Abstract
Bulky adducts are DNA lesions generated in response to environmental agents including benzo[a]pyrene (a combustion product) and solar ultraviolet radiation. Error-prone replication of adducted DNA can cause mutations, which may result in cancer. To minimize the detrimental effects of bulky adducts and other DNA lesions, S-phase checkpoint mechanisms sense DNA damage and integrate DNA repair with ongoing DNA replication. The essential protein kinase Chk1 mediates the S-phase checkpoint, inhibiting initiation of new DNA synthesis and promoting stabilization and recovery of stalled replication forks. Here we review the mechanisms by which Chk1 is activated in response to bulky adducts and potential mechanisms by which Chk1 signaling inhibits the initiation stage of DNA synthesis. Additionally, we discuss mechanisms by which Chk1 signaling facilitates bypass of bulky lesions by specialized Y-family DNA polymerases, thereby attenuating checkpoint signaling and allowing resumption of normal cell cycle progression.
Introduction
Cells are continuously exposed to endogenous and exogenous sources of DNA damage. In normal cells, mechanisms termed `cell cycle checkpoints' monitor genome integrity and are activated in response to DNA damage. Upon activation, checkpoints co-ordinate cell cycle progression with DNA repair, thereby maintaining genetic stability (1). Failure to properly integrate the biological responses after DNA damage and to accurately duplicate the human genome results in genetic instability, a hallmark of cancer (2,3).
Bulky adducts are forms of DNA damage that result from exposure to ubiquitous environmental agents such as Polycyclic Aromatic Hydrocarbons (PAH, typified by benzo[a]pyrene or B[a]P) and solar ultraviolet (UV) radiation. Appropriate activation of checkpoints following acquisition of bulky adducts is likely important for protecting against tumorigenesis. In this article we review checkpoint signaling events induced in response to bulky DNA adducts during the S-phase of the cell cycle. There is growing evidence that in addition to inhibiting cell cycle progression, the S-phase checkpoint promotes repair of DNA damage. In particular, it appears that Trans-Lesion DNA Synthesis (TLS, a major mechanism of post-replication repair) and the S-phase checkpoint are tightly co-ordinated. Here we emphasize recent findings that efficient TLS requires S-phase checkpoint signaling, and that TLS in turn facilitates attenuation of the S-phase checkpoint and resumption of normal cell cycle progression after acquisition of DNA damage.
Bulky adducts and environmental carcinogenesis
This review is concerned with checkpoint responses to types of DNA damage generically termed `DNA bulky adducts'. The ubiquitous pollutant B[a]P and solar UVC radiation are two major sources of bulky adducts, whose effects on checkpoint signaling have been widely studied.
B[a]P is generated as a byproduct of incomplete combustion and is found in cigarette smoke, burnt coal, diesel exhaust fumes and even natural foods (4). B[a]P is a pro-carcinogen that undergoes metabolic activation by cytochrome P450 (CYP)-dependent oxidation and epoxide hydrolysis to form the highly reactive intermediate (+)-r-7,t-8-dihydroxy-t-9, 10-epoxy-7, 8, 9, 10-tetrahydrobenzo[a]pyrene (BPDE) (5). Tumorigenicity studies have demonstrated that BPDE is the `ultimate carcinogen' which results from B[a]P metabolism and causes lung, breast and other tumors in mice (6,7). BPDE binds covalently to DNA, reacting primarily with the N2 amino group of guanine to form a BPDE-DNA (BPDE-N2-dG) adduct (8,9). Error-prone replication of un-repaired BPDE-N2-dG adducts can result in mutations and the mutational properties of this species have been documented extensively (10–12).
Activation of oncogenes or inactivation of tumor suppressor genes as a consequence of BPDE-induced mutations can contribute to multi-step chemical carcinogenesis. In lung cancer patients, cigarette smoking correlates in a dose-related manner with an increase in p53 mutations, which are mainly G→T transversions (13–16). Interestingly, in B[a]P-treated cells the distribution of BPDE-N2-dG adducts in the p53 tumor suppressor gene correlates closely with p53 mutational `hotspots' found in lung tumors from cancer patients (17,18). These studies provide compelling evidence that environmentally-induced BPDE-N2-dG adducts inactivate p53 via mutagenesis, thereby contributing to lung cancer in humans.
Solar UV radiation contributes to basal and squamous cell carcinomas of the skin and cutaneous malignant melanoma in humans (19). The carcinogenic effect of UV is attributed to its ability to produce pro-mutagenic DNA lesions. These include the predominant cyclobutane pyrimidine dimers (CPD) formed by linkage of two adjacent pyrimidines and to a lesser extent pyrimidine (6-4) pyrimidone photoproducts [(6-4)PPs] (20). The mutation spectrum for p53 in skin cancers corresponds to sites of slow repair of CPD lesions, suggesting that CPDs are responsible for p53 mutations in UV-induced tumors (21,22).
Cell Cycle Checkpoints
Un-repaired DNA damage can cause mutations and poses a serious threat to genome stability. Therefore, cells possess multiple mechanisms to minimize the detrimental effects of DNA damage. Cell cycle checkpoints are signal transduction pathways that respond to DNA damage by eliciting transient delays in the cell cycle. The resulting cell cycle delays integrate DNA repair with cell cycle progression and are thought to be important for maintaining genomic stability (1). In response to genotoxins, the DNA damage-induced signaling pathways prevent entry into S-phase (the G1/S checkpoint), slow progression through S-phase (the intra-S or S-phase checkpoint), and block entry in to mitosis (the G2/M checkpoint). Components of checkpoint signaling pathways are highly conserved in eukaryotes. DNA damage is detected by sensors (e.g. ATR/ATRIP, 9-1-1 complex, ATM), with the aid of mediators (e.g. BRCA1, 53BP1, Claspin). Sensors act upstream of transducers (e.g. Chk1, Chk2) which in turn regulate effectors (p53, Cdc25A) that interact with cell cycle machinery to inhibit cell cycle progression (23). The molecular anatomy of G1 and G2/M checkpoint signaling pathways has been reviewed extensively elsewhere (1,23). This article is concerned specifically with responses to DNA damage during the S-phase of the cell cycle.
Responses to DNA damage acquired during S-phase
Acquisition of DNA damage during S-phase elicits three important responses: (I) Inhibition of firing of late origins, resulting in cessation of DNA synthesis, most often termed the `S-phase checkpoint' or the `intra-S-phase checkpoint' (reviewed in (3)). (II) Stabilization of stalled replication forks, considered to be the crucial function of S-phase checkpoint signaling (24). (III) Inhibition of entry into mitosis in the presence of un-replicated DNA, a mechanism also termed the `replication checkpoint' (25,26). The precise mechanisms by which the S-phase checkpoint inhibits late origins and stabilizes stalled replication forks are poorly understood and will be discussed in this review.
To understand the effects of bulky adducts on DNA replication, it is useful first to consider the organization of DNA synthesis during S phase in eukaryotic cells. DNA synthesis initiates at numerous loci termed `origins of replication' that are distributed throughout the genome. Origins of DNA replication are separated physically, with different subsets of origins initiating DNA synthesis (`firing') at different times during S-phase (27,28). In general, euchromatin and transcribed regions are replicated early in S-phase whereas heterochromatin, telomeres and centromeres are late-replicating. The basis for temporal regulation of origin firing during S phase are not fully understood but appears to involve chromatin modification (29,30).
Replication origins must be controlled to ensure that the entire genome is replicated efficiently and accurately only once during S phase (31). Eukaryotic cells achieve this by dividing replication into two distinct phases. During the first phase, replication proteins Orc1-6, Cdc6, Cdt1 and Mcm2–7 (a helicase complex) assemble at origins of replication to form pre-Replication Complexes (pre-RCs). Pre-RC assembly, also termed `licensing', begins in telophase and is completed after passage through G1.
The second phase of replication begins at the onset of S-phase, when the concerted actions of Cyclin Dependent Kinase (CDK) and Dbf4/Drf1-Dependent Kinase Cdc7 (also termed DDK), result in initiation of DNA synthesis (32,33). CDK and DDK activities promote the recruitment of Mcm10, Cdc45 and other initiation factors to pre-RCs. Loading of Cdc45 is crucial for subsequent unwinding of origin DNA by the Mcm2–7 helicase, recruitment of Pol α/primase, and synthesis of the RNA primer that initiates DNA replication (34). This RNA/DNA primer is required for the loading of Proliferating Cell Nuclear Antigen (PCNA)-bound DNA Polymerases δ and ε which perform processive elongation of leading and lagging strands (35–37). After initiation (also termed `firing'), the Mcm2–7 helicase continues to unwind duplex DNA in preparation for semi-conservative replication by DNA polymerases. Cdc45 migrates with Mcm2–7 and the GINS complex (comprising Psf1, Psf2, Psf3, and Sld5) which is required for elongation (38,39). Therefore, Cdc45 is involved in both elongation and initiation steps of DNA synthesis.
The mechanisms by which the concerted actions of CDK and DDK induce initiation are incompletely understood. Recent studies using S. cerevisiae have identified Sld3 (a Cdc45-binding protein) and Sld2 as essential CDK targets (40–42). Phosphorylated Sld2 and Sld3 associate with Dpb11 and the resulting complex may facilitate interactions between initiation factors including Cdc45 and GINS (40,42). The prime target of DDK appears to be the Mcm2–7 helicase complex. N-terminal phosphorylation of Mcm2, Mcm4, and Mcm6 promotes association of Cdc45 with chromatin and is important for S-phase progression (43–45).
The effects of DNA damage on DNA synthesis have been studied for several decades using velocity sedimentation to separate replication intermediates by size. Such analyses have characterized the effects of various genotoxin treatments on the size distribution and elongation rates of bulk and newly-synthesized DNA. Consequently, velocity sedimentation studies have provided considerable information on the different mechanisms by which DNA damage can affect DNA synthesis. A major conclusion from such studies is that different doses of genotoxins (including BPDE, UV, and others) inhibit the different stages of DNA synthesis via distinct mechanisms. For example, low concentrations of BPDE (<100 nM) or UV (1–2 J/m2) transiently inhibit the formation of small nascent DNAs (representing initiation events) but do not affect global elongation of large DNA chains (representing ongoing replicons) (46–48). Experimentally interfering with checkpoint signaling pathways can ablate the effects of low doses of BPDE and UV on initiation of replication (49). Therefore, the inhibition of DNA synthesis induced by low doses of BPDE or UV is mediated by the S-phase checkpoint. Cells eventually recover from the S-phase checkpoint and do not experience significant loss of viability. In contrast, higher concentrations of BPDE (200–600 nM) or UV (>2 J/m2) inhibit both initiation and elongation steps of DNA replication (46–48) resulting in a global block to DNA synthesis which results in loss of viability. A strong checkpoint signal is generated in response to high genotoxin concentrations. However, inhibition of DNA synthesis in response to high doses of genotoxins results from global stalling of DNA polymerases and is not mediated by checkpoint signaling.
More recently DNA fiber labeling strategies have been developed as a powerful new approach to analyze the dynamics of DNA replication during the S phase checkpoint (50,51). Consistent with results from velocity sedimentation studies this technique has demonstrated dose-dependent effects of genotoxins on initiation and elongation stages of DNA synthesis (51).
Many studies of DNA damage signaling are conducted with high doses of genotoxins. For example UVC doses of 5–100 J/m2 are commonly used to activate DNA damage signaling pathways (52–54). Therefore, it is not clear whether all the DNA damage responses identified in such experiments are relevant to the S-phase checkpoint (i.e. inhibition of initiation) or whether they represent responses to replication blocks (inhibition of elongation).
Initiation of checkpoint signaling and Chk1 activation
It is now firmly established that the protein kinase ATR and its downstream effector Chk1 are activated in response to bulky adducts (and other lesions). Considerable evidence indicates that Chk1 mediates the S-phase checkpoint. For instance, over-expression of catalytically-inactive (putative dominant negative) Chk1 and treatment with the Chk1 inhibitor UCN-01 were found to compromise the BPDE- and UV-induced S-phase checkpoints (49,55). A caveat of those studies is that biochemical evidence for dominant-negative activity of kinase-dead Chk1 has never been presented. Furthermore, UCN-01 appears to inhibit additional kinases that may participate in damage signaling and checkpoint control (56). However, results of more recent studies that used siRNA to specifically ablate Chk1 expression do indicate that Chk1 mediates the bulky adduct-induced S-phase checkpoint (51). Mechanisms of ATR/Chk1 activation have been reviewed in detail elsewhere (57–60) and will be summarized briefly below.
