Abstract
All organisms have multiple DNA polymerases specialized for translesion DNA synthesis (TLS) on damaged DNA templates. Mammalian TLS DNA polymerases include Pol η, Pol ι, Pol κ and Rev1 (all classified as ‘Y-family’ members) and Pol ζ (a ‘B-family’ member). Y-family DNA polymerases have highly structured catalytic domains; however, some of these proteins adopt different structures when bound to DNA (such as archaeal Dpo4 and human Pol κ), while others maintain similar structures independently of DNA binding (such as archaeal Dbh and S. cerevisiae Pol η). DNA binding-induced structural conversions of TLS polymerases depend on flexible regions present within the catalytic domains. In contrast, non-catalytic regions of Y-family proteins, which contain multiple domains and motifs for interactions with other proteins, are predicted to be mostly unstructured, except for short regions corresponding to ubiquitin-binding domains. In this review we discuss how the organization of structured and unstructured regions in TLS polymerases is relevant to their regulation and function during lesion bypass.
1) Historical background
For stable transmission of genetic information over generations, chromosomal and mitochondrial DNAs need to be replicated with extreme accuracy. Consistent with the requirement for replication accuracy, the three E. coli DNA polymerases I, II, and III (discovered in 1954discovered in 1970 and 1971, respectively) were found to exhibit high fidelity partly owing to the proof-reading function of intrinsic 3′–5′ exonuclease activities (Kornberg & Baker, 1991). However, over very long time periods, genomes must adapt and acquire the changes that drive evolution. Thus, accumulation of point mutations in duplicated genes facilitates acquisition of new diversified functions for gene products (“No mutation, no evolution”). Under stressful conditions, it is particularly advantageous for mutation rates to increase, thereby giving rise to mutants likely to be better adapted to new environments (Radman, 1999).
Genetic studies in E. coli revealed that such an increase in mutation frequency requires active products of certain genes, so-called SOS-inducible genes such as umuC and dinB (alternatively named dinP) whose products show significant similarity to each other (for reviews, see Friedberg et al., 2006). Based on the presumption that E. coli cells have only three DNA polymerases with proofreading function, the products of umuC (together with umuD) and dinB were assumed to interact with the replicative DNA polymerase III (Pol III) so as to decrease its fidelity when encountering a lesion on the template DNA. Similarly, genetic studies in S. cerevisiae (Sc) indicated that several genes (designated REV after reversion-less phenotype) are involved in both inducible and spontaneous mutagenesis (for a review, see Lawrence, 2004). The yeast genes required for mutagenesis include REV1, REV3 and REV7. The REV1 gene product shows similarity to the E. coli UmuC and DinB proteins, and the REV3 gene product resembles the catalytic subunit of the replicative DNA polymerase δ. Furthermore, sequence analysis of S. cerevisiae genome revealed that the yeast has another gene homologous to the E. coli umuC and dinB, namely RAD30 (McDonald et al, 1997; Roush et al., 1998).
In 1996, Lawrence and his colleagues showed that the Rev3 and Rev7 proteins form an enzyme complex (designated Pol ζ for the sixth DNA polymerase found in S. cerevisiae) which performs replicative bypass of cis-syn T-T cyclobutane pyrimidine dimer (CPD), albeit inefficiently (Nelson et al., 1996a). Additionally, these workers showed that the yeast Rev1 protein has an activity that inserts dCMP opposite an abasic site in template DNA (dCMP transferase activity) (Nelson et al., 1996b). However, since the yeast Rev1 protein is much larger than the UmuC and DinB proteins (985 versus 422 and 351 amino acid residues respectively), it was unclear at the time whether or not the region of Rev1 resembling UmuC and DinB proteins corresponded to the dCMP transferase catalytic domain. In 1999, in vitro studies with purified DinB and UmuC (with or without UmuD′, an active form of UmuD) proteins revealed that each of the proteins exhibits a DNA polymerase activity that is devoid of 3′–5′ exonuclease activity. DinB and UmuC were designated Pol IV and Pol V, respectively (Reuven et al., 1999; Tang et al., 1999; Wagner et al., 1999). Thus, the fourth and fifth E. coli DNA polymerases were identified over 25 years following discovery of the third replicative DNA polymerase III.
In the same year, the yeast Rad30 protein was found to bypass T-T CPD very efficiently and quite accurately by inserting two As opposite the dimer and the product was designated Pol η (the seventh DNA polymerase found in S. cerevisiae) (Johnson et al., 1999a). More importantly in relationship to human diseases, the gene responsible to a cancer-prone syndrome, xeroderma pigmentosum variant (XP-V) was found to code for a human counterpart of Pol η (Masutani et al, 1999b; Johnson et al., 1999b). Furthermore, mammals have another homologue of the yeast Rad30 protein (designated Pol ι, Tissier et al., 2000) as well as a homologue of the E. coli DinB protein (designated Pol κ, Ohashi et al., 2000). Thus, together with a Rev1 homologue (Gibbs et al., 2000), mammals have four similar DNA polymerases lacking proofreading function. These newly identified enzymes, all of which participate in translesion DNA synthesis (TLS), were classified as Y-family DNA polymerases in order to distinguish them from hitherto known families of DNA polymerases (A, B, C and X) (Ohmori et al, 2001).
