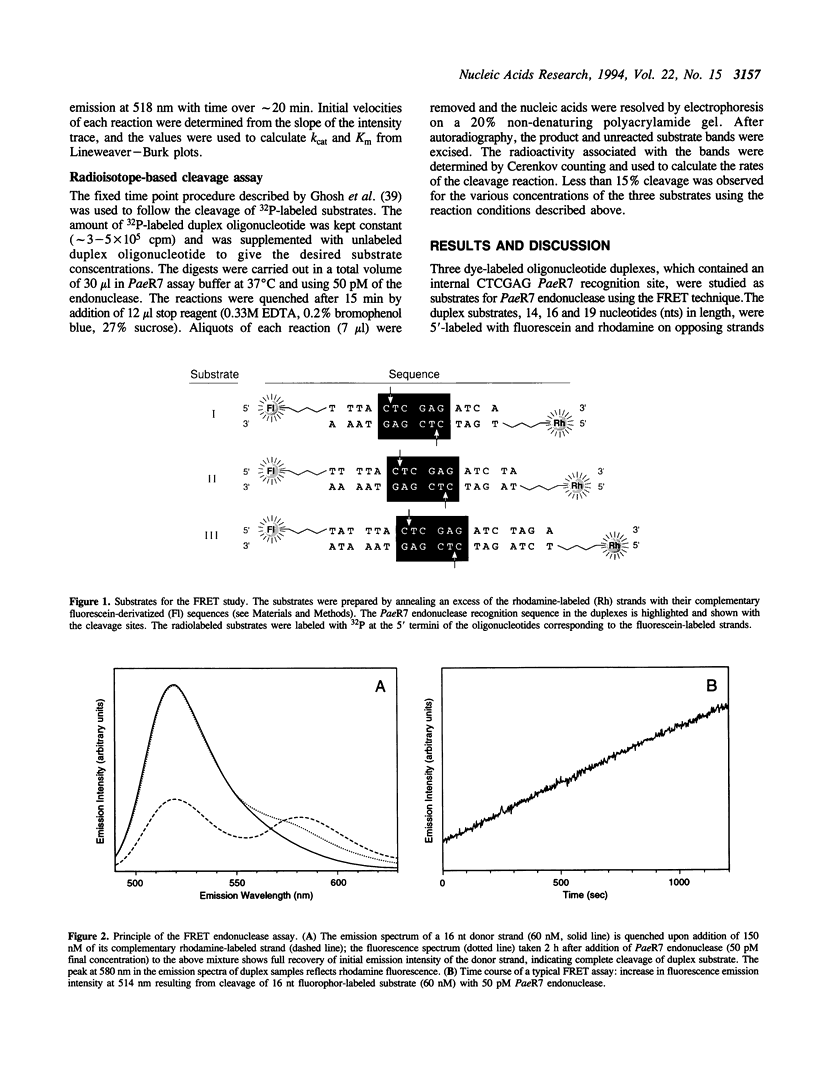
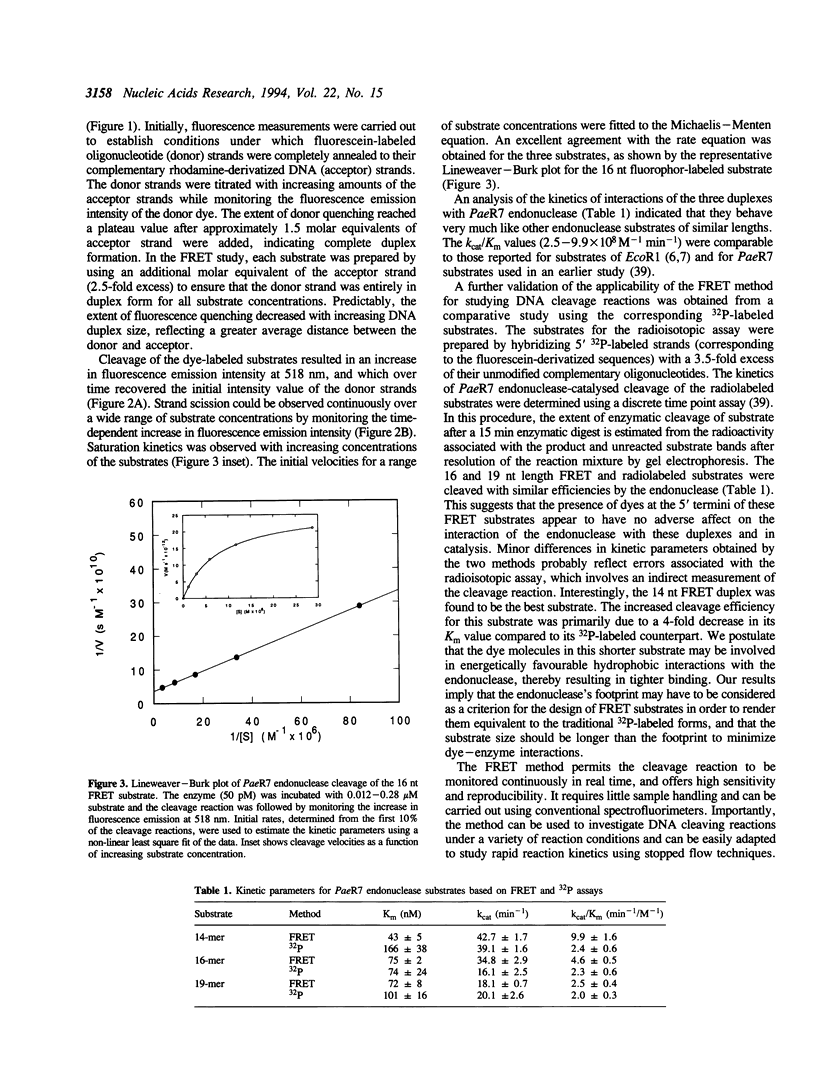

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Agrawal S., Christodoulou C., Gait M. J. Efficient methods for attaching non-radioactive labels to the 5' ends of synthetic oligodeoxyribonucleotides. Nucleic Acids Res. 1986 Aug 11;14(15):6227–6245. doi: 10.1093/nar/14.15.6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen D. J., Benkovic S. J. Resonance energy transfer measurements between substrate binding sites within the large (Klenow) fragment of Escherichia coli DNA polymerase I. Biochemistry. 1989 Dec 12;28(25):9586–9593. doi: 10.1021/bi00451a006. [DOI] [PubMed] [Google Scholar]
- Alves J., Rüter T., Geiger R., Fliess A., Maass G., Pingoud A. Changing the hydrogen-bonding potential in the DNA binding site of EcoRI by site-directed mutagenesis drastically reduces the enzymatic activity, not, however, the preference of this restriction endonuclease for cleavage within the site-GAATTC-. Biochemistry. 1989 Mar 21;28(6):2678–2684. doi: 10.1021/bi00432a047. [DOI] [PubMed] [Google Scholar]
- Beardsley K., Cantor C. R. Studies of transfer RNA tertiary structure by singlet-singlet energy transfer. Proc Natl Acad Sci U S A. 1970 Jan;65(1):39–46. doi: 10.1073/pnas.65.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D., White R. L., Skolnick M., Davis R. W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet. 1980 May;32(3):314–331. [PMC free article] [PubMed] [Google Scholar]
- Cardullo R. A., Agrawal S., Flores C., Zamecnik P. C., Wolf D. E. Detection of nucleic acid hybridization by nonradiative fluorescence resonance energy transfer. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8790–8794. doi: 10.1073/pnas.85.23.8790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg R. M. Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol. 1992;211:353–388. doi: 10.1016/0076-6879(92)11020-j. [DOI] [PubMed] [Google Scholar]
- Clegg R. M., Murchie A. I., Zechel A., Lilley D. M. Observing the helical geometry of double-stranded DNA in solution by fluorescence resonance energy transfer. Proc Natl Acad Sci U S A. 1993 Apr 1;90(7):2994–2998. doi: 10.1073/pnas.90.7.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J. P., Hagerman P. J. Analysis of fluorescence energy transfer in duplex and branched DNA molecules. Biochemistry. 1990 Oct 2;29(39):9261–9268. doi: 10.1021/bi00491a022. [DOI] [PubMed] [Google Scholar]