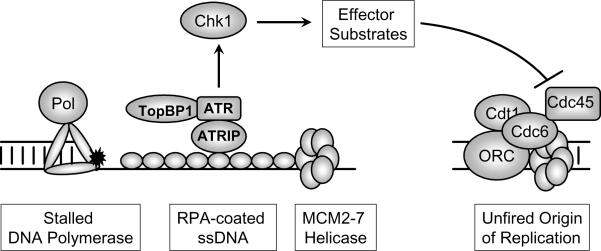
Studies using replication-competent Xenopus nuclei have provided considerable insight into the proximal events that initiate S-phase checkpoint signaling. It appears that checkpoint signaling is not triggered directly by damaged DNA, but instead results from uncoupling of the replicative helicase and polymerase activities (61) by fork-stalling DNA lesions. Thus, when polymerase movement is blocked by various lesions (such as bulky adducts), replicative helicase activity continues to unwind DNA ahead of the stalled polymerase. Uncoupling of helicase activity from replication fork movement results in excessive accumulation of single-stranded DNA (ssDNA) which is coated by Replication Protein A (RPA). RPA-coated ssDNA plays a central role in initiating S-phase checkpoint signaling (57) (Fig. 1). The ATR kinase, which exists as a heterodimeric complex with ATR-Interacting Protein (ATRIP), is re-localized to stalled replication forks or sites of DNA damage (62,63). ATRIP interacts directly with RPA and has been proposed as a likely candidate for recruiting ATR to stalled forks (63). ATR and ATRIP form oligomeric complexes in cells and ATRIP oligomerization is necessary for its binding to ATR and for recruiting ATR to nuclear foci (64). Although important for recruitment of ATR to sites of damage, RPA-ssDNA does not play a direct role in stimulating ATR kinase activity (65). Instead, recent work shows that genotoxin-induced ATR activation requires TopBP1 (topoisomerase IIβ-binding protein) (66,67). TopBP1 (the human counterpart of S. cerevisiae Dpb11) is a BRCT-containing protein that interacts transiently with ATR in an ATRIP-dependent manner and stimulates ATR kinase activity. In contrast with TopBP1 which is a general activator of ATR kinase, the mediator protein Claspin selectively promotes phosphorylation of Chk1 by ATR (60,68). Claspin associates with the kinase domain of Chk1 and is required for ATR-dependent Chk1 phosphorylation and activation. Claspin recruits Chk1 to ATR but disengages once phosphorylation and activation of Chk1 by ATR have taken place.
Fig. 1. S-phase checkpoint signaling results from uncoupling of replicative helicase and polymerase activities.
Bulky adducts and other DNA lesions (shown in black) cause stalling of the replicative DNA polymerase (illustrated here in association the PCNA trimer). Continued unwinding of DNA by the Mcm2–7 replicative helicase complex generates excessive ssDNA which is coated by RPA. ATRIP-ATR and the 9-1-1 complex (not shown) are recruited independently to the stalled replication fork. TopBP1 stimulates ATR kinase activity (no association between TopBP1 and RPA-ssDNA is implied). Chk1 is an ATR substrate and is activated by phosphorylation. ATR-mediated Chk1 activation also requires the mediator protein Claspin (not shown). Chk1 signaling inhibits initiation of DNA synthesis at late-firing origins of replication, as described in the text and in Fig. 2.
Chk1 activation by ATR also requires the putative Rad9-Rad1-Hus1 (`9-1-1') sliding clamp (69,70) and its clamp loader Rad17. 9-1-1 is recruited to chromatin independently of the ATR-ATRIP complex (71). RPA-coated ssDNA may contribute to 9-1-1 recruitment since RPA-ssDNA and a recessed DNA strand end stimulate 9-1-1 loading onto plasmid DNA in vitro (72). It has been suggested that the 9-1-1 complex enables ATR to recognize its substrates (71) but the precise role of 9-1-1 in Chk1 activation is not fully understood.
Recent studies suggest that Timeless (Tim, the mammalian homologue of S. cerevisiae Tof1), which together with its binding partners Tipin and Claspin form the fork protection complex (FPC), is also required to generate the checkpoint signal (73–76). The Tim-Tipin complex associates with the p34 subunit of RPA (77), possibly providing a mechanism for DNA damage-induced recruitment of Tim-Tipin to sites of DNA damage. The precise roles of Tim and Tipin in checkpoint signaling are unclear, but these proteins might regulate the chromatin loading of Claspin (74), thereby contributing to Chk1 activation by ATR.
Although many details of initiation of checkpoint signaling have yet to be resolved, it appears that Chk1 activation is necessary for the S-phase checkpoint response to various forms of DNA damage including BPDE adducts (55,78), CPD (49) and other lesions. However, the downstream effectors of Chk1 responsible for inhibition of initiation at unfired origins of replication remain elusive. Candidate effectors of the Chk1-mediated S-phase checkpoint are considered below.
Relevant targets of Chk1 in the S-phase checkpoint
The cell cycle regulators Cdc25A, DDK, and Cdc45 have all been described as targets of Chk1 signaling. Therefore, possible roles for Cdc25A, DDK, and Cdc45 in the bulky adduct-induced S-phase checkpoint are discussed below.
1. Cdc25A/CDK2
The mechanism by which Chk1 inhibits DNA synthesis at unfired origins has been studied extensively in the context of IR-induced DNA Double-Strand Breaks (DSBs) (79–82). There is considerable evidence that the tyrosine phosphatase Cdc25A is targeted for degradation by DSB-induced S-phase checkpoint signaling (79,80,82). Cdc25A contributes to the activation of CDK2 and is necessary for normal S-phase progression. Current models suggest that DSB-induced Cdc25A degradation and the resulting decline in CDK2 activity lead to reduced origin firing (59).
IR-induced degradation of Cdc25A is thought to involve Chk1 and the unrelated checkpoint kinase Chk2. Basal turnover of Cdc25A requires ATR, Claspin and Chk1 (81,83). C-terminal phosphorylation of Cdc25A by Chk1 (together with N-terminal phosphorylation by an unknown kinase) directs Cdc25A to a Skp1/Cullin/F-box (SCF) complex containing the F-box protein β-TrCP (82,84,85). It is thought that the DSB-induced intra-S-phase checkpoint targets Cdc25A through Chk2-mediated amplification of Chk1-dependent signals (81). Thus, loss of Chk1 results in accumulation of hypo-phosphorylated Cdc25A and failure to degrade Cdc25A after IR. The decrease in CDK2 activity resulting from IR-induced Cdc25A degradation is likely to contribute to the S-phase checkpoint. Consistent with a Cdc25A/CDK2-mediated mechanism for the S-phase checkpoint, association of Cdc45 with origins of replication (a late CDK2-dependent event in initiation of DNA replication), is inhibited in response to IR concomitant with Cdc25A degradation (86). Negative regulation of Cdc45 following IR treatment was attributed to the ATM/Chk2/Cdc25A pathway and no role for Chk1 was demonstrated (86). However, Chk1 inhibition also appears to increase Cdc45 loading via stabilization of Cdc25A (87).
Similar to IR-induced DSB, high doses (10 J/m2) of UVC-induced CPD can induce Cdc25A degradation (54). However, two recent studies showed that low levels of bulky adducts (BPDE, UV lesions) activate the S-phase checkpoint without causing global elongation blocks but do not elicit changes in Cdc25A or CDK2 (78,88). Moreover, Cdc25A and CDK2 levels and activities are not limiting for DNA synthesis when the S-phase checkpoint is active (78). In those studies, decreases in Cdc25A were only observed in response to high doses of BPDE and UV that inhibit elongation (78,88). Taken together, it appears that the Cdc25A degradation pathway is responsive to DSB but that bulky adduct-induced S-phase checkpoints are Cdc25A /CDK2-independent.
It is unclear why bulky adduct-induced (Chk1-mediated) inhibition of DNA synthesis occurs without changes in Cdc25A or CDK2 activity. One possible explanation is that inhibition of Cdc25A by Chk1 requires a critical threshold level (or duration) of Chk1 signaling that exceeds the transient and modest Chk1 response induced by low doses of BPDE and UV. High doses of BPDE and UV that inhibit elongation induce a higher level and more sustained phosphorylation of Chk1 than is evident in cells treated with low doses that inhibit only initiation (55). It is also possible that additional signaling events induced by high doses of BPDE contribute to Cdc25A downregulation. For example, Chk2 is phosphorylated concomitantly with high dose BPDE-induced elongation blocks, but not after treatment with low doses of BPDE that activate the S-phase checkpoint (55). Since the DSB-induced intra-S-phase checkpoint involves Chk2-mediated amplification of Chk1-dependent events (81), efficient Cdc25A degradation might require threshold levels of adducts or secondary forms of DNA damage (i.e. DSBs) that elicit more extensive Chk1 and Chk2 signaling. Repair of DSB involves 5'-3' nucleolytic resection of flush ends, thereby exposing ssDNA that could lead to ATR/Chk1 activation (89). Therefore, the recombination substrate (ssDNA) generated in response to high levels of bulky adducts could contribute to Chk1 activation via a mechanism not involving uncoupled helicase and polymerase activities.
Regardless of the basis for mechanistic differences between DSB- and bulky adduct-induced S-phase checkpoints, it is clear that changes in Cdc25A/CDK activity do not mediate cessation of DNA synthesis in response to bulky lesions. Therefore, it is important to identify alternative targets of Chk1 that mediate the bulky adduct-induced S-phase checkpoint.
2. Cdc7
The Cdc7 protein kinase together with its activating partners Dbf4 and Drf1 is termed Dbf4/Drf1-Dependent Kinase (DDK) (90). During normal S-phase DDK acts at individual origins of replication to promote initiation of DNA synthesis (91). It has long been known that cdc7Δ yeast strains are hypersensitive to hydroxyurea (HU, a ribonucleotide diphosphate reductase inhibitor) and various genotoxins including UV (92). Therefore, DDK may be required for a normal DNA damage response. Analysis of the role of Cdc7 in S-phase checkpoints has been complicated because CDC7 is an essential gene. Therefore, work with Cdc7 mutants has been conducted using conditional mutants not a cdc7-null. Moreover, Cdc7 plays a key role in establishing the replication forks that are necessary for initiation of the S-phase checkpoint signal. Therefore, DDK could play an indirect role in checkpoint signaling. Nevertheless, a number of studies have suggested a role for Cdc7 as a target of S-phase checkpoints. For example, in S. pombe, Hsk1 (Cdc7) undergoes a Cds1 (the functional counterpart of hChk1 and S. cerevisiae Rad53)-dependent phosphorylation in response to HU, and Hsk1 is phosphorylated by Cds1 in vitro (93). Dbf4 (Dfp1 in S. pombe) also undergoes a Cds1/Rad53-dependent phosphorylation after HU treatment that results in decreased DDK kinase activity (94–96). Moreover, Dbf4 is removed from chromatin in a Rad53-dependent manner following HU treatment (97). Therefore, inhibitory phosphorylation of DDK is likely to prevent initiation at late origins of replication. Interestingly, hDbf4 contains consensus sequences for phosphorylation by Chk1 (90), and Heffernan et al. recently showed Chk1-dependent phosphorylation of Dbf4 from mammalian cells (88). Therefore, a conserved checkpoint kinase-dependent mechanism may target DDK in yeast and humans.