Low fidelity TLS polymerases represent a “double-edged sword”. As evidenced by the fact that the XP-V patients lacking active Pol η are predisposed to skin cancer due to high incidence of mutations, the activity of human Pol η (hPol η) is required for decreasing mutations induced by UV-irradiation. However, owing to intrinsic low fidelity, the action of Pol η or any other TLS polymerase inevitably confers an increase in the frequencies of mutations (McCulloch et al., 2004). Nevertheless, higher organisms benefit from replication errors during ‘somatic hypermutations’ (SHMs) that occur mainly at defined loci in the genes encoding immunoglobulin in immune cells. Indeed, Pol η has been shown to play a role in SHMs (for a review, see Weill & Reynaud, 2008). In addition to Y-family DNA polymerases, other error-prone DNA polymerases have been identified. For example, Pol μ and λ, which together with Pol β are classified as X-family polymerases, are involved in non-homologous end joining in immune cells. Thus, error-prone DNA polymerases have important specialized roles in vivo.
In this review, we discuss structure-and-function relationship of Y-family DNA polymerases, focusing on motifs and domains that are important for interplay among Y-family DNA polymerases and their interactions with other proteins. The structures and functions of catalytic domains are more comprehensively discussed in other reviews (Prakash et al., 2005; Yang and Woodgate, 2007). We now know that each TLS polymerase exhibits a characteristic pattern for bypassing different DNA lesions. A number of structural analyses of Y-family enzyme catalytic domains have deepened our understandings of their unique activities. All the structures of Y-family DNA polymerases have basic right hand-like architecture consisted of ‘thumb’, ‘palm’ and ‘finger’ domains with one additional domain termed ‘LF’ (little finger), ‘PAD’ (polymerase-associated domain) or ‘wrist’. Y-family DNA polymerases have multiple motif sequences in common (Ohmori et al., 1995; Kulaeva et al., 1996). The five common motif sequences reside around the active sites in the tertiary structures near the incoming substrate site and the primer terminus. However, the regions of the active sites, residing near the lesion-containing DNA template, are variable among the Y-family polymerases, consistent with the notion that each TLS polymerase copes with different species of DNA lesions.
Although a wealth of information is now available on the catalytic functions of TLS polymerases, very little is known about how these enzymes are recruited to sites of DNA damage in vivo. Our understanding of mechanisms of TLS polymerase recruitment was advanced considerably by the finding that the Rad6-Rad18 complex (an E2-E3 ubiquitin ligase) modifies PCNA (proliferating cell nuclear antigen), the sliding clamp for DNA polymerases, via mono-ubiquitination in cells that acquire DNA damage (Hoege et al., 2002; Stelter & Ulrich, 2003). Consistent with a role for mono-ubiquitinated PCNA (mUb-PCNA) in regulating TLS polymerase recruitment, each of the four Y-family polymerases has one or two copies of ubiquitin-binding domain (UBD) (Bienko et al., 2005). Thus, direct interactions between mUb-PCNA and the UBD motifs may provide a mechanism for initial recognition of stalled replication forks by TLS polymerases. Additionally, Pol η, Pol ι and Pol κ possess PCNA-interacting protein (PIP)-box sequences, further consistent with a PCNA-based mode of recruitment to sites of DNA damage. A PIP-box has not been identified in Rev1, although some workers have suggested that mouse Rev1 binds PCNA via its N-terminal BRCT motif (Guo et al., 2006a). Thus it is possible that BRCT motif may serve as PIP box substitutes in the context of Rev1. Another unique feature of Rev1 is that it interacts via its C-terminal domain (CTD) with Pol η, Pol ι and Pol κ (Guo et al., 2003; Ohashi et al., 2004; Tissier et al., 2004). The Rev1-interacting region (RIR) of Pol η, Pol ι and Pol κ is defined by short sequences in which the presence of two consecutive phenylalanine (F) residues is critical (Ohashi et al., 2009). The Rev1-interaction is essential, at least for appropriate function of Pol κ. It is also known that Rev1-CTD interacts with the Rev7 accessory subunit of Pol ζ (Murakumo et al., 2001), which is another TLS enzyme classified as a ‘B-family’ DNA polymerase and believed to function mainly at the extension step after a DNA polymerase has inserted a base opposite DNA lesion.
Binding partners for Pol η have also been identified including Pol ι (Kannouche et al., 2002), Msh2 (Wilson et al., 2005), Rad18 (Watanabe et al., 2004), Rad51 (Mcllwraith et al., 2005) and many other proteins (Yuasa et al., 2006), in addition to PCNA and Rev1-CTD. How are such interactions with many proteins accomplished and regulated? It seems likely that N-terminal halves of Pol η and other Y-family polymerases have tight tertiary structure suited for their respective catalytic functions, but the C-terminal halves involved for transient interactions with other proteins are mostly disordered. In general, intrinsically disordered proteins or inherently unfolded proteins are very common in eukaryotes, but less so in prokaryotes (see recent reviews, Fink, 2005; Dunker et al., 2008). One advantage of disorder may be that multiple metastable conformations allow recognition of several targets with high specificity and low affinity. For any given protein it is more difficult to verify that a particular region is intrinsically disordered than to solve an ordered structure. Therefore, we necessarily rely on bioinformatics for predicting the presence of unstructured regions. Since information for protein folding is determined by primary amino acid sequence, information of non-folding should also be specified by amino acid sequence. In fact, it is known that compared to sequences of ordered proteins, intrinsically disordered segments and proteins have significantly higher levels of certain amino acids (E, K, R, G, Q, S and P) and lower levels of others (I, L, V, W, F, Y, C and N). Recently, a very useful method (DISOPRED2, http://bioinf.cs.ucl.ac.uk/disopred/) was developed for predicting disordered regions based on the primary sequence of proteins (Ward et al., 2004). Employing this prediction program, we discuss structure-and-function relationships in Y-family polymerases.
