Abstract
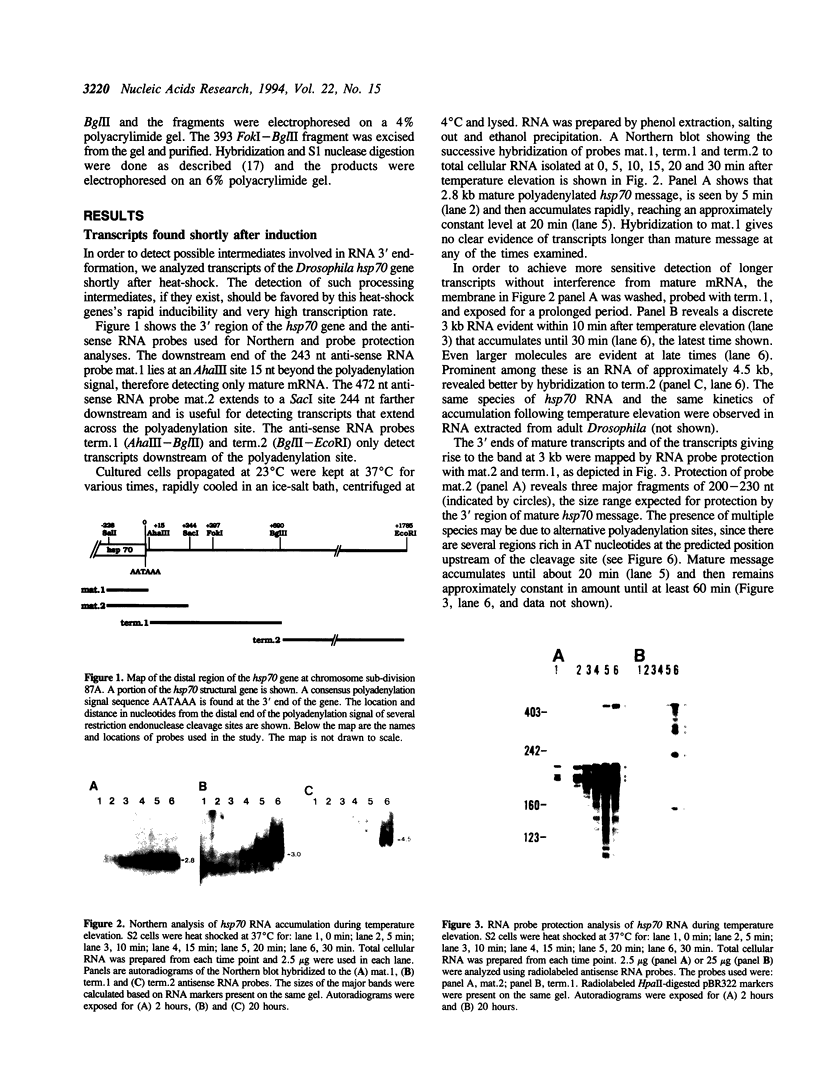
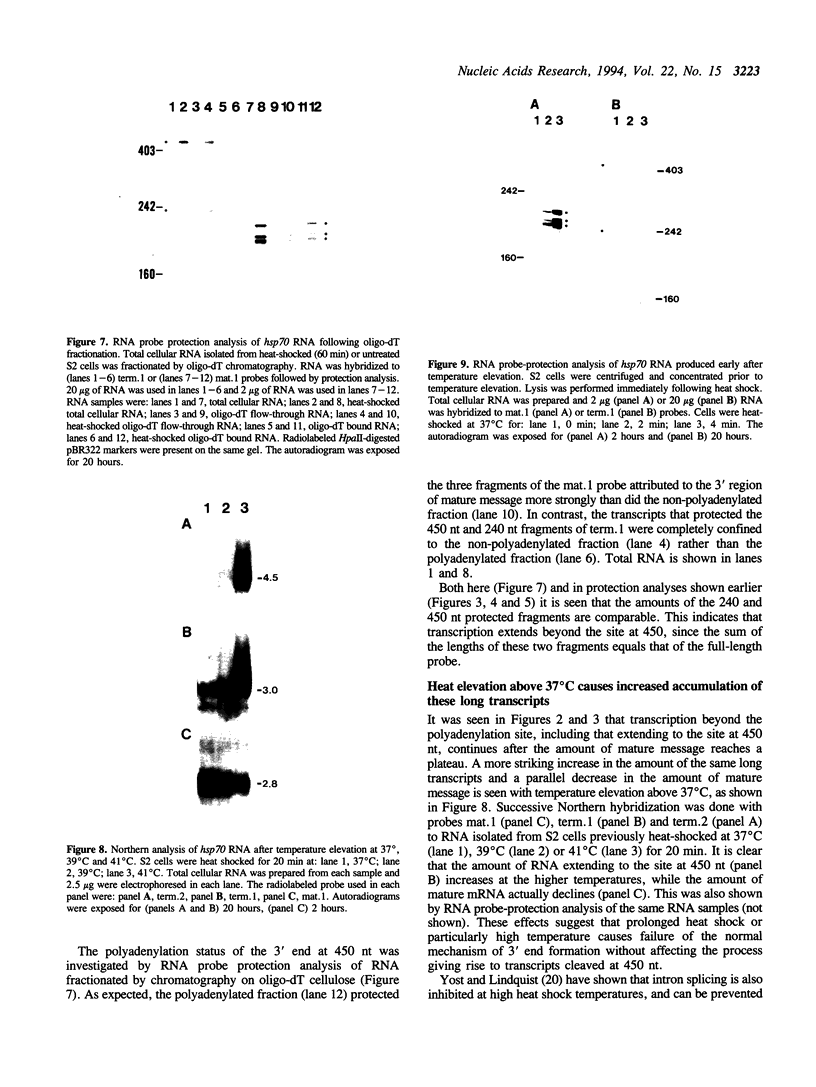
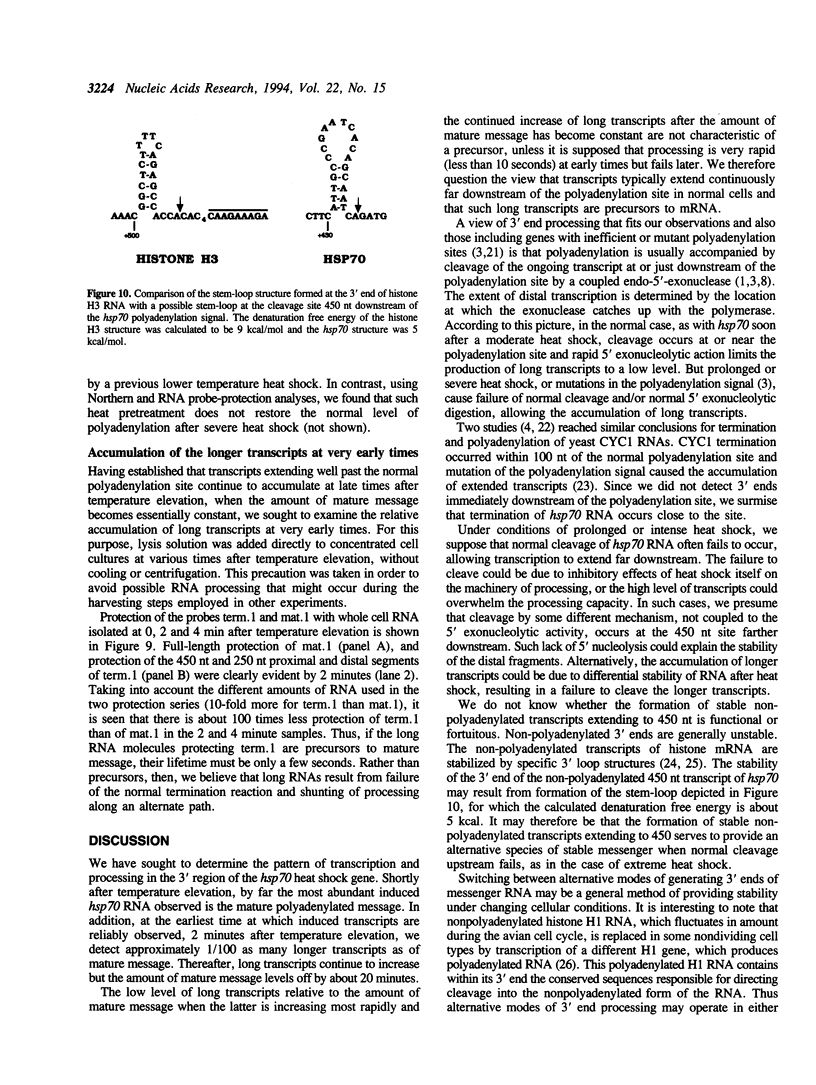
Transcription downstream of the polyadenylation site was studied in the Drosophila hsp70 gene, whose high level of transcription in response to temperature elevation facilitates detection of rare and possibly short-lived transcripts. Transcription downstream of the polyadenylation site was detected both in cultured cells and in intact animals. Even shortly after temperature elevation the extended nonpolyadenylated RNAs were rare relative to mature message, and their level continued to increase following temperature elevation even after the amount of mature message stopped increasing. The extended transcripts therefore are unlikely to be message precursors. Although continuous transcripts were detected extending as far as 2 kb downstream of the normal polyadenylation site, the predominant extended transcript was 0.45 kb long, apparently produced by cleavage of longer transcripts. Its amount relative to mature message increased with the duration and severity of heat-shock. As is the case in nonpolyadenylated histone mRNA, there is a potential stem-loop structure just upstream of the cleavage site. These data and other lines of evidence suggest that this extended transcript results from an alternative mode of stable 3'-end formation.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ashfield R., Enriquez-Harris P., Proudfoot N. J. Transcriptional termination between the closely linked human complement genes C2 and factor B: common termination factor for C2 and c-myc? EMBO J. 1991 Dec;10(13):4197–4207. doi: 10.1002/j.1460-2075.1991.tb04998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly S., Manley J. L. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes Dev. 1988 Apr;2(4):440–452. doi: 10.1101/gad.2.4.440. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Bonner J. Preparation, molecular weight, base composition, and secondary structure of giant nuclear ribonucleic acid. Biochemistry. 1973 Jun 5;12(12):2330–2338. doi: 10.1021/bi00736a023. [DOI] [PubMed] [Google Scholar]
- Hyman L. E., Moore C. L. Termination and pausing of RNA polymerase II downstream of yeast polyadenylation sites. Mol Cell Biol. 1993 Sep;13(9):5159–5167. doi: 10.1128/mcb.13.9.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola T. K., Kane C. M. RNA polymerase: regulation of transcript elongation and termination. FASEB J. 1991 Oct;5(13):2833–2842. doi: 10.1096/fasebj.5.13.1916107. [DOI] [PubMed] [Google Scholar]
- Kirsh A. L., Groudine M., Challoner P. B. Polyadenylation and U7 snRNP-mediated cleavage: alternative modes of RNA 3' processing in two avian histone H1 genes. Genes Dev. 1989 Dec;3(12B):2172–2179. doi: 10.1101/gad.3.12b.2172. [DOI] [PubMed] [Google Scholar]
- Lanoix J., Acheson N. H. A rabbit beta-globin polyadenylation signal directs efficient termination of transcription of polyomavirus DNA. EMBO J. 1988 Aug;7(8):2515–2522. doi: 10.1002/j.1460-2075.1988.tb03099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Freund R., Schweber M., Wensink P. C., Meselson M. Sequence organization and transcription at two heat shock loci in Drosophila. Proc Natl Acad Sci U S A. 1978 Nov;75(11):5613–5617. doi: 10.1073/pnas.75.11.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowry K. L., Oh R., Steitz J. A. Each of the conserved sequence elements flanking the cleavage site of mammalian histone pre-mRNAs has a distinct role in the 3'-end processing reaction. Mol Cell Biol. 1989 Jul;9(7):3105–3108. doi: 10.1128/mcb.9.7.