Perhaps the most convincing evidence that inhibition of Cdc7/Dbf4 via checkpoint signaling mediates the S-phase checkpoint in vertebrate cells comes from a biochemical study with Xenopus extracts. Gautier and colleagues showed that the association between Dbf4 and Cdc7 was perturbed in an ATR-dependent manner after treatment with etoposide (a topoisomerase II inhibitor) (98). Moreover, the addition of recombinant Dbf4 enabled DNA synthesis in etoposide-treated nuclei, demonstrating that Dbf4 was limiting for DNA synthesis after acquisition of etoposide-induced DNA damage (98). Similarly, Heffernan et al. recently showed that ectopically-expressed Dbf4 abrogates the S-phase checkpoint response to UVC in primary human cells (88). Therefore, high levels of DDK activity may overcome the checkpoint-mediated inhibition of initiation in some instances.
However, other recent studies using human cancer cell lines have found no major role for DDK in the S-phase checkpoint. Liu et al. reported that inhibition of initiation or elongation following BPDE treatment did not affect levels or chromatin association of Cdc7 in mammalian cells (78). In that study, the association of Cdc7 with Dbf4 was unaffected by BPDE concentrations that inhibited initiation (although inhibition of elongation in response to high-dose (600nM) BPDE was associated with loss of the Cdc7-Dbf4 interaction) (78). In contrast with etoposide-treated Xenopus nuclei (98), or UV-treated primary human cells (88) ectopic expression of Dbf4 (alone or in combination with Cdc7) did not prevent inhibition of DNA synthesis in response to BPDE. Clearly it is important to determine why Dbf4 expression abrogates the S-phase checkpoint in primary human cells but not in cancer cell lines. Consistent with the findings of Liu et al., a recent report from the Santocanale laboratory found that Cdc7-Dbf4 and Cdc7-Drf1 complexes were stable in HU- or etoposide-treated HeLa cells (99). Therefore, unlike etoposide-treated Xenopus nuclei (98), genotoxin treatment and replication stress do not dissociate DDK complexes and Dbf4 levels are not rate-limiting for DNA synthesis in BPDE-treated cancer cell lines (78,99). Interestingly, recent studies suggest that immunodepletion of Dbf4 from Xenopus egg extracts fails to inhibit DNA synthesis (100,101). Therefore, Dbf4 may not be the sole target of S-phase checkpoint signaling in etoposide-treated Xenopus extracts.
Using replication-competent Xenopus extracts, Dunphy and colleagues have shown that Drf1, an alternative binding partner of Cdc7, associates with chromatin in response to DNA damage and replication stress (102). It is unclear whether the concentrations of genotoxins used in that study affected initiation or elongation. Nevertheless, those workers suggested that Drf1 might be involved in S-phase checkpoint responses, and that its association with chromatin could provide a mechanism for inhibition of DNA synthesis. ASKL1a and ASKL1c are splice variants of the mammalian Drf1 homologue (103). In contrast with results from Xenopus extracts, Liu et al. found no increase in association of ASKL1a or ASKL1c with chromatin in response to bulky adducts. Furthermore, high level expression of ASKL1a or ASKL1c did not affect DNA synthesis in control or BPDE-treated mammalian cells (78). Therefore, there is no evidence that Dbf4 and ASKL1 have major roles in the bulky adduct-induced S-phase checkpoint in mammalian cells. However, it is possible that Dbf4 and ASKL1/Drf1 are involved in S-phase checkpoint responses to other forms of DNA damage. Alternatively it is possible that DDK plays a role in recovery from S-phase checkpoints rather than checkpoint activation. Potentially, DDK-mediated stabilization of stalled replication forks or DDK-dependent activation of unfired origins could constitute recovery or restart mechanisms. Inefficient checkpoint recovery or defective restart mechanisms following fork stalling could explain the hyper-sensitivity of Cdc7 and Dbf4 mutants to DNA damage (95).
3. Cdc45
Studies in mammalian cells using the Chk1 inhibitor UCN-01, catalytically-inactive (putative dominant-negative) Chk1, and siRNA suggest that Chk1 signaling inhibits the initiation step of DNA synthesis (49,51,104). Moreover, new origin firing was detected in aphidicolin-arrested Chk1−/− chicken DT40 cells (but not in Chk1+/+ cells), also consistent with a role for Chk1 in repressing initiation events (105). Therefore, Liu et al. investigated the effects of Chk1 signaling on regulation of initiation factors in mammalian cells. These workers found that BPDE inhibits the association of Cdc45 with chromatin concomitantly with activation of the S-phase checkpoint (78). Chromatin association of other replication factors (including Mcm10) was unaffected by BPDE, possibly indicating that checkpoint signaling inhibits initiation of replication at a step distal to Mcm10 recruitment but prior to Cdc45 loading. The dissociation of Cdc45 from Mcm7 after BPDE treatment might provide a mechanism for negative regulation of initiation of DNA synthesis by DNA damage signaling. As already noted, IR-induced DSB also inhibit the chromatin-association of Cdc45, although the effect of IR on Cdc45 response results from changes in Cdc25A stability and loss of CDK activity (59). Therefore, the IR- and bulky lesion-induced S-phase checkpoints both target Cdc45 but via distinct mechanisms.
Potentially the Chk1-mediated loss of Mcm-associated Cdc45 might reflect decreased loading of Cdc45 onto the pre-RC or may result from increased removal of Cdc45 from initiation complexes. Since CDK and DDK remain active during the S-phase checkpoint (78,88), the cellular environment is permissive for Cdc45 loading. Therefore a DNA damage-induced Cdc45 'unloading' mechanism is feasible. Direct phosphorylation of Cdc45 by Chk1 could interfere with its loading or might promote its unloading. Phosphorylation of mammalian Cdc45 has not been studied during an unperturbed cell cycle, or in the DNA damage response. Appropriate experiments are needed to test the role of Cdc45 phosphorylation in normal DNA replication and during S-phase checkpoints.
Alternatively, Chk1-mediated phosphorylation could interfere with Cdc45 loading by targeting a Cdc45-associated protein, for example components of the Mcm2–7 complex,. Several investigators have identified phosphorylation sites in Mcm2–7 complex members including Mcm2, Mcm3, and Mcm4 (44,106). The significance of DNA damage-induced Mcm helicase phosphorylation is unclear. One possibility is that Mcm2–7 phosphorylation precludes its association with Cdc45, thereby interfering with initiation of DNA synthesis.
DNA damage-induced proteolysis of Cdc45 represents another speculative mechanism that could account for the effect of bulky lesions on levels of Mcm2-7 complex-bound Cdc45 when the S-phase checkpoint is active. Proteolysis of Cdc45 has not been studied previously in mammalian cells. However, DNA damage-induced proteolysis of replication factors is not unprecedented. For example, the replication licensing factor Cdt1 is subject to DNA damage-induced degradation (107). Since replication licensing occurs during telophase and G1, Cdt1 degradation is unlikely to be involved in S-phase checkpoints (which result from decreased initiation). However, putative checkpoint-induced mechanisms for degradation of Cdc45 (or of other initiation factors) could explain the observed loss of chromatin-bound Cdc45. Proteins are frequently targeted for ubiquitin-mediated proteolysis via phosphorylation. Therefore, putative (Chk1-mediated) phosphorylation- and ubiquitination-based mechanisms of Cdc45 regulation during the S-phase checkpoint are not mutually exclusive. Clearly further work is necessary to understand the regulation of Cdc45 both during normal cell cycle and in the DNA damage response.
Attenuation of checkpoint signaling by Trans-Lesion Synthesis
It is well accepted that checkpoint signaling inhibits cell cycle progression in response to DNA damage. The mechanisms by which checkpoints are activated and checkpoint signaling delays cell cycle progression have been studied extensively. However, mechanisms that eventually attenuate checkpoint signaling after genotoxin treatment have received relatively little attention. Recent studies suggest that specialized DNA polymerases collectively termed the TLS polymerases play a key role in the resumption of DNA synthesis after acquisition of bulky UV- and BPDE-induced lesions (108–110). As discussed below, regulation of TLS polymerases appears to be tightly coordinated with S-phase checkpoint signaling events in mammalian cells.
Trans-Lesion Synthesis (TLS) DNA polymerases
In contrast with DNA polymerases such as Pol delta and Pol epsilon that perform bulk replicative DNA synthesis during S-phase, TLS polymerases have low fidelity and low processivity on undamaged DNA. However, because of a 'relaxed' active site that can accommodate helix-distorting lesions, TLS enzymes can bypass various forms of DNA damage with reduced accuracy compared with replicative polymerases (111). In eukaryotes, the main TLS polymerases are Pol eta (η), Pol iota (ι), Pol kappa (κ) and REV1 (all belonging to the Y-family), and Pol zeta (ζ, belonging to the B-family). Polζ comprises a catalytic subunit (REV3) and a non-catalytic subunit (REV7). Collectively, these enzymes are responsible for bypass synthesis of various DNA lesions.
The biochemical properties and substrate-specificities of purified TLS polymerases have been studied extensively. It is now clear that each TLS polymerase displays a preference for bypass of specific lesions in vitro. For example, Polη is unique among eukaryotic DNA polymerases in its ability to replicate templates containing cis-syn thymine-thymine (T-T) dimers (the major DNA lesions generated by UV radiation) very efficiently and accurately by inserting two 'A's opposite the lesion (112). While Polκ is unable to bypass cis-syn T-T dimers, the enzyme is able to bypass benzo[a]pyrene-adducted guanines, inserting the correct C opposite the bulky lesions (113). In contrast, Polη bypasses BPDE-adducted guanines inefficiently by inserting A or G in preference to C in vitro (114).
The phenotypes associated with loss of specific TLS polymerases in vivo are consistent with the biochemical properties of those polymerases in vitro. Most notably, xeroderma pigmentosum variant patients with defects in Polη (encoded by the XPV gene) are extremely prone to sunlight-induced skin cancers (115,116), demonstrating a physiological role for Polη in response to UV-induced DNA damage. Polκ-deficient individuals have not been identified. However, Polk-deficient mutant mouse embryonic stem (ES) cells are highly sensitive to B[a]P-induced mutagenesis and genotoxicity (117), further suggesting a role for Polκ in cellular responses to B[a]P-adducted DNA. Furthermore, in Polk-deficient mutant mouse ES cells, the G-to-T mutations become much more predominant than in the wild-type cells, thus implicating Polη in the error-prone bypass of BPDE-adducted guanines in the absence of Polκ (117).
Defects in Polκ or Polη might impair completion of DNA replication at sites damaged by BPDE or UV respectively. Alternatively, lesion bypass might occur in an error-prone manner that is mediated by an inappropriate polymerase. For example, in a recent study Livneh and colleagues showed that lesion bypass of B[a]P-adducted plasmid DNA was reduced by 30% in Polk-deficient mouse fibroblasts relative to wild-type cells, but that the residual bypass (possibly mediated by Polη or other TLS polymerases) was 20% more error prone (118). When the TLS process is completed, the DNA region containing a bulky DNA lesion that blocked the replication machinery becomes accessible for excision repair mechanisms to eliminate the lesion. Mutations can be fixed during excision repair, for example, by insertion of T opposite A that was mis-incorporated opposite BPDE-adducted guanine. Clearly, failure to complete replication, and error-prone replication represent possible causes of genetic instability and are likely to contribute to tumorigenesis.
Defects in TLS polymerase activities may result in prolonged checkpoint signaling in response to bulky DNA lesions. For example, BPDE-induced Chk1 activation and S-phase arrest are more persistent in a Polk−/− background relative to Polk+/+ cells (109). This implies that Polη cannot substitute Polκ for bypassing BPDE-adducts in vivo. Conversely, the UV-induced S-phase checkpoint is persistent in XPV cells (although Polη-deficiency does not affect the kinetics of the BPDE-induced S-phase checkpoint). These results suggest that TLS is important for eliminating stalled replication forks and reversing the uncoupling between polymerase and helicase that triggers S-phase checkpoint signaling. Interestingly, the persistent S-phase arrests resulting from BPDE and UV treatment of Polκ- and Polη-deficient cells respectively are associated with activation of DSB-induced checkpoint signaling pathways including p95, ATM, and Chk2 (109,119,120). These findings may suggest that stalled replication forks resulting from TLS polymerase-deficiency are prone to collapse, thereby generating secondary forms of DNA damage such as DSB (Fig. 3). As discussed earlier, DSB repair involves resection of flush ends, thereby exposing ssDNA that could activate ATR/Chk1 (89). Therefore, the persistent activation of Chk1 in TLS-polymerase-deficient cells might be partly due to ssDNA formed during DSB processing, in addition to the ssDNA generated by uncoupled replicative helicase and polymerase activities.