This review is separated into two halves. In the first half, we describe structural aspects of each subgroup proteins among Y-family polymerases, trying to explain why they show different patterns of lesion bypass. Also, we focus on the differences in the structures of DNA-bound and -unbound forms, pointing out flexibility in structures of TLS polymerases. In the second half, we discuss roles of protein-protein interactions, comparing binding motifs and domains present in Pol η, Pol ι, Pol κ and Rev1 proteins. Among the five subfamilies in the Y-family DNA polymerases, the UmuC subfamily proteins are present only in bacteria and they are not discussed here. We first discuss the DinB/Pol κ subfamily proteins that are present in all three kingdoms of life, bacteria, archaea and eukaryotes. Then we discuss the Pol η, Pol ι and Rev1 subfamilies that are present only in eukaryotes.
2) Flexible structures of Y-family DNA polymerases
DinB/Pol κ subfamily, enzymes conserved in bacteria, archaea and eukaryotes
The DinB/Pol κ subgroup proteins are ubiquitously present from bacteria to humans, but notably absent in the completely sequenced genomes of Saccharomyces cerevisiae and Drosophila melanogaster. The E. coli DinB protein was shown to have DNA polymerase activity (designated DNA polymerase IV, or Pol IV), independently of accessory proteins such as UmuD’ and RecA which are required for UmuC-dependent DNA polymerase activity (designated Pol V). The structural aspects of Dpo4 (DNA polymerase IV) from the thermophilic archeon Sulfolobus solfataricus have been studied more extensively than E. coli Pol IV. To date, three different types of the Dpo4 structures have been reported, which are complexes with DNA and substrate (Ling et al., 2001), apoenzyme (Wong et al., 2008), and a complex with PCNA (Xing et al., 2009). PCNA in archaea and eukaryotes or β-clamp in bacteria is a sliding clamp that encircles double-stranded DNA and tethers DNA polymerases to the primer terminus, thereby functioning as DNA polymerase processivity factor. Y-family polymerases are low-processivity enzymes, and their activities are probably restricted to limited regions near sites of DNA damage. Most (although may not all) Y-family polymerases possess motif(s) for binding to PCNA or β-clamp.
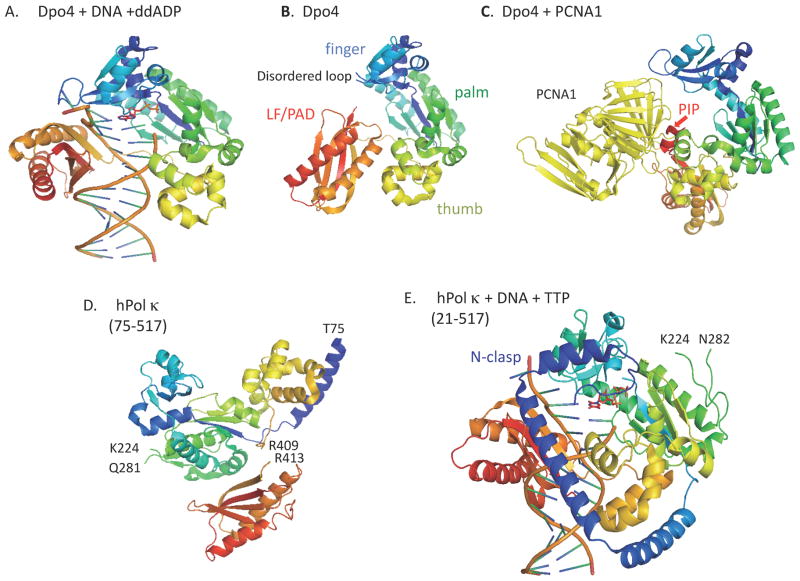
The crystal structures of Dpo4, first reported as a ternary complex with DNA and substrate, revealed that Dpo4 has some unique features, while having a right hand-like shape consisting of finger, thumb and palm domains, as observed for replicative DNA polymerases (Ling et al., 2001; see Fig. 1A). The finger and thumb domains are significantly small, resulting in an open and spacious active site that accommodates various DNA lesions. In addition, the catalytic cores of Dpo4 and other Y-family polymerases have an additional domain called “little finger (LF)” (Ling et al., 2001), “polymerase-associated domain (PAD)” (Trincao et al., 2001) or “wrist” (Silvian et al., 2001), which together with the other three catalytic core domains, constitutes a DNA-binding cleft. The open active site enables bypass of various DNA lesions, but together with the absence of proofreading exonuclease activity renders Y-family polymerases error-prone, especially when copying undamaged DNA templates. Subsequently, the Dpo4 apoenzyme structure was solved for a construct lacking the C-terminal 10 residues (342–351 out of the total 352 residues) that were disordered in the above ternary complex. The apoenzyme structure revealed that Dpo4 undergoes a global conformational change with a large rotation (131°) of the LF domain relative to the three other domains upon DNA binding (Wong et al., 2008, see Fig. 1B). Another difference between the apoenzyme and the ternary complex is that a loop (residues 34–39, see Fig. 1B) in the finger domain in the apo structure is disordered due to the absence of contacts made by the LF domain and DNA as observed in the DNA-bound structure.