- Czworkowski J., Odom O. W., Hardesty B. Fluorescence study of the topology of messenger RNA bound to the 30S ribosomal subunit of Escherichia coli. Biochemistry. 1991 May 14;30(19):4821–4830. doi: 10.1021/bi00233a026. [DOI] [PubMed] [Google Scholar]
- Eis P. S., Millar D. P. Conformational distributions of a four-way DNA junction revealed by time-resolved fluorescence resonance energy transfer. Biochemistry. 1993 Dec 21;32(50):13852–13860. doi: 10.1021/bi00213a014. [DOI] [PubMed] [Google Scholar]
- Eshaghpour H., Dieterich A. E., Cantor C. R., Crothers D. M. Singlet-singlet energy transfer studies of the internal organization of nucleosomes. Biochemistry. 1980 Apr 29;19(9):1797–1805. doi: 10.1021/bi00550a011. [DOI] [PubMed] [Google Scholar]
- Fairclough R. H., Cantor C. R. The use of singlet-singlet energy transfer to study macromolecular assemblies. Methods Enzymol. 1978;48:347–379. doi: 10.1016/s0076-6879(78)48019-x. [DOI] [PubMed] [Google Scholar]
- Fliess A., Wolfes H., Rosenthal A., Schwellnus K., Blöcker H., Frank R., Pingoud A. Role of thymidine residues in DNA recognition by the EcoRI and EcoRV restriction endonucleases. Nucleic Acids Res. 1986 Apr 25;14(8):3463–3474. doi: 10.1093/nar/14.8.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florentin D., Sassi A., Roques B. P. A highly sensitive fluorometric assay for "enkephalinase," a neutral metalloendopeptidase that releases tyrosine-glycine-glycine from enkephalins. Anal Biochem. 1984 Aug 15;141(1):62–69. doi: 10.1016/0003-2697(84)90425-1. [DOI] [PubMed] [Google Scholar]
- Ghosh S. S., Musso G. F. Covalent attachment of oligonucleotides to solid supports. Nucleic Acids Res. 1987 Jul 10;15(13):5353–5372. doi: 10.1093/nar/15.13.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S. S., Obermiller P. S., Kwoh T. J., Gingeras T. R. Analysis of substrate specificity of the PaeR7 endonuclease: effect of base methylation on the kinetics of cleavage. Nucleic Acids Res. 1990 Sep 11;18(17):5063–5068. doi: 10.1093/nar/18.17.5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedroc D. P., Khan R., Barnhart K. Site-specific 1,N6-ethenoadenylated single-stranded oligonucleotides as structural probes for the T4 gene 32 protein-ssDNA complex. Biochemistry. 1991 Aug 20;30(33):8230–8242. doi: 10.1021/bi00247a020. [DOI] [PubMed] [Google Scholar]
- Goodchild J. Conjugates of oligonucleotides and modified oligonucleotides: a review of their synthesis and properties. Bioconjug Chem. 1990 May-Jun;1(3):165–187. doi: 10.1021/bc00003a001. [DOI] [PubMed] [Google Scholar]