Fig. 3. Hypothetical mechanisms for checkpoint-dependent PCNA mono-ubiquitination and polymerase switching in mammalian cells.
Chk1 signaling could promote PCNA mono-ubiquitination and TLS via Rad18 recruitment or activation, negative regulation of USP1 (or other DUBs), or by a combination of both mechanisms. Although not shown in the figure, 9-1-1 represents an alternative sliding clamp for TLS polymerases.
A recent study in S. cerevisiae demonstrated that defective TLS does not affect fork progression following UV-irradiation, but results in accumulation of ssDNA gaps behind the replication fork (121). This suggests that following replication fork stalling DNA synthesis resumes due to re-initiation events on the leading strand downstream of the stalled fork. If TLS operates similarly in mammalian cells, the presence of ssDNA gaps behind the fork may also account for the persistence of checkpoint signaling in TLS-deficient cells after genotoxin treatment. Therefore, the prolongation of checkpoint activation observed in TLS-deficient mammalian cells is not necessarily attributable to persistent uncoupling of helicase and polymerase activities. Nevertheless, regardless of the mechanism of persistent Chk1 activation in Polκ/Polη-deficient cells that acquire bulky adducts, TLS polymerases are clearly required for normal checkpoint recovery (108–110).
Mechanisms of recruitment of TLS Polymerases
The process by which stalled replicative polymerases encountering DNA lesions are replaced by TLS enzymes is termed 'polymerase switching'. The molecular basis of the polymerase switch was first described for S. cerevisiae Rad30 and its mammalian homologue Polη. When replication forks stall at sites of DNA damage, PCNA (which functions as the processivity factor for replicative DNA polymerases), is mono-ubiquitinated on Lysine 164 (122–125). The three TLS polymerases Polη, Polι and Polκ possess PCNA-interacting motifs (termed `PIP boxes') that mediate direct associations with unmodified PCNA (126–131). REV1, another Y-family member, does not possess a typical PIP box. However, Friedberg and colleagues reported that the BRCT domain located at the N-terminal region of REV1 might instead mediate its interaction with PCNA (132). Mono-ubiquitination of PCNA is believed to increase its affinity for TLS polymerases (123–125). In fact, a recent study demonstrated the existence of novel ubiquitin-binding motifs (termed UBZ for Polη and Polκ and UBM for Polι and REV1) (133). The ubiquitin-binding motifs, as well as the conserved PIP sequences were shown to be necessary for efficient recruitment of Polη, Polι and REV1 to PCNA following genotoxin treatments (133–135). Therefore, it appears likely that a common mechanism involving UBM/Z motifs recruits TLS polymerases to monoubiquitinated PCNA (mUb-PCNA) at stalled replication forks. A recent study indicated that ubiquitin-fusion at the N- or C-terminus of PCNA can efficiently substitute for modification of K164 in increasing affinity for Polη (131). This suggests that the ubiquitin domain is required to attract Polη to modified PCNA, but not for directing a configuration of the bound TLS enzyme for the primer terminus. An alternative explanation is that PCNA monoubiquitination lowers its affinity for replicative polymerases (136,137); however, to date, there is no firm experimental evidence to support the assertion.
PCNA mono-ubiquitination at K164 is mediated by the complex of Rad6 (an E2 ubiquitin-conjugating enzyme) and Rad18 (an E3 ubiquitin ligase). Rad18 also interacts with Polη to facilitate recruitment to stalled replication forks (125,138). When lesion bypass is completed the replicative polymerase needs to regain access to the primer terminus. It remains to be seen whether or not PCNA must be de-ubiquitinated to facilitate dissociation of the TLS polymerase and re-association of the replicative polymerase. In this connection, it is still unknown whether all three subunits of PCNA in the stalled replication fork need to be modified to recruit a TLS enzyme (as suggested in (124)) or whether modification of one or two subunits is sufficient. If modification of a single PCNA subunit can recruit a TLS enzyme, the replicative polymerase might remain bound to one of the two other subunits. Thus, rotation around the DNA axis of a PCNA trimer with two attached polymerases could constuitute the switching process.
How does DNA damage trigger PCNA modification?
The mechanism(s) that stimulate PCNA mono-ubiquitination following acquisition of DNA damage are the subject of intensive investigation. Increased PCNA ubiquitination or decreased PCNA de-ubiquitination (or both) could account for the accumulation of mUb-PCNA in genotoxin-treated cells. Rad18 is re-distributed to sites of DNA damage, possibly suggesting that Rad18 is a target for regulation in DNA damage-induced PCNA ubiquitination (125). A recent study indicates that the SAF-A/B, Acinus and PIAS (SAP) domain of Rad18 is required for the re-distribution of Rad18 to sites of DNA damage (139). The SAP domain is thought to mediate protein-protein interactions and/or DNA binding (140). A Rad18-independent pathway for PCNA mono-ubiquitination was recently identified in RAD18-deficient DT40 cells (141). Therefore, it is possible that E3 ligases other than Rad18 also contribute to PCNA ubiquitination in vivo.
It has also been suggested that changes in PCNA de-ubiquitinating activities account for DNA damage-induced PCNA ubiquitination. D'Andrea and colleagues reported that USP1 is a de-ubiquitinating enzyme (DUB) for mUb-PCNA (52). USP1 is functionally inactivated in response to DNA damage via an autocleavage mechanism and the resulting loss of USP1 might contribute to increasing the level of mUb-PCNA (52). However, the doses of UVC needed to elicit USP1 degradation are very high (30–100 J/m2), result in global elongation blocks, and far exceed the low doses (~1 J/m2) required to induce PCNA ubiquitination in intact cells. Therefore it is possible that USP1 plays a major role in PCNA ubiquitination only in response to very high levels of DNA damage or when there is global inhibition of DNA synthesis.
Interestingly, USP1 is also a DUB for mono-ubiquitinated FANCD2, the putative effector of the Fanconi Anemia (FA) pathway that is known to be important for inter-strand cross-link repair (142). USP1 cleavage might represent a mechanism for coordinating the TLS and FA pathways. There is no reported evidence that the FA pathway is involved in checkpoint responses to bulky adducts. Therefore, it is possible that PCNA ubiquitination resulting from USP1 cleavage represents a response to genotoxins that preferentially activate the FA pathway. Nevertheless, USP1 provides an interesting precedent for control of PCNA ubiquitination via regulated DUB activity. Changes in alternative PCNA-directed DUB activities may mediate PCNA ubiquitination in response to bulky adducts.
Since PCNA ubiquitination is stimulated in response to DNA damage it is possible that Rad18 and USP1 (or other PCNA-directed E3 ligases and DUBs) are regulated by DNA damage-induced checkpoint signaling. Bi et al. showed that BPDE-induced PCNA mono-ubiquitination and recruitment of Polκ is sensitive to ablation of ATR/RPA (by siRNA) or Chk1 inhibition (using dominant negative Chk1) (108). Therefore, the ATR/Chk1 pathway appears to contribute to efficient PCNA mono-ubiquitination in mammalian cells.
In contrast with results obtained with mammalian cells, UV-induced PCNA mono-ubiquitination and checkpoint activation in S. pombe represent two independent responses to DNA damage (143). Similarly, a recent study by the Cimprich lab using Xenopus cell free extracts suggests that genotoxin-induced PCNA mono-ubiquitination is checkpoint-independent (144). These workers also concluded that PCNA ubiquitination and ATR activation are separate responses to single-stranded DNA generated by uncoupling of replicative helicase and DNA polymerase activities. Cimprich and colleagues suggested that the effect of checkpoint inhibition on PCNA mono-ubiquitination in the study of Bi et al. (108) were indirect, resulting from toxicity and reduced replication in ATR/Chk1-deficient cells. While this could be argued for ATR and RPA (whose ablation inhibits DNA synthesis), Chk1 inhibition actually stimulates basal rates of DNA synthesis (87), most likely via increased initiation (51,105). Therefore, the inhibitory effect of Chk1-deficiency on PCNA ubiquitination and TLS polymerase recruitment cannot be attributed to reduced DNA replication. Instead, Chk1 might promote TLS via direct or indirect regulation of PCNA-directed E3 ligase(s) or DUBs (Fig. 4). Clearly further work is necessary to elucidate the putative relationship between Chk1 signaling and TLS in mammalian cells. Interestingly, it has long been suggested that S-phase checkpoint signaling stabilizes stalled replication forks (24). As already noted, in TLS polymerase- or Rad18-deficient cells, bulky adducts elicit increased DNA DSB signaling and loss of viability, hallmarks of fork collapse (108,109). It is possible therefore that the stabilizing effect of Chk1 on stalled replication fork is mediated at least in part via stimulation of PCNA ubiquitination and recruitment of TLS enzymes. Alternatively, it is possible that the role of Chk1 in TLS is to stabilize the stalled forks, allowing interactions between PCNA and TLS polymerases to take place.
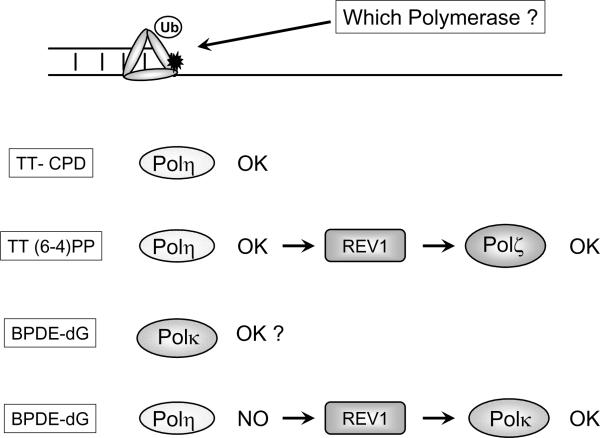
Fig. 4. Hypothetical role of REV1 in lesion-specific recruitment of Y-family polymerases in mammalian cells.
According to this model, Polη is the first TLS polymerase recruited to forks stalled by any DNA lesion. For thymine dimers (TT-CPD), Polη is sufficient to perform the insertion and extension steps of TLS and no additional polymerases are necessary. For (6-4) photoproducts (TT (6-4)PP), Polη inserts one of the 4dXMPs opposite 3'-T while Polι inserts the correct A, but Polζ is required for extension and completion lesion bypass. REV1 may facilitate the exchange of Polη for Polζ. For BPDE adducts (BPDE-dG), Polκ carries out both insertion and extension steps of TLS, whereas Polη cannot perform lesion bypass. If Polη is recruited to BPDE-stalled forks, REV1 may facilitate the exchange of Polη for Polκ, thereby enabling lesion bypass.
Other workers have also suggested a role for checkpoint proteins in TLS polymerase regulation. Kai and Wang showed that the 9-1-1 complex recruits DinB (S. pombe Polκ) by direct interactions with Hus1p (145). Interaction between Polκ and 9-1-1 has not been demonstrated in vertebrate cells. However, because efficient Polκ recruitment in mammalian cells requires Chk1 signaling (108), and since BPDE-induced Chk1 activation requires 9-1-1 (69) there is an indirect requirement for Hus1 in Polκ regulation in mammalian cells. Therefore, mammalian cells and S. pombe both require 9-1-1 for efficient Polκ recruitment.
Interestingly, in S. cerevisiae, 9-1-1 associates with Polζ and is required for Polζ-dependent spontaneous mutagenesis (146). Therefore, 9-1-1 provides a potential mechanism for checkpoint-dependent regulation of TLS polymerases. Sugimoto and colleagues recently showed a requirement for MEC1 (the S. cerevisiae ATR homologue) in localization of Polζ and Rev1 to HO-endonuclease-induced DSB (147). Although the recruitment of Polζ-Rev1 to DSB is Rad18-independent (and therefore differs from bulky adduct-dependent Polκ/η regulation) the study is consistent with a more general role for checkpoint signaling in TLS polymerase function. Taken together, emerging evidence suggests that regulation of TLS enzymes by checkpoint signaling is conserved in eukaryotes in response to bulky lesions as well as other forms of DNA damage.