Figure 1. Catalytic domain structures of Y-family polymerases.
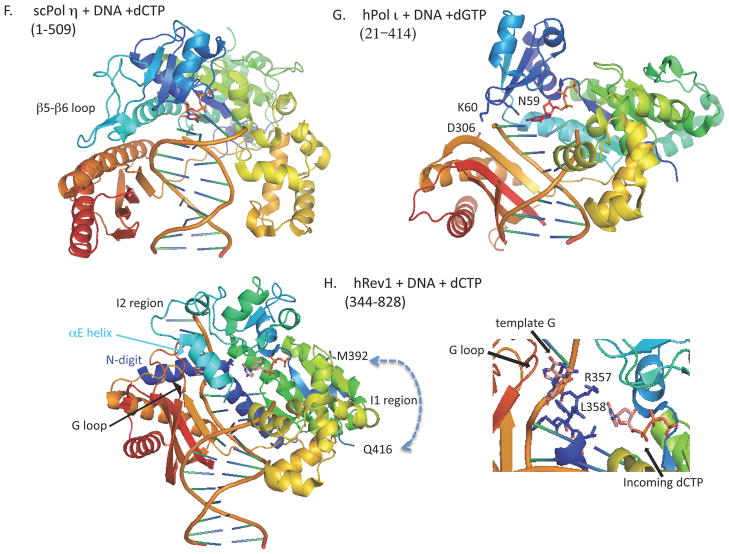
All molecular graphics images were prepared using MacPyMOL (http://www.pymol.org/), based on the PDB entries indicated in the parenthesis. Y-family proteins are rainbow-colored; N-terminal regions are shown in blue and C-terminal regions in red. A. Dpo4+DNA (1JX4), B. Apo-Dpo4 (2RDI), C. Dpo4+PCNA1 (3FDS), D. Apo-hPol κ (1T94), E. hPol κ+DNA+TTP (2OH2), F. scPol η+DNA+dCTP (2R8J), G. hPol ι+DNA+dGTP (3GV8), H. hRev1+DNA+dCTP (3GQC). In H, some portions of hRev1 around the template G and the incoming dCTP are enlarged. When truncated forms were used for structural analysis, the regions analyzed are denoted in the parenthesis below each of the proteins.
At this point, it may be useful to compare the Dpo4 structures with those of Dbh (DinB homolog) from Sulfolobus acidocaldarius (previously described as Sulfolobus solfataricus P1), which has 54% amino acid identity with Dpo4. The Dbh structures were first described as apo forms of the N-terminal catalytic fragment (2–216) (Zhou et al., 2001) and the full-length protein (1–355) (Silvian et al., 2001). In the apo structure of the full-length Dbh, the C-terminal domain (“wrist”) projects from the end of the palm opposite the finger, differently from the extended form of the Dpo4 apoprotein. This is due to the interdomain contact mediated by the β9 strand of the wrist and the β5 strand of the palm (Silvian et al., 2001). In the apo-Dbh structure, the C-terminal 10 residues and a loop region (35–39) in the finger domain are disordered, similarly as in the apo-Dpo4 structure (see Fig. 1B for the apo-Dpo4 structure). However, unlike Dpo4, the C-terminal wrist domain of Dbh does not undergo a large conformational change upon binding DNA, just rotating by 15° to 19° relative to the unliganded structure (Wilson & Pata, 2008). A large gap remaining between the finger and LF domains provides ample space to accommodate an extrahelical base in the template DNA chain, which permits misalignment of the template leading to deletion formation (Wilson & Pata, 2008). The E. coli DinB frequently makes single base deletions in runs of identical bases in vivo (Kim et al., 1997) and Dbh shows a higher rate of single base deletion in vitro than Dpo4 (Potapova et al., 2002; Boudsocq et al, 2004). Thus, the position of the LF domain relative to the catalytic core domains appears to explain some functional differences observed between Dpo4 and Dbh.
While most eukaryotic and archaeal PCNA proteins are homotrimeric rings, three PCNA homologues (PCNA1, PCNA2 and PCNA3) exist in Sulfolobus solfataricus and the functional form of PCNA in the archaeon is a heterotrimer consisted of the three homologues. Interestingly, Dpo4 interacts with PCNA1 alone, mainly through the C-terminal sequence 342-EAIGLDKFFDT-352 (the conserved residues are underlined) that is similar to the consensus sequence of PIP-box Qxx(M,L,I)xxFF (x is any residue) (Warbrick, 1998). Structural analysis of Dpo4 in complex with the PCNA1-PCNA2 heterodimer (Dpo4-p12) revealed that the PCNA-bound Dpo4 is in an extended conformation, in which the LF domain is rotated from the other three domains, differently from the apoprotein (Xing et al., 2009, see Fig. 1C). In the Dpo4-p12 complex, the LF is at the top of the front ring surface, being dissociated from the thumb, palm and finger domains that are located at the side-surface of PCNA ring. Some residues in the finger, thumb and LF domains are involved in conformation-dependent interactions with PCNA1, in addition to the main interaction by the C-terminal PIP-box. In the PCNA-bound structure, the DNA-binding cleft is disrupted because the LF stays near the central cavity of the PCNA ring structure, but the other three domains are kept away from it.