- Halford S. E., Goodall A. J. Modes of DNA cleavage by the EcoRV restriction endonuclease. Biochemistry. 1988 Mar 8;27(5):1771–1777. doi: 10.1021/bi00405a058. [DOI] [PubMed] [Google Scholar]
- Hochstrasser R. A., Chen S. M., Millar D. P. Distance distribution in a dye-linked oligonucleotide determined by time-resolved fluorescence energy transfer. Biophys Chem. 1992 Dec;45(2):133–141. doi: 10.1016/0301-4622(92)87005-4. [DOI] [PubMed] [Google Scholar]
- Ishii J. K., Ghosh S. S. Bead-based sandwich hybridization characteristics of oligonucleotide-alkaline phosphatase conjugates and their potential for quantitating target RNA sequences. Bioconjug Chem. 1993 Jan-Feb;4(1):34–41. doi: 10.1021/bc00019a005. [DOI] [PubMed] [Google Scholar]
- Jeltsch A., Fritz A., Alves J., Wolfes H., Pingoud A. A fast and accurate enzyme-linked immunosorbent assay for the determination of the DNA cleavage activity of restriction endonucleases. Anal Biochem. 1993 Sep;213(2):234–240. doi: 10.1006/abio.1993.1415. [DOI] [PubMed] [Google Scholar]
- Malfroy B., Burnier J. New substrates for enkephalinase (neutral endopeptidase) based on fluorescence energy transfer. Biochem Biophys Res Commun. 1987 Feb 27;143(1):58–66. doi: 10.1016/0006-291x(87)90629-2. [DOI] [PubMed] [Google Scholar]
- Matayoshi E. D., Wang G. T., Krafft G. A., Erickson J. Novel fluorogenic substrates for assaying retroviral proteases by resonance energy transfer. Science. 1990 Feb 23;247(4945):954–958. doi: 10.1126/science.2106161. [DOI] [PubMed] [Google Scholar]
- McLaughlin L. W., Benseler F., Graeser E., Piel N., Scholtissek S. Effects of functional group changes in the EcoRI recognition site on the cleavage reaction catalyzed by the endonuclease. Biochemistry. 1987 Nov 17;26(23):7238–7245. doi: 10.1021/bi00397a007. [DOI] [PubMed] [Google Scholar]
- Moser H. E., Dervan P. B. Sequence-specific cleavage of double helical DNA by triple helix formation. Science. 1987 Oct 30;238(4827):645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- Murchie A. I., Clegg R. M., von Kitzing E., Duckett D. R., Diekmann S., Lilley D. M. Fluorescence energy transfer shows that the four-way DNA junction is a right-handed cross of antiparallel molecules. Nature. 1989 Oct 26;341(6244):763–766. doi: 10.1038/341763a0. [DOI] [PubMed] [Google Scholar]