How do cells ensure specific recruitment of appropriate TLS enzymes for bypass of bulky adducts?
The TLS process must be tightly regulated to restrict the activity of error-prone polymerases and ensure genomic stability. As already noted, all Y-family TLS polymerases possess ubiquitin-binding motifs (UBZ and UBM) that are thought to mediate interactions with ubiquitinated PCNA (133). Moreover, a large number of genotoxins (including, but not limited to bulky adducts) induce PCNA ubiquitination. Hydroxyl urea (HU), which decreases the dNTP pool size by inhibiting the rNDP reductase, also induces PCNA ubiquitination and focus formation of Polκ(148), while there is no obvious effect of Polκ-deficiency on recovery from HU-induced replication blocks (109). For accurate replication of damaged DNA it is important that only appropriate TLS enzyme(s) perform bypass synthesis of their preferred lesions and that TLS enzymes inappropriately bound to Ub-PCNA are removed. Lesion bypass by inappropriate polymerases (e.g. Polη-mediated bypass of BPDE adducts) could result in reduced fidelity of replication and mutagenesis. How the appropriate enzyme is recruited for each type of damage remains an interesting and open question. In this regard, the finding that Polη, Polι, Polκ and REV7 all interact with a C-terminal region of REV1 suggests that REV1 plays a special role in the TLS process (149–151).
One possibility is that REV1 facilitates the exchange of polymerases that carry out the sequential insertion and extension steps of TLS, as already discussed by others (111). Following the initial insertion across a lesion, an extending enzyme such as Polζ is required for completion of TLS. For instance, Polι is able to insert the correct A opposite the 3'-T of a T-T (6–4) photoproduct, but further extension requires an additional DNA polymerase (152). REV1 may facilitate the exchange between Polι and Polζ, via interaction with Polι and the REV7 subunit of Polζ.
We further speculate that REV1 may help eliminate TLS enzymes inappropriately bound to Ub-PCNA. Polη is likely the first TLS polymerase to be recruited to any lesion because it associates directly with the Rad6-Rad18 complex (138). Thus, once PCNA at the stalled replication fork is mono-ubiquitninated, Rad6-Rad18-associated Polη is the preferred TLS polymerase for binding to the modified PCNA. Initial recruitment of Polη to stalled forks could be advantageous because Polη is a versatile enzyme that is able to bypass many different DNA lesions, correctly in many cases (153). However, Polη forms nuclear foci in response to various lesions, including ones it cannot bypass such as BPDE (109). Therefore, recruitment of Polη in response to BPDE lesions would necessitate its subsequent exchange for Polκ. Such a polymerase exchange process might be dependent upon REV1 which interacts with both Polκ and Polη (Fig. 4).
PCNA Poly-ubiquitination
In addition to promoting TLS (which is error-prone), the budding yeast Rad6–Rad18 ubiquitin-conjugating complex regulates an error-free pathway of post-replication repair (PRR) which involves PCNA poly-ubiquitination. Error-free PRR is mediated via Rad5 (a SWI/SNF family ATPase and E3 ubiquitin ligase) together with Mms2-Ubc13 (E2 and E2 variant complex) (122). Thus, following acquisition of DNA damage, the mono-ubiquitinated Lysine 164 residue of PCNA is poly-ubiquitinated by the Rad5-Mms2-Ubc13 complex which generates a lysine 63-linked ubiquitin chain. Rad5-mediated PCNA poly-ubiquitination promotes an error-free PRR mechanism whose details are poorly understood. A recent study by Zhang and Lawrence suggest that the Rad5-mediated error-free pathway involves recombination between partially-replicated sister strands, although it does not depend on Rad52 which is essentially required for most DNA recombination events in budding yeast (154). Thus, a template-switching mechanism may be employed in which the newly-synthesized daughter strand of the undamaged complementary sequence is used as a template for error-free lesion bypass.
Until recently, the idea that PCNA is poly-ubiquitinated in mammalian cells was the subject of considerable debate, in part because a mammalian Rad5 homologue had not been identified (155). However, several groups have now demonstrated that PCNA is poly-ubiquitinated in various cultured mammalian cell lines (156,157). Moreover, the Myung and Haracska laboratories independently identified the putative tumor suppressor SHPRH as a human orthologue of yeast Rad5 (157,158). Haracska and colleagues showed that purified SHPRH promotes Rad6–Rad18-dependent PCNA poly-ubiquitination in vitro (158). The Myung group demonstrated that hSHPRH is necessary for DNA damage-induced PCNA poly-ubiquitination in response to methylmethane sulfonate (MMS) and that SHPRH ablation confers increased MMS-sensitivity (as measured by clonogenic survival) and chromosome breakage (157). Therefore, similar to yRad5, mammalian SHPRH appears to suppress genomic stability via PCNA poly-ubiquitination and error-free recombination-based tolerance mechanisms.
Although not yet tested, a role for SHPRH and PCNA poly-ubiquitination in error-free bypass of bulky adducts such as BPDE and UV lesions appears very likely. Moreover, since Rad18-mediated PCNA mono-ubiquitination requires ATR/Chk1 activities, it is likely that SHPRH-mediated PCNA poly-ubiquitination will share a similar dependence on checkpoint kinase signaling. It remains to be determined how cells choose between error-prone (Rad18/TLS polymerase) or error-free (SHPRH/recombination)-based mechanisms to recover replication forks that are stalled by bulky lesions. Cells lacking Polκ or Polη fail to recover from S-phase checkpoints induced by BPDE and UV respectively. It is unclear why SHPRH-based error-free mechanisms do not compensate for defective bypass of bulky adducts in the absence of the lesion-specific error-prone TLS enzyme. Potentially SHPRH-mediated PCNA poly-ubiquitination in vivo might occur only secondarily to TLS polymerase recruitment or might represent a low-efficiency, low-capacity pathway relative to TLS. Another possibility is that PCNA poly-ubiquitination occurs only when replication is persistently blocked due to the presence of a lesion on the template for the leading strand synthesis. Clearly further work is necessary to elucidate the molecular basis for selection of error-prone or error-free pathways for bypass of bulky adducts and the relative contribution of each mechanism to recovery from bulky adduct-induced S-phase checkpoints.
Conclusions and Perspectives
Here we have summarized mechanisms of activation and attenuation of bulky lesion induced S-phase checkpoints. Similarities and differences between bulky adduct-induced and other S-phase checkpoints have been discussed. Clearly more work is needed to understand mechanisms by which Cdc45 and perhaps other initiation factors are regulated by Chk1 signaling. Although it is well accepted that S-phase checkpoint signaling regulates the initiation step of DNA synthesis, several recent reports also indicate that rates of chain elongation are also controlled by DNA damage-induced checkpoint signaling (76,159). Additional work is necessary to identify checkpoint targets that mediate the effects of DNA damage on the elongation phase of DNA synthesis. It is now becoming clear that the process of TLS is intimately linked to checkpoint activation, and that TLS polymerase-mediated lesion bypass is necessary for elimination of stalled forks and attenuation of DNA damage signaling. However, further studies are necessary to define the precise mechanisms that couple ATR/Chk1 signaling with PCNA ubiquitination and TLS polymerase recruitment. Additionally, it is crucial to determine the mechanisms that ensure recruitment of appropriate TLS polymerases to their preferred DNA lesions and to understand how cells decide between error-prone (TLS) and error-free (SHPRH-mediated) mechanisms of lesion bypass. In addition to providing insight into mechanisms of tumor suppression, a detailed understanding of S-phase checkpoints might also be useful in the clinic. For example, abrogation of Chk1-mediated checkpoints in p53-deficient cancer cells can elicit mitotic catastrophe and apoptosis. Therefore, there is considerable interest in exploiting checkpoint kinases as possible therapeutic targets in p53-deficient cancers (160,161). Defects in TLS polymerases or Rad18 also sensitize cells to genotoxin-induced killing. It is possible that small molecule inhibition of TLS enzymes represents an additional strategy for sensitizing cancer cells to existing genotoxic chemotherapies.
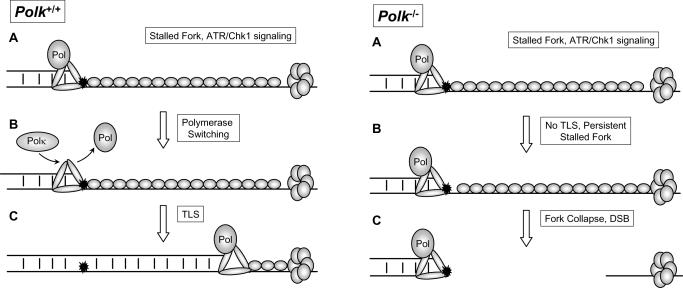
Fig. 2. Model describing effect of Polk status on BPDE-induced S-phase checkpoint signaling.
In Polk+/+ cells (left panel) BPDE-adducted DNA (shown in black) uncouples the replicative DNA polymerase (Pol) and helicase activities thereby generating ssDNA and initiating ATR/Chk1 signaling (A). A polymerase switch replaces the stalled DNA polymerase with Polκ (B). Polκ-mediated bypass of the adduct lesion allows recovery of the stalled replication fork and attenuates S-phase checkpoint signaling.
In Polk−/− cells (right panel), failure to bypass BPDE-adducted DNA results in persistently stalled forks (B). Eventual fork collapse generates DSB (C) and elicits ATM/Chk2 signaling.
Acknowledgements
Cited work from the authors' laboratories was supported by NIH grant ES09558 to C.V. and by grants (17013041 and 16370077) from the Japanese Ministry of Education, Culture, Sports, Science and Technology to H.O.