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne B. I., Guarente L. Transcription by RNA polymerase II induces changes of DNA topology in yeast. Genes Dev. 1988 Jun;2(6):766–772. doi: 10.1101/gad.2.6.766. [DOI] [PubMed] [Google Scholar]
- Russo P., Sherman F. Transcription terminates near the poly(A) site in the CYC1 gene of the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8348–8352. doi: 10.1073/pnas.86.21.8348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Ito R., Baek K. H., Agarwal K. A specific DNA sequence controls termination of transcription in the gastrin gene. Mol Cell Biol. 1986 Apr;6(4):1032–1043. doi: 10.1128/mcb.6.4.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider I. Cell lines derived from late embryonic stages of Drosophila melanogaster. J Embryol Exp Morphol. 1972 Apr;27(2):353–365. [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Vasserot A. P., Schaufele F. J., Birnstiel M. L. Conserved terminal hairpin sequences of histone mRNA precursors are not involved in duplex formation with the U7 RNA but act as a target site for a distinct processing factor. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4345–4349. doi: 10.1073/pnas.86.12.4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wensink P. C., Finnegan D. J., Donelson J. E., Hogness D. S. A system for mapping DNA sequences in the chromosomes of Drosophila melanogaster. Cell. 1974 Dec;3(4):315–325. doi: 10.1016/0092-8674(74)90045-2. [DOI] [PubMed] [Google Scholar]
- Yost H. J., Lindquist S. RNA splicing is interrupted by heat shock and is rescued by heat shock protein synthesis. Cell. 1986 Apr 25;45(2):185–193. doi: 10.1016/0092-8674(86)90382-x. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]