The above results predict at least two major conformational changes in the Dpo4 structure upon binding to PCNA and DNA, respectively. Two flexible hinge regions (hinge 1 and 2) were identified in Dpo4. The hinge 1 (residues 234–243) is at the linker located between LF and the other three catalytic core domains (thumb, palm and finger), which affects the position and orientation of LF relative to the core domains. The hinge 2 exists at the C-terminus of LF and confers different orientations of LF and other domains relative to the PCNA ring. Such flexibility appears to be critical for Dpo4 to adopt different conformations even when bound to PCNA; a ‘carrier configuration’ not competing with the replicative DNA polymerase for binding to the primer terminus and an ‘active configuration’ engaged for DNA synthesis when the replicative enzyme is stalled at the site of DNA damage (Xing et al., 2009).
Eukaryotic Pol κ homologues are much larger than Dpo4 that is essentially consisted of catalytic domains, with extensions at both N- and C-termini that confer additional domains for interaction with DNA and other proteins. The N-terminal extension of hPol κ (~100 residues in length, called the “N-clasp”) encircles DNA together with the palm, finger, thumb and LF/PAD domains, thus explaining why the N-terminal portion is important for binding to DNA (Lone et al., 2007, see Fig. 1E). The structure of the apo protein was determined with a form lacking the N-clasp (hPol κ69–526), in which the PAD tucks under and behind the palm domain, not forming a DNA binding cleft with the other three domains (Uljion et al., 2004, see Fig. 1D). In a ternary complex of a longer form (hPol κ19–526) with DNA and substrate, the PAD docks directly in the major groove of DNA, moving ~50 A from the position in the apoenzyme (Lone et al., 2007). This suggests that the linker region between LF/PAD and the other catalytic core domains is flexible in hPol κ, as described above for Dpo4. Thus, we conclude that Dpo4 and hPol κ are “convertible” between two different forms, DNA-bond and unbound ones.
Whether Pol κ functions solely as an “extender” to extend from the nucleotide inserted opposite a lesion by another polymerase (Prakash et al., 2005) or also as an “inserter” for certain species of DNA lesion (Ohmori et al., 2004) is a controversial issue. Several groups have shown that hPol κ is capable of inserting the correct dCMP opposite N2-benzo[a]pyrene diol epoxide (BPDE)-adducted dG (N2-BPDE-dG) adducts, while very inefficiently inserting a base opposite N6-BPDE-dA adducts (Zhang et al., 2000; Rechkoblit et al., 2002, Suzuki et al., 2002). Choi et al. (2006) showed that hPolκ could bypass bulky N2-alkyl dG adducts of increasing size accurately, but the efficiencies of dCMP insertion opposite such N2-dG lesions were still lower when compared with that opposite non-damaged dG template. In contrast, Jarosz et al. (2006) reported that mouse Pol κ bypassed N2-furfuryl-dG adduct 2-fold more efficiently than non-damaged dG. Determining the structure of a ternary complex of hPol κ19–526, DNA (not containing any lesion) and a substrate (dTTP), Lone et al. (2007) found that hPol κ has a constricted active site, not large enough to accommodate two damaged bases simultaneously. These workers proposed that the “extender” activity of hPol κ could be explained by the constricted active site and DNA encirclement by N-clasp. In contrast, from modeling studies based on the same structure, Jia et al. (2008) argued that the N-clasp of hPol κ could favor base-paring of dCTP with N2-BPDE-dG adduct and disfavor any dNTP incorporation opposite N6-BPDE-dA adduct.
The three-dimensional structure of hPol κ19–526 is very similar to that of Dpo4, and the two proteins are mostly superimposable, the biggest difference being the presence of N-clasp in hPol κ19–526. Nevertheless, Dpo4 and hPol κ19–526 show clear difference in the bypass of 7,8-dihydro-8-oxoguanine (8-oxoG); Dpo4 incorporates dCTP more efficiently than dATP opposite 8-oxoG (Rechkoblit et al., 2006; Zang et al., 2006), but hPol κ incorporates dATP more efficiently than dCTP (Zhang et al, 2000; Haracska et al., 2002; Jaloszynski et al., 2005, Irimia et al., 2009). Two groups recently succeeded in solving the structures of hPol κ19–526 inserting dATP opposite 8-oxoG (Carpio et al., 2009; Irimia et al., 2009) and found that the syn conformation of 8-oxoG for Hoogsteen base-paring with incoming dATP is stabilized by the Met135 residue in the finger domain of hPol κ. Within the active site of Dpo4, the anti conformation of 8-oxoG for base-pairing with dCTP is stabilized mainly due to the presence of the Arg322 residue in the LF/PAD domain, but the corresponding Leu508 residue in the LF/PAD of hPol κ allows 8-oxoG to keep the syn conformation. Thus, error-free or prone bypass of a DNA lesion by TLS polymerases appears to be determined by subtle differences in the structures surrounding the substrate and the damaged template.