References
- 1.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 2.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 3.Nyberg KA, Michelson RJ, Putnam CW, Weinert TA. Toward maintaining the genome: DNA damage and replication checkpoints. Annu Rev Genet. 2002;36:617–656. doi: 10.1146/annurev.genet.36.060402.113540. [DOI] [PubMed] [Google Scholar]
- 4.Baum EJ. Occurrence and surveillance of polycyclic aromatic hydrocarbons. Academic Press; NY: 1978. [Google Scholar]
- 5.Conney AH. Induction of microsomal enzymes by foreign chemicals and carcinogenesis by polycyclic aromatic hydrocarbons: G. H. A. Clowes Memorial Lecture. Cancer Res. 1982;42:4875–4917. [PubMed] [Google Scholar]
- 6.Thakker DR, Yagi H, Lu AY, Levin W, Conney AH. Metabolism of benzo[a]pyrene: conversion of (+/−)-trans-7,8-dihydroxy-7,8-dihydrobenzo[a]pyrene to highly mutagenic 7,8-diol-9,10-epoxides. Proc Natl Acad Sci U S A. 1976;73:3381–3385. doi: 10.1073/pnas.73.10.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kapitulnik J, Wislocki PG, Levin W, Yagi H, Jerina DM, Conney AH. Tumorigenicity studies with diol-epoxides of benzo(a)pyrene which indicate that (+/−)-trans-7beta,8alpha-dihydroxy-9alpha,10alpha-epoxy-7,8,9,10-tetrahydr obenzo(a)pyrene is an ultimate carcinogen in newborn mice. Cancer Res. 1978;38:354–358. [PubMed] [Google Scholar]
- 8.Dipple A. DNA adducts of chemical carcinogens. Carcinogenesis. 1995;16:437–441. doi: 10.1093/carcin/16.3.437. [DOI] [PubMed] [Google Scholar]
- 9.Dipple A, Khan QA, Page JE, Ponten I, Szeliga J. DNA reactions, mutagenic action and stealth properties of polycyclic aromatic hydrocarbon carcinogens (review) Int J Oncol. 1999;14:103–111. doi: 10.3892/ijo.14.1.103. [DOI] [PubMed] [Google Scholar]
- 10.Shukla R, Liu T, Geacintov NE, Loechler EL. The major, N2-dG adduct of (+)-anti-B[a]PDE shows a dramatically different mutagenic specificity (predominantly, G --> A) in a 5'-CGT-3' sequence context. Biochemistry. 1997;36:10256–10261. doi: 10.1021/bi970541+. [DOI] [PubMed] [Google Scholar]
- 11.Hanrahan CJ, Bacolod MD, Vyas RR, Liu T, Geacintov NE, Loechler EL, Basu AK. Sequence specific mutagenesis of the major (+)-anti-benzo[a]pyrene diol epoxide-DNA adduct at a mutational hot spot in vitro and in Escherichia coli cells. Chem Res Toxicol. 1997;10:369–377. doi: 10.1021/tx9601925. [DOI] [PubMed] [Google Scholar]
- 12.Kozack RE, Shukla R, Loechler EL. A hypothesis for what conformation of the major adduct of (+)-anti-B[a]PDE (N2-dG) causes G-->T versus G-->A mutations based upon a correlation between mutagenesis and molecular modeling results. Carcinogenesis. 1999;20:95–102. doi: 10.1093/carcin/20.1.95. [DOI] [PubMed] [Google Scholar]
- 13.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 14.Krawczak M, Cooper DN. p53 mutations, benzo[a]pyrene and lung cancer. Mutagenesis. 1998;13:319–320. doi: 10.1093/mutage/13.4.319. [DOI] [PubMed] [Google Scholar]
- 15.Rodin SN, Rodin AS. Human lung cancer and p53: the interplay between mutagenesis and selection. Proc Natl Acad Sci U S A. 2000;97:12244–12249. doi: 10.1073/pnas.180320897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hainaut P, Pfeifer GP. Patterns of p53 G-->T transversions in lung cancers reflect the primary mutagenic signature of DNA-damage by tobacco smoke. Carcinogenesis. 2001;22:367–374. doi: 10.1093/carcin/22.3.367. [DOI] [PubMed] [Google Scholar]
- 17.Denissenko MF, Pao A, Tang M, Pfeifer GP. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science. 1996;274:430–432. doi: 10.1126/science.274.5286.430. [DOI] [PubMed] [Google Scholar]
- 18.Denissenko MF, Venkatachalam S, Ma YH, Wani AA. Site-specific induction and repair of benzo[a]pyrene diol epoxide DNA damage in human H-ras protooncogene as revealed by restriction cleavage inhibition. Mutat Res. 1996;363:27–42. doi: 10.1016/0921-8777(95)00059-3. [DOI] [PubMed] [Google Scholar]
- 19.Woodhead AD, Setlow RB, Tanaka M. Environmental factors in nonmelanoma and melanoma skin cancer. J Epidemiol. 1999;9:S102–114. doi: 10.2188/jea.9.6sup_102. [DOI] [PubMed] [Google Scholar]
- 20.Pfeifer GP, You YH, Besaratinia A. Mutations induced by ultraviolet light. Mutat Res. 2005;571:19–31. doi: 10.1016/j.mrfmmm.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 21.Tornaletti S, Rozek D, Pfeifer GP. The distribution of UV photoproducts along the human p53 gene and its relation to mutations in skin cancer. Oncogene. 1993;8:2051–2057. [PubMed] [Google Scholar]
- 22.You YH, Szabo PE, Pfeifer GP. Cyclobutane pyrimidine dimers form preferentially at the major p53 mutational hotspot in UVB-induced mouse skin tumors. Carcinogenesis. 2000;21:2113–2117. doi: 10.1093/carcin/21.11.2113. [DOI] [PubMed] [Google Scholar]
- 23.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 24.Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, Newlon CS, Foiani M. The DNA replication checkpoint response stabilizes stalled replication forks. Nature. 2001;412:557–561. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- 25.Brown EJ. The ATR-independent DNA replication checkpoint. Cell Cycle. 2003;2:188–189. [PubMed] [Google Scholar]
- 26.Brown EJ, Baltimore D. Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes Dev. 2003;17:615–628. doi: 10.1101/gad.1067403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rhind N. DNA replication timing: random thoughts about origin firing. Nat Cell Biol. 2006;8:1313–1316. doi: 10.1038/ncb1206-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lucas I, Feng W. The essence of replication timing: determinants and significance. Cell Cycle. 2003;2:560–563. [PubMed] [Google Scholar]
- 29.Gilbert DM. In search of the holy replicator. Nat Rev Mol Cell Biol. 2004;5:848–855. doi: 10.1038/nrm1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilbert DM. Replication timing and transcriptional control: beyond cause and effect. Curr Opin Cell Biol. 2002;14:377–383. doi: 10.1016/s0955-0674(02)00326-5. [DOI] [PubMed] [Google Scholar]
- 31.Blow JJ, Dutta A. Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Biol. 2005;6:476–486. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takeda DY, Dutta A. DNA replication and progression through S phase. Oncogene. 2005;24:2827–2843. doi: 10.1038/sj.onc.1208616. [DOI] [PubMed] [Google Scholar]
- 33.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 34.Kukimoto I, Igaki H, Kanda T. Human CDC45 protein binds to minichromosome maintenance 7 protein and the p70 subunit of DNA polymerase alpha. Eur J Biochem. 1999;265:936–943. doi: 10.1046/j.1432-1327.1999.00791.x. [DOI] [PubMed] [Google Scholar]
- 35.Hubscher U, Maga G, Spadari S. Eukaryotic DNA polymerases. Annu Rev Biochem. 2002;71:133–163. doi: 10.1146/annurev.biochem.71.090501.150041. [DOI] [PubMed] [Google Scholar]
- 36.Tsurimoto T, Stillman B. Replication factors required for SV40 DNA replication in vitro. II. Switching of DNA polymerase alpha and delta during initiation of leading and lagging strand synthesis. J Biol Chem. 1991;266:1961–1968. [PubMed] [Google Scholar]
- 37.Tsurimoto T, Stillman B. Replication factors required for SV40 DNA replication in vitro. I. DNA structure-specific recognition of a primer-template junction by eukaryotic DNA polymerases and their accessory proteins. J Biol Chem. 1991;266:1950–1960. [PubMed] [Google Scholar]
- 38.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2–7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci U S A. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aparicio T, Ibarra A, Mendez J. Cdc45-MCM-GINS, a new power player for DNA replication. Cell Div. 2006;1:18. doi: 10.1186/1747-1028-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zegerman P, Diffley JF. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2006 doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- 41.Masumoto H, Muramatsu S, Kamimura Y, Araki H. S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature. 2002;415:651–655. doi: 10.1038/nature713. [DOI] [PubMed] [Google Scholar]
- 42.Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2006 doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- 43.Sheu YJ, Stillman B. Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol Cell. 2006;24:101–113. doi: 10.1016/j.molcel.2006.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masai H, Taniyama C, Ogino K, Matsui E, Kakusho N, Matsumoto S, Kim JM, Ishii A, Tanaka T, Kobayashi T, et al. Phosphorylation of MCM4 by Cdc7 Kinase Facilitates Its Interaction with Cdc45 on the Chromatin. J Biol Chem. 2006;281:39249–39261. doi: 10.1074/jbc.M608935200. [DOI] [PubMed] [Google Scholar]
- 45.Cho WH, Lee YJ, Kong SI, Hurwitz J, Lee JK. CDC7 kinase phosphorylates serine residues adjacent to acidic amino acids in the minichromosome maintenance 2 protein. Proc Natl Acad Sci U S A. 2006;103:11521–11526. doi: 10.1073/pnas.0604990103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cordeiro-Stone M, Boyer JC, Smith BA, Kaufmann WK. Effect of benzo[a]pyrene-diol-epoxide-I on growth of nascent DNA in synchronized human fibroblasts. Carcinogenesis. 1986;7:1775–1781. doi: 10.1093/carcin/7.10.1775. [DOI] [PubMed] [Google Scholar]
- 47.Kaufmann WK, Cleaver JE. Mechanisms of inhibition of DNA replication by ultraviolet light in normal human and xeroderma pigmentosum fibroblasts. J Mol Biol. 1981;149:171–187. doi: 10.1016/0022-2836(81)90297-7. [DOI] [PubMed] [Google Scholar]
- 48.Cleaver JE, Kaufmann WK, Kapp LN, Park SD. Replicon size and excision repair as factors in the inhibition and recovery of DNA synthesis from ultraviolet damage. Biochim Biophys Acta. 1983;739:207–215. doi: 10.1016/0167-4781(83)90031-3. [DOI] [PubMed] [Google Scholar]
- 49.Heffernan TP, Simpson DA, Frank AR, Heinloth AN, Paules RS, Cordeiro-Stone M, Kaufmann WK. An ATR- and Chk1-dependent S checkpoint inhibits replicon initiation following UVC-induced DNA damage. Mol Cell Biol. 2002;22:8552–8561. doi: 10.1128/MCB.22.24.8552-8561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Merrick CJ, Jackson D, Diffley JF. Visualization of altered replication dynamics after DNA damage in human cells. J Biol Chem. 2004;279:20067–20075. doi: 10.1074/jbc.M400022200. [DOI] [PubMed] [Google Scholar]
- 51.Chastain PD, 2nd, Heffernan TP, Nevis KR, Lin L, Kaufmann WK, Kaufman DG, Cordeiro-Stone M. Checkpoint regulation of replication dynamics in UV-irradiated human cells. Cell Cycle. 2006;5:2160–2167. doi: 10.4161/cc.5.18.3236. [DOI] [PubMed] [Google Scholar]
- 52.Huang TT, Nijman SM, Mirchandani KD, Galardy PJ, Cohn MA, Haas W, Gygi SP, Ploegh HL, Bernards R, D'Andrea AD. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol. 2006;8:339–347. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- 53.Bendjennat M, Boulaire J, Jascur T, Brickner H, Barbier V, Sarasin A, Fotedar A, Fotedar R. UV irradiation triggers ubiquitin-dependent degradation of p21(WAF1) to promote DNA repair. Cell. 2003;114:599–610. doi: 10.1016/j.cell.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 54.Agner J, Falck J, Lukas J, Bartek J. Differential impact of diverse anticancer chemotherapeutics on the Cdc25A-degradation checkpoint pathway. Exp Cell Res. 2005;302:162–169. doi: 10.1016/j.yexcr.2004.08.035. [DOI] [PubMed] [Google Scholar]
- 55.Guo N, Faller DV, Vaziri C. Carcinogen-induced S-phase arrest is Chk1 mediated and caffeine sensitive. Cell Growth Differ. 2002;13:77–86. [PubMed] [Google Scholar]
- 56.Reinhardt HC, Aslanian AS, Lees JA, Yaffe MB. p53-Deficient Cells Rely on ATM- and ATR-Mediated Checkpoint Signaling through the p38MAPK/MK2 Pathway for Survival after DNA Damage. Cancer Cell. 2007;11:175–189. doi: 10.1016/j.ccr.2006.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cortez D. Unwind and slow down: checkpoint activation by helicase and polymerase uncoupling. Genes Dev. 2005;19:1007–1012. doi: 10.1101/gad.1316905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bartek J, Mailand N. TOPping up ATR activity. Cell. 2006;124:888–890. doi: 10.1016/j.cell.2006.02.029. [DOI] [PubMed] [Google Scholar]