- Ng M., Auld D. S. A fluorescent oligopeptide energy transfer assay with broad applications for neutral proteases. Anal Biochem. 1989 Nov 15;183(1):50–56. doi: 10.1016/0003-2697(89)90170-x. [DOI] [PubMed] [Google Scholar]
- Ozaki H., McLaughlin L. W. The estimation of distances between specific backbone-labeled sites in DNA using fluorescence resonance energy transfer. Nucleic Acids Res. 1992 Oct 11;20(19):5205–5214. doi: 10.1093/nar/20.19.5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. J. Restriction enzymes and their isoschizomers. Nucleic Acids Res. 1990 Apr 25;18 (Suppl):2331–2365. doi: 10.1093/nar/18.suppl.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L. M., Fung S., Hunkapiller M. W., Hunkapiller T. J., Hood L. E. The synthesis of oligonucleotides containing an aliphatic amino group at the 5' terminus: synthesis of fluorescent DNA primers for use in DNA sequence analysis. Nucleic Acids Res. 1985 Apr 11;13(7):2399–2412. doi: 10.1093/nar/13.7.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stryer L. Fluorescence energy transfer as a spectroscopic ruler. Annu Rev Biochem. 1978;47:819–846. doi: 10.1146/annurev.bi.47.070178.004131. [DOI] [PubMed] [Google Scholar]
- Stöcker W., Ng M., Auld D. S. Fluorescent oligopeptide substrates for kinetic characterization of the specificity of Astacus protease. Biochemistry. 1990 Nov 13;29(45):10418–10425. doi: 10.1021/bi00497a018. [DOI] [PubMed] [Google Scholar]
- Theriault G., Roy P. H., Howard K. A., Benner J. S., Brooks J. E., Waters A. F., Gingeras T. R. Nucleotide sequence of the PaeR7 restriction/modification system and partial characterization of its protein products. Nucleic Acids Res. 1985 Dec 9;13(23):8441–8461. doi: 10.1093/nar/13.23.8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlenbeck O. C., Borer P. N., Dengler B., Tinoco I., Jr Stability of RNA hairpin loops: A 6 -C m -U 6 . J Mol Biol. 1973 Feb 5;73(4):483–496. doi: 10.1016/0022-2836(73)90095-8. [DOI] [PubMed] [Google Scholar]
- Waters T. R., Connolly B. A. Continuous spectrophotometric assay for restriction endonucleases using synthetic oligodeoxynucleotides and based on the hyperchromic effect. Anal Biochem. 1992 Jul;204(1):204–209. doi: 10.1016/0003-2697(92)90162-z. [DOI] [PubMed] [Google Scholar]
- Yang C. H., Söll D. Studies of transfer RNA tertiary structure of singlet-singlet energy transfer. Proc Natl Acad Sci U S A. 1974 Jul;71(7):2838–2842. doi: 10.1073/pnas.71.7.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]