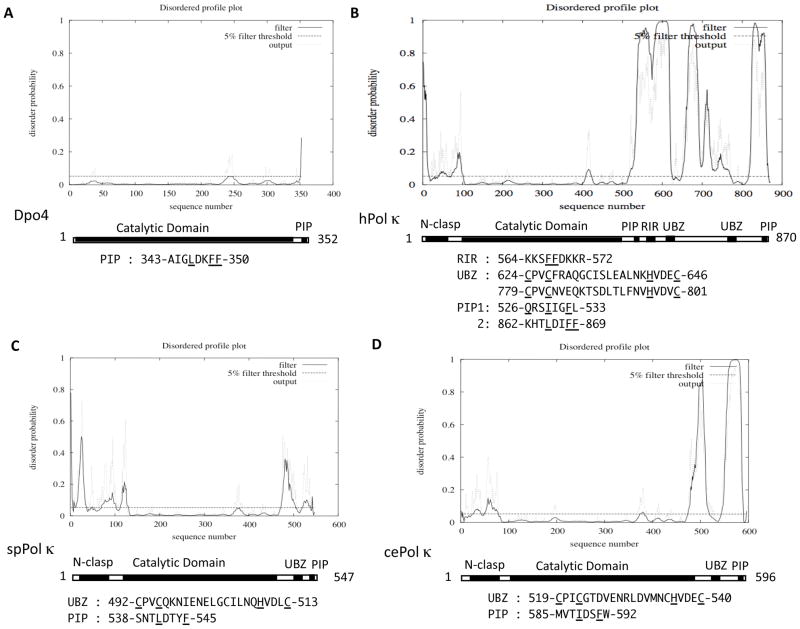
The disorder profiles of Dpo4 and hPol κ obtained by using the DISOPRED2 program are shown in Fig. 2A and B, respectively. Fig. 2A suggests that the entire region of Dpo4 has very low disorder probabilities, except for the C-terminal PIP-box region. As described above, the structure of DNA-bound Dpo4 indicated that only a small portion at the C-terminus is disordered. Therefore, the structure of apo-Dpo4 molecule was determined by crystallizing a truncated form that lacks the ten C-terminal residues (342–351). Subsequent analysis of the Dpo4-p12 complex structure showed that upon binding to PCNA1, the C-terminal PIP-box is well structured, except for the very last residue Thr-352. It should be noted that a small peak around 240 in Fig. 2A corresponds to the hinge 1 in Dpo4 (residues 234–243). Fig. 2B suggests that the catalytic domain (100~520) of hPol κ has low disorder probabilities throughout the entire region, similarly to Dpo4. Here again, a small peak around amino acid 410 corresponds to the flexible linker region between LF/PAD and the three other catalytic core domains. Thus far, several different forms of hPol κ (apoprotein or complexes with damaged or undamaged DNA) have been structurally analyzed, all of which indicate that an internal region (225~281) is unstructured. However, the disorder profile shown in Fig. 2B does not predict the presence of such an unstructured region, implying that the prediction by the DISOPRED2 program cannot detect every unstructured region. Because hPol κ69–526 was the only N-clasp-deficient construct successfully crystallized (Lone et al., 2007), the N-clasp region was suggested to be disordered in the absence of DNA. Fig. 2B shows that the N-terminal region up to 100 has higher disorder probabilities, with a peak around 90 probably corresponding to the linker region between the N-clasp and the thumb domain.
Figure 2. Disordered profile plots of Dpo4 (A), hPol κ (B), spPol κ (C) and cePol κ (D).
Each of the plots was obtained from the DISOPRED2 server (http://bioinf.ucl.ac.uk/diospred/), after inputting the entire primary sequence of the respective protein. Amino acid sequences of domains and motifs are shown below the plot, in which conserved residues are underlined.
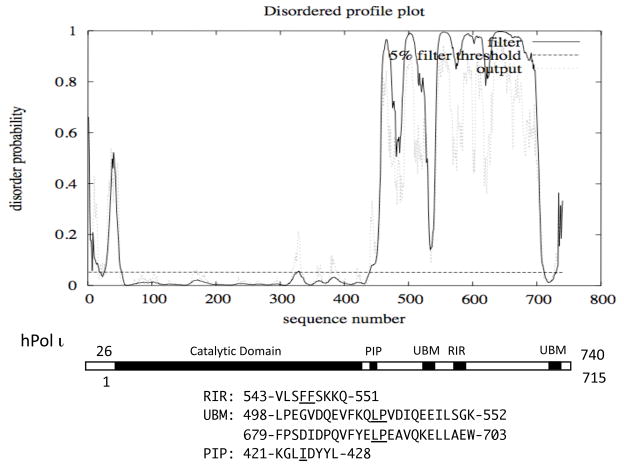
In contrast to the rigid catalytic domain, the C-terminal half of hPol κ is predicted to have multiple regions of high or low disorder probabilities. The two “valley” regions around amino acids 630 and 790 with very low disorder probabilities correspond to the ubiquitin-binding zinc-finger (UBZ) domains. The consensus sequence of Rev1-interacting region (RIR) is denoted by xxxFFyyyy (x, no specific residue and y, no specific residue but not proline) (Ohashi et al., 2009). The hPol κ sequence has two sites containing FF, one at 567–568 in RIR and the other at 868–869 in the C-terminal PIP-box sequence. In contrast to the two UBZs, the RIR site around FF567–568 appears to be embedded in one of unstructured regions. Although the disordered profile plot suggests that the extreme C-terminal region of hPol κ is “ordered”, it is likely that the C-terminal PIP-box assumes the characteristic 310 helix structure upon binding to PCNA (see Hishiki et al., 2009), as described above for the C-terminal PIP-box sequence in Dpo4. It is noted that in the DISOPRED2 prediction, false assignment of order can occur as a result of stabilizing interactions by other macromolecules. Very recently, another PIP-box-like sequence (526-QRSIIGFL-533) was found to have a PCNA-binding activity by yeast two-hybrid assay (our unpublished observation). Since this sequence is located immediately downstream of the catalytic domain (a similar arrangement to the sole PIP-box of hPol ι and the PIP1 site of hPolη as described below), the internal PIP-box of hPol κ is designated PIP1 and the C-terminal PIP-box PIP2.