- 59.Bartek J, Lukas C, Lukas J. Checking on DNA damage in S phase. Nat Rev Mol Cell Biol. 2004;5:792–804. doi: 10.1038/nrm1493. [DOI] [PubMed] [Google Scholar]
- 60.Chini CC, Chen J. Claspin, a regulator of Chk1 in DNA replication stress pathway. DNA Repair (Amst) 2004;3:1033–1037. doi: 10.1016/j.dnarep.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 61.Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 63.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 64.Ball HL, Cortez D. ATRIP oligomerization is required for ATR-dependent checkpoint signaling. J Biol Chem. 2005;280:31390–31396. doi: 10.1074/jbc.M504961200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ball HL, Myers JS, Cortez D. ATRIP binding to replication protein A-single-stranded DNA promotes ATR-ATRIP localization but is dispensable for Chk1 phosphorylation. Mol Biol Cell. 2005;16:2372–2381. doi: 10.1091/mbc.E04-11-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kumagai A, Dunphy WG. How cells activate ATR. Cell Cycle. 2006;5:1265–1268. doi: 10.4161/cc.5.12.2834. [DOI] [PubMed] [Google Scholar]
- 67.Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 68.Liu S, Bekker-Jensen S, Mailand N, Lukas C, Bartek J, Lukas J. Claspin operates downstream of TopBP1 to direct ATR signaling towards Chk1 activation. Mol Cell Biol. 2006;26:6056–6064. doi: 10.1128/MCB.00492-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weiss RS, Leder P, Vaziri C. Critical role for mouse Hus1 in an S-phase DNA damage cell cycle checkpoint. Mol Cell Biol. 2003;23:791–803. doi: 10.1128/MCB.23.3.791-803.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weiss RS, Matsuoka S, Elledge SJ, Leder P. Hus1 acts upstream of chk1 in a mammalian DNA damage response pathway. Curr Biol. 2002;12:73–77. doi: 10.1016/s0960-9822(01)00626-1. [DOI] [PubMed] [Google Scholar]
- 71.Zou L, Cortez D, Elledge SJ. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 2002;16:198–208. doi: 10.1101/gad.950302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ellison V, Stillman B. Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5' recessed DNA. PLoS Biol. 2003;1:E33. doi: 10.1371/journal.pbio.0000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Unsal-Kacmaz K, Mullen TE, Kaufmann WK, Sancar A. Coupling of human circadian and cell cycles by the timeless protein. Mol Cell Biol. 2005;25:3109–3116. doi: 10.1128/MCB.25.8.3109-3116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chou DM, Elledge SJ. Tipin and Timeless form a mutually protective complex required for genotoxic stress resistance and checkpoint function. Proc Natl Acad Sci U S A. 2006;103:18143–18147. doi: 10.1073/pnas.0609251103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yoshizawa-Sugata N, Masai H. Human Tim/Timeless-interacting protein, Tipin, is required for efficient progression of S phase and DNA replication checkpoint. J Biol Chem. 2006 doi: 10.1074/jbc.M605596200. [DOI] [PubMed] [Google Scholar]
- 76.Unsal-Kacmaz K, Chastain PD, Qu PP, Minoo P, Cordeiro-Stone M, Sancar A, Kaufmann WK. The human Tim/Tipin complex coordinates an intra-S checkpoint response to UV that slows replication fork displacement. Mol Cell Biol. 2007 doi: 10.1128/MCB.02190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gotter AL, Suppa C, Emanuel BS. Mammalian TIMELESS and Tipin are Evolutionarily Conserved Replication Fork-associated Factors. J Mol Biol. 2006 doi: 10.1016/j.jmb.2006.10.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu P, Barkley LR, Day T, Bi X, Slater DM, Alexandrow MG, Nasheuer HP, Vaziri C. The Chk1-mediated S-phase checkpoint targets initiation factor Cdc45 via a Cdc25a/Cdk2-independent mechanism. J Biol Chem. 2006 doi: 10.1074/jbc.M602982200. [DOI] [PubMed] [Google Scholar]
- 79.Mailand N, Falck J, Lukas C, Syljuasen RG, Welcker M, Bartek J, Lukas J. Rapid destruction of human Cdc25A in response to DNA damage. Science. 2000;288:1425–1429. doi: 10.1126/science.288.5470.1425. [DOI] [PubMed] [Google Scholar]
- 80.Falck J, Mailand N, Syljuasen RG, Bartek J, Lukas J. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature. 2001;410:842–847. doi: 10.1038/35071124. [DOI] [PubMed] [Google Scholar]
- 81.Sorensen CS, Syljuasen RG, Falck J, Schroeder T, Ronnstrand L, Khanna KK, Zhou BB, Bartek J, Lukas J. Chk1 regulates the S phase checkpoint by coupling the physiological turnover and ionizing radiation-induced accelerated proteolysis of Cdc25A. Cancer Cell. 2003;3:247–258. doi: 10.1016/s1535-6108(03)00048-5. [DOI] [PubMed] [Google Scholar]
- 82.Jin J, Shirogane T, Xu L, Nalepa G, Qin J, Elledge SJ, Harper JW. SCFbeta-TRCP links Chk1 signaling to degradation of the Cdc25A protein phosphatase. Genes Dev. 2003;17:3062–3074. doi: 10.1101/gad.1157503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sorensen CS, Syljuasen RG, Lukas J, Bartek J. ATR, Claspin and the Rad9-Rad1-Hus1 complex regulate Chk1 and Cdc25A in the absence of DNA damage. Cell Cycle. 2004;3:941–945. [PubMed] [Google Scholar]
- 84.Busino L, Chiesa M, Draetta GF, Donzelli M. Cdc25A phosphatase: combinatorial phosphorylation, ubiquitylation and proteolysis. Oncogene. 2004;23:2050–2056. doi: 10.1038/sj.onc.1207394. [DOI] [PubMed] [Google Scholar]
- 85.Busino L, Donzelli M, Chiesa M, Guardavaccaro D, Ganoth D, Dorrello NV, Hershko A, Pagano M, Draetta GF. Degradation of Cdc25A by beta-TrCP during S phase and in response to DNA damage. Nature. 2003;426:87–91. doi: 10.1038/nature02082. [DOI] [PubMed] [Google Scholar]
- 86.Falck J, Petrini JH, Williams BR, Lukas J, Bartek J. The DNA damage-dependent intra-S phase checkpoint is regulated by parallel pathways. Nat Genet. 2002;30:290–294. doi: 10.1038/ng845. [DOI] [PubMed] [Google Scholar]
- 87.Syljuasen RG, Sorensen CS, Hansen LT, Fugger K, Lundin C, Johansson F, Helleday T, Sehested M, Lukas J, Bartek J. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol Cell Biol. 2005;25:3553–3562. doi: 10.1128/MCB.25.9.3553-3562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Heffernan TP, Unsal-Kacmaz K, Heinloth AN, Simpson DA, Paules RS, Sancar A, Cordeiro-Stone M, Kaufmann WK. Cdc7/Dbf4 and the human S checkpoint response to UVC. J Biol Chem. 2007 doi: 10.1074/jbc.M611292200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.West SC. Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 90.Kim JM, Yamada M, Masai H. Functions of mammalian Cdc7 kinase in initiation/monitoring of DNA replication and development. Mutat Res. 2003;532:29–40. doi: 10.1016/j.mrfmmm.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 91.Bousset K, Diffley JF. The Cdc7 protein kinase is required for origin firing during S phase. Genes Dev. 1998;12:480–490. doi: 10.1101/gad.12.4.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sclafani RA. Cdc7p-Dbf4p becomes famous in the cell cycle. J Cell Sci. 2000;113(Pt 12):2111–2117. doi: 10.1242/jcs.113.12.2111. [DOI] [PubMed] [Google Scholar]
- 93.Snaith HA, Brown GW, Forsburg SL. Schizosaccharomyces pombe Hsk1p is a potential cds1p target required for genome integrity. Mol Cell Biol. 2000;20:7922–7932. doi: 10.1128/mcb.20.21.7922-7932.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Brown GW, Kelly TJ. Cell cycle regulation of Dfp1, an activator of the Hsk1 protein kinase. Proc Natl Acad Sci U S A. 1999;96:8443–8448. doi: 10.1073/pnas.96.15.8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Takeda T, Ogino K, Matsui E, Cho MK, Kumagai H, Miyake T, Arai K, Masai H. A fission yeast gene, him1(+)/dfp1(+), encoding a regulatory subunit for Hsk1 kinase, plays essential roles in S-phase initiation as well as in S-phase checkpoint control and recovery from DNA damage. Mol Cell Biol. 1999;19:5535–5547. doi: 10.1128/mcb.19.8.5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Weinreich M, Stillman B. Cdc7p-Dbf4p kinase binds to chromatin during S phase and is regulated by both the APC and the RAD53 checkpoint pathway. Embo J. 1999;18:5334–5346. doi: 10.1093/emboj/18.19.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pasero P, Duncker BP, Schwob E, Gasser SM. A role for the Cdc7 kinase regulatory subunit Dbf4p in the formation of initiation-competent origins of replication. Genes Dev. 1999;13:2159–2176. doi: 10.1101/gad.13.16.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Costanzo V, Shechter D, Lupardus PJ, Cimprich KA, Gottesman M, Gautier J. An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol Cell. 2003;11:203–213. doi: 10.1016/s1097-2765(02)00799-2. [DOI] [PubMed] [Google Scholar]
- 99.Tenca P, Brotherton D, Montagnoli A, Rainoldi S, Albanese C, Santocanale C. Cdc7 is an active kinase in human cancer cells undergoing replication stress. J Biol Chem. 2007;282:208–215. doi: 10.1074/jbc.M604457200. [DOI] [PubMed] [Google Scholar]
- 100.Takahashi TS, Walter JC. Cdc7-Drf1 is a developmentally regulated protein kinase required for the initiation of vertebrate DNA replication. Genes Dev. 2005;19:2295–2300. doi: 10.1101/gad.1339805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Petersen P, Chou DM, You Z, Hunter T, Walter JC, Walter G. Protein phosphatase 2A antagonizes ATM and ATR in a Cdk2- and Cdc7-independent DNA damage checkpoint. Mol Cell Biol. 2006;26:1997–2011. doi: 10.1128/MCB.26.5.1997-2011.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yanow SK, Gold DA, Yoo HY, Dunphy WG. Xenopus Drf1, a regulator of Cdc7, displays checkpoint-dependent accumulation on chromatin during an S-phase arrest. J Biol Chem. 2003;278:41083–41092. doi: 10.1074/jbc.M307144200. [DOI] [PubMed] [Google Scholar]
- 103.Yoshizawa-Sugata N, Ishii A, Taniyama C, Matsui E, Arai K, Masai H. A second human Dbf4/ASK-related protein, Drf1/ASKL1, is required for efficient progression of S and M phases. J Biol Chem. 2005;280:13062–13070. doi: 10.1074/jbc.M411653200. [DOI] [PubMed] [Google Scholar]
- 104.Miao H, Seiler JA, Burhans WC. Regulation of cellular and SV40 virus origins of replication by Chk1-dependent intrinsic and UVC radiation-induced checkpoints. J Biol Chem. 2003;278:4295–4304. doi: 10.1074/jbc.M204264200. [DOI] [PubMed] [Google Scholar]
- 105.Zachos G, Rainey MD, Gillespie DA. Chk1-deficient tumour cells are viable but exhibit multiple checkpoint and survival defects. Embo J. 2003;22:713–723. doi: 10.1093/emboj/cdg060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cortez D, Glick G, Elledge SJ. Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc Natl Acad Sci U S A. 2004;101:10078–10083. doi: 10.1073/pnas.0403410101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hu J, McCall CM, Ohta T, Xiong Y. Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat Cell Biol. 2004;6:1003–1009. doi: 10.1038/ncb1172. [DOI] [PubMed] [Google Scholar]
- 108.Bi X, Barkley LR, Slater DM, Tateishi S, Yamaizumi M, Ohmori H, Vaziri C. Rad18 Regulates DNA Polymerase {kappa} and Is Required for Recovery from S-Phase Checkpoint-Mediated Arrest. Mol Cell Biol. 2006;26:3527–3540. doi: 10.1128/MCB.26.9.3527-3540.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bi X, Slater DM, Ohmori H, Vaziri C. DNA polymerase kappa is specifically required for recovery from the benzo[a]pyrene-dihydrodiol epoxide (BPDE)-induced S-phase checkpoint. J Biol Chem. 2005;280:22343–22355. doi: 10.1074/jbc.M501562200. [DOI] [PubMed] [Google Scholar]
- 110.Bomgarden RD, Lupardus PJ, Soni DV, Yee MC, Ford JM, Cimprich KA. Opposing effects of the UV lesion repair protein XPA and UV bypass polymerase eta on ATR checkpoint signaling. Embo J. 2006;25:2605–2614. doi: 10.1038/sj.emboj.7601123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 112.Masutani C, Araki M, Yamada A, Kusumoto R, Nogimori T, Maekawa T, Iwai S, Hanaoka F. Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. Embo J. 1999;18:3491–3501. doi: 10.1093/emboj/18.12.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ohmori H, Ohashi E, Ogi T. Mammalian Pol kappa: regulation of its expression and lesion substrates. Adv Protein Chem. 2004;69:265–278. doi: 10.1016/S0065-3233(04)69009-7. [DOI] [PubMed] [Google Scholar]
- 114.Chiapperino D, Kroth H, Kramarczuk IH, Sayer JM, Masutani C, Hanaoka F, Jerina DM, Cheh AM. Preferential misincorporation of purine nucleotides by human DNA polymerase eta opposite benzo[a]pyrene 7,8-diol 9,10-epoxide deoxyguanosine adducts. J Biol Chem. 2002;277:11765–11771. doi: 10.1074/jbc.M112139200. [DOI] [PubMed] [Google Scholar]
- 115.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 116.Johnson RE, Kondratick CM, Prakash S, Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 117.Ogi T, Shinkai Y, Tanaka K, Ohmori H. Polkappa protects mammalian cells against the lethal and mutagenic effects of benzo[a]pyrene. Proc Natl Acad Sci U S A. 2002;99:15548–15553. doi: 10.1073/pnas.222377899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Avkin S, Goldsmith M, Velasco-Miguel S, Geacintov N, Friedberg EC, Livneh Z. Quantitative analysis of translesion DNA synthesis across a benzo[a]pyrene-guanine adduct in mammalian cells: the role of DNA polymerase kappa. J Biol Chem. 2004;279:53298–53305. doi: 10.1074/jbc.M409155200. [DOI] [PubMed] [Google Scholar]
- 119.Cleaver JE, Laposa RR, Limoli CL. DNA replication in the face of (In)surmountable odds. Cell Cycle. 2003;2:310–315. [PubMed] [Google Scholar]
- 120.Limoli CL, Laposa R, Cleaver JE. DNA replication arrest in XP variant cells after UV exposure is diverted into an Mre11-dependent recombination pathway by the kinase inhibitor wortmannin. Mutat Res. 2002;510:121–129. doi: 10.1016/s0027-5107(02)00257-9. [DOI] [PubMed] [Google Scholar]
- 121.Lopes M, Foiani M, Sogo JM. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 122.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 123.Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 124.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 125.Watanabe K, Tateishi S, Kawasuji M, Tsurimoto T, Inoue H, Yamaizumi M. Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. Embo J. 2004;23:3886–3896. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Haracska L, Johnson RE, Unk I, Phillips B, Hurwitz J, Prakash L, Prakash S. Physical and functional interactions of human DNA polymerase eta with PCNA. Mol Cell Biol. 2001;21:7199–7206. doi: 10.1128/MCB.21.21.7199-7206.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Haracska L, Johnson RE, Unk I, Phillips BB, Hurwitz J, Prakash L, Prakash S. Targeting of human DNA polymerase iota to the replication machinery via interaction with PCNA. Proc Natl Acad Sci U S A. 2001;98:14256–14261. doi: 10.1073/pnas.261560798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Haracska L, Unk I, Johnson RE, Phillips BB, Hurwitz J, Prakash L, Prakash S. Stimulation of DNA synthesis activity of human DNA polymerase kappa by PCNA. Mol Cell Biol. 2002;22:784–791. doi: 10.1128/MCB.22.3.784-791.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Prakash S. A single domain in human DNA polymerase iota mediates interaction with PCNA: implications for translesion DNA synthesis. Mol Cell Biol. 2005;25:1183–1190. doi: 10.1128/MCB.25.3.1183-1190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Vidal AE, Kannouche P, Podust VN, Yang W, Lehmann AR, Woodgate R. Proliferating cell nuclear antigen-dependent coordination of the biological functions of human DNA polymerase iota. J Biol Chem. 2004;279:48360–48368. doi: 10.1074/jbc.M406511200. [DOI] [PubMed] [Google Scholar]
- 131.Parker JL, Bielen AB, Dikic I, Ulrich HD. Contributions of ubiquitin- and PCNA-binding domains to the activity of Polymerase eta in Saccharomyces cerevisiae. Nucleic Acids Res. 2007;35:881–889. doi: 10.1093/nar/gkl1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Guo C, Sonoda E, Tang TS, Parker JL, Bielen AB, Takeda S, Ulrich HD, Friedberg EC. REV1 protein interacts with PCNA: significance of the REV1 BRCT domain in vitro and in vivo. Mol Cell. 2006;23:265–271. doi: 10.1016/j.molcel.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 133.Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- 134.Plosky BS, Vidal AE, Fernandez de Henestrosa AR, McLenigan MP, McDonald JP, Mead S, Woodgate R. Controlling the subcellular localization of DNA polymerases iota and eta via interactions with ubiquitin. Embo J. 2006;25:2847–2855. doi: 10.1038/sj.emboj.7601178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Guo C, Tang TS, Bienko M, Parker JL, Bielen AB, Sonoda E, Takeda S, Ulrich HD, Dikic I, Friedberg EC. Ubiquitin-binding motifs in REV1 protein are required for its role in the tolerance of DNA damage. Mol Cell Biol. 2006;26:8892–8900. doi: 10.1128/MCB.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Haracska L, Torres-Ramos CA, Johnson RE, Prakash S, Prakash L. Opposing effects of ubiquitin conjugation and SUMO modification of PCNA on replicational bypass of DNA lesions in Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:4267–4274. doi: 10.1128/MCB.24.10.4267-4274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Haracska L, Unk I, Prakash L, Prakash S. Ubiquitylation of yeast proliferating cell nuclear antigen and its implications for translesion DNA synthesis. Proc Natl Acad Sci U S A. 2006;103:6477–6482. doi: 10.1073/pnas.0510924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yuasa MS, Masutani C, Hirano A, Cohn MA, Yamaizumi M, Nakatani Y, Hanaoka F. A human DNA polymerase eta complex containing Rad18, Rad6 and Rev1; proteomic analysis and targeting of the complex to the chromatin-bound fraction of cells undergoing replication fork arrest. Genes Cells. 2006;11:731–744. doi: 10.1111/j.1365-2443.2006.00974.x. [DOI] [PubMed] [Google Scholar]
- 139.Nakajima S, Lan L, Kanno S, Usami N, Kobayashi K, Mori M, Shiomi T, Yasui A. Replication-dependent and -independent responses of RAD18 to DNA damage in human cells. J Biol Chem. 2006;281:34687–34695. doi: 10.1074/jbc.M605545200. [DOI] [PubMed] [Google Scholar]
- 140.Aravind L, Koonin EV. SAP - a putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci. 2000;25:112–114. doi: 10.1016/s0968-0004(99)01537-6. [DOI] [PubMed] [Google Scholar]
- 141.Simpson LJ, Ross AL, Szuts D, Alviani CA, Oestergaard VH, Patel KJ, Sale JE. RAD18-independent ubiquitination of proliferating-cell nuclear antigen in the avian cell line DT40. EMBO Rep. 2006;7:927–932. doi: 10.1038/sj.embor.7400777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Nijman SM, Huang TT, Dirac AM, Brummelkamp TR, Kerkhoven RM, D'Andrea AD, Bernards R. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell. 2005;17:331–339. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 143.Frampton J, Irmisch A, Green CM, Neiss A, Trickey M, Ulrich HD, Furuya K, Watts FZ, Carr AM, Lehmann AR. Postreplication repair and PCNA modification in Schizosaccharomyces pombe. Mol Biol Cell. 2006;17:2976–2985. doi: 10.1091/mbc.E05-11-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chang DJ, Lupardus PJ, Cimprich KA. Monoubiquitination of proliferating cell nuclear antigen induced by stalled replication requires uncoupling of DNA polymerase and mini-chromosome maintenance helicase activities. J Biol Chem. 2006;281:32081–32088. doi: 10.1074/jbc.M606799200. [DOI] [PubMed] [Google Scholar]
- 145.Kai M, Wang TS. Checkpoint activation regulates mutagenic translesion synthesis. Genes Dev. 2003;17:64–76. doi: 10.1101/gad.1043203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sabbioneda S, Minesinger BK, Giannattasio M, Plevani P, Muzi-Falconi M, Jinks-Robertson S. The 9-1-1 checkpoint clamp physically interacts with polzeta and is partially required for spontaneous polzeta-dependent mutagenesis in Saccharomyces cerevisiae. J Biol Chem. 2005;280:38657–38665. doi: 10.1074/jbc.M507638200. [DOI] [PubMed] [Google Scholar]
- 147.Hirano Y, Sugimoto K. ATR homolog Mec1 controls association of DNA polymerase zeta-Rev1 complex with regions near a double-strand break. Curr Biol. 2006;16:586–590. doi: 10.1016/j.cub.2006.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bergoglio V, Bavoux C, Verbiest V, Hoffmann JS, Cazaux C. Localisation of human DNA polymerase kappa to replication foci. J Cell Sci. 2002;115:4413–4418. doi: 10.1242/jcs.00162. [DOI] [PubMed] [Google Scholar]
- 149.Ohashi E, Murakumo Y, Kanjo N, Akagi J, Masutani C, Hanaoka F, Ohmori H. Interaction of hREV1 with three human Y-family DNA polymerases. Genes Cells. 2004;9:523–531. doi: 10.1111/j.1356-9597.2004.00747.x. [DOI] [PubMed] [Google Scholar]
- 150.Tissier A, Kannouche P, Reck MP, Lehmann AR, Fuchs RP, Cordonnier A. Co-localization in replication foci and interaction of human Y-family members, DNA polymerase pol eta and REVl protein. DNA Repair (Amst) 2004;3:1503–1514. doi: 10.1016/j.dnarep.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 151.Guo C, Fischhaber PL, Luk-Paszyc MJ, Masuda Y, Zhou J, Kamiya K, Kisker C, Friedberg EC. Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. Embo J. 2003;22:6621–6630. doi: 10.1093/emboj/cdg626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions. Nature. 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 153.Masutani C, Kusumoto R, Iwai S, Hanaoka F. Mechanisms of accurate translesion synthesis by human DNA polymerase eta. Embo J. 2000;19:3100–3109. doi: 10.1093/emboj/19.12.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhang H, Lawrence CW. The error-free component of the RAD6/RAD18 DNA damage tolerance pathway of budding yeast employs sister-strand recombination. Proc Natl Acad Sci U S A. 2005;102:15954–15959. doi: 10.1073/pnas.0504586102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wood RD, Mitchell M, Sgouros J, Lindahl T. Human DNA repair genes. Science. 2001;291:1284–1289. doi: 10.1126/science.1056154. [DOI] [PubMed] [Google Scholar]
- 156.Chiu RK, Brun J, Ramaekers C, Theys J, Weng L, Lambin P, Gray DA, Wouters BG. Lysine 63-polyubiquitination guards against translesion synthesis-induced mutations. PLoS Genet. 2006;2:e116. doi: 10.1371/journal.pgen.0020116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Motegi A, Sood R, Moinova H, Markowitz SD, Liu PP, Myung K. Human SHPRH suppresses genomic instability through proliferating cell nuclear antigen polyubiquitination. J Cell Biol. 2006;175:703–708. doi: 10.1083/jcb.200606145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Unk I, Hajdu I, Fatyol K, Szakal B, Blastyak A, Bermudez V, Hurwitz J, Prakash L, Prakash S, Haracska L. Human SHPRH is a ubiquitin ligase for Mms2-Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen. Proc Natl Acad Sci U S A. 2006;103:18107–18112. doi: 10.1073/pnas.0608595103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wang X, Guan J, Hu B, Weiss RS, Iliakis G, Wang Y. Involvement of Hus1 in the chain elongation step of DNA replication after exposure to camptothecin or ionizing radiation. Nucleic Acids Res. 2004;32:767–775. doi: 10.1093/nar/gkh243. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 160.Zhou BB, Sausville EA. Drug discovery targeting Chk1 and Chk2 kinases. Prog Cell Cycle Res. 2003;5:413–421. [PubMed] [Google Scholar]
- 161.Zhou BB, Bartek J. Targeting the checkpoint kinases: chemosensitization versus chemoprotection. Nat Rev Cancer. 2004;4:216–225. doi: 10.1038/nrc1296. [DOI] [PubMed] [Google Scholar]