While the disorder profile of the E. coli DinB is very similar to that of Dpo4, those of the S. pombe and C. elegans Pol κ homologues (spPol κ and cePol κ, 547 and 596 residues in the total, respectively) are of intermediate disorder when compared with prokaryotic and human DinB/Pol κ homologs, as shown in Fig. 2C and 2D. Both spPolκ and cePol κ have N-terminal expansions similar to hPol κ and C-terminal extensions much shorter than that of hPol κ. Both spPol κ and cePol κ have a single copy of UBZ, as well as a PIP-box sequence at the C-terminus.
Pol η subfamily, versatile enzymes for correct bypass of a variety of DNA lesions
Pol η homologues are found from S. cerevisiae (encoded by the RAD30 gene) to humans (encoded by the XPV gene), almost in every eukaryotic organism whose genome sequence has been determined, but not in prokaryotes. The E. coli Pol V (encoded by the umuC gene) and its homologues are considered to be functional counterparts in bacteria, because Pol V and Pol η are able to cope with a wide variety of DNA lesions. Pol η is unique because it is able to replicate through T-T CPD, by inserting two As opposite the lesion at the same efficiency and accuracy as that with undamaged template DNA. Furthermore, it is able to bypass a variety of DNA lesions by inserting correct base(s) opposite them at different efficiencies depending on species of DNA lesions (Masutani et al., 2000). Nevertheless, Pol η is not always error-free and sometimes commits error-prone bypass of certain DNA lesions (for more detailed descriptions of Pol η-mediated error-prone bypass see Vaisman et al., 2004).
The most characteristic feature of Pol η is its ability to replicate through UV-induced CPDs efficiently and accurately (Johnson et al., 1999a, b; Masutani et al., 1999a), suggesting that the active site should be large enough to accommodate such cross-links. However, Pol η is unable to bypass through another major UV-induced adduct (6-4) photoproducts [(6-4)PP], while it incorporates one of the four dXTPs opposite the 3′-T of T-T (6-4)PP, not extending further (Masutani et al., 1999a). In general, TLS is efficient for those DNA lesions (such as CPDs) which have small distortion in the DNA structure still allowing Watson-Crick base-paring, but it is inefficient for those [such as (6-4)PP] making large distortion, most of which should be rapidly removed by nucleotide excision repair (Masutani et al., 1999b).
The structure of hPol η, as either an apoprotein or a complex with DNA, has not been reported. Therefore we discuss structure-function aspects of hPol η based on the reported structures of S. cerevisiae Pol η (scPol η, see Fig. 1F). The catalytic core structure of scPol η apoprotein was first determined with a truncated form lacking the C-terminal 119 amino acid residues that contained the ubiquitin-binding domain (termed UBZ) and the PIP-box (Trincao et al., 2001). Subsequently, ternary complexes of scPol η with DNA containing cisplatin-induced 1,2-d(GG) adducts and dCTP were solved (Alt et al., 2007). Comparison of the ternary complexes with the apoprotein revealed small motions of the thumb and PAD domains toward DNA resulting in more intimate interaction with DNA (Alt et al., 2007). This implies that the catalytic core of scPol η apoprotein has a tight structure, in contrast to Dpo4 and hPol κ that undergo large conformational changes upon DNA binding. While scPol η also has a relatively long linker region (15 aa in length) between the thumb and LF/PAD domains, it is not as flexible as the hinge 1 (10 aa) in Dpo4, probably due to the close contacts between the finger and PAD domains in the apoprotein structure. Thus, we may conclude that scPolη has a tightly “preassembled” structure regardless of DNA binding, and does not interconvert between ‘carrier’ and ‘active’ conformations as proposed above for Dpo4. However, it is unknown whether hPol η is “preassembled” or “convertible” type. In scPol η, the long loop region between the β5 and β6 sheets in the finger domain (see Fig. 1F) is apparently involved in the contact between the finger and PAD domains, but the sequence corresponding to the loop is much shorter in hPol η (Trincao et al., 2001).
It is generally believed that high-fidelity replicative DNA polymerases make errors much less frequently than TLS polymerases devoid of proofreading function. There is one exceptional case for bypass of 8-oxoG. Replicative DNA polymerases replicate through 8-oxoG by inserting dAMP more frequently than dCMP (Shibutani et al., 1991), but some TLS polymerases, for example Dpo4 as described above, insert dCMP in preference to dAMP opposite 8-oxoG. The enzyme scPol η exhibits 20-fold higher efficiency for incorporation of dCMP over dAMP opposite 8-oxoG, which contributes to preventing spontaneous G:C to T:A transversions caused by 8-oxoG in ogg1-defective mutants of S. cerevisiae (Haracska et al., 2000). In the absence of the Ogg1 glycosylase to remove 8-oxoG paired with C, replicative polymerases have increased chances to encounter 8-oxoG and replicate past it by frequently inserting A opposite the lesion. The resulting 8-oxoG/A is recognized by the mismatch repair (MMR) system, which removes a DNA fragment containing A in pairing with 8-oxoG. If the subsequent DNA synthesis is mediated by scPol η, error-free bypass across 8-oxoG contributes to the suppression of G:C to T:A mutations. Interestingly, similar to other modes of TLS, this process also depends on mono-ubiquitination of PCNA by the Rad6-Rad18 complex and on the UBZ and PIP-box functions in scPol η (de Padula et al., 2004; Auffret van der Kemp et al., 2009). If unmodified PCNA recruits Pol δ to fill a gap containing 8-oxoG, dAMP may be incorporated again opposite the lesion. Thus, error-free or prone DNA synthesis is not simply determined by the presence of a DNA lesion, rather it depends on the combination of a lesion and DNA polymerase involved in its bypass.
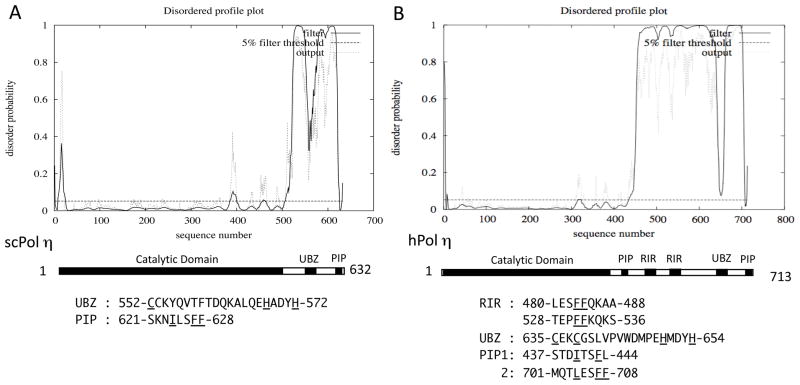
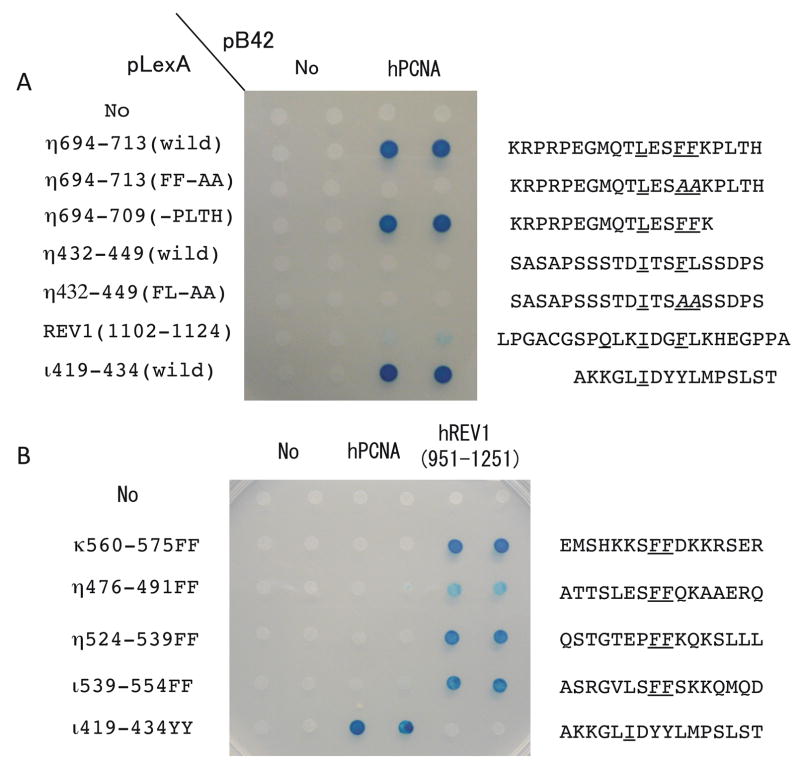
As shown in Fig. 3A and B, the N-terminal catalytic domains of scPol η and hPol η have low disorder probabilities, whereas the C-terminal regions of both are predicted to be mostly disordered, except for the regions corresponding to the single UBZ domain and the PIP-box at the C-terminus. Acharya et al. (2008) suggested that hPol η might have a PIP-box (437-STDITSFL-444, designated PIP1) immediately downstream of the catalytic domain, which is distantly related to the PIP-box consensus sequence Qxx(I,L,M)xxFF. However, we could not see any signal for PCNA-binding by PIP1 when the 430–449 sequence of hPol η was examined for interaction with PCNA by yeast two-hybrid assay (Fig. 4A). As expected, we detected strong signals for PCNA-binding with the C-terminal PIP-box sequence of hPol η (designated PIP2) and the PIP-box sequence of hPol ι under the same conditions. Furthermore, we could not see any significant increase on the PCNA-interaction even when we introduced L444F substitution to make the PIP1 more similar to the consensus sequence of PIP-box (data not shown). Nevertheless, we cannot exclude a possibility that the PIP1 of hPol η may require some additional sequence to exhibit its PCNA-binding activity. The region near the C-terminus corresponding to the PIP2 sequence is predicted to be “ordered”; however, the region is expected to be “disordered” without binding to PCNA, as discussed above for the C-terminal PIP-box sequences of Dpo4 and hPol κ.
Figure 3.
Disordered profile plots of scPol η (A) and hPol η (B).
Figure 4. Yeast two-hybrid assay for binding to PCNA and hRev1-CTD.
Each segment of hPol η, hPol ι, hPol κ or hRev1 was inserted into pLexA (BD) vector and the entire region of PCNA or a region (951–1251) of hRev1 was inserted into pB42AD vector. Amino acid sequences of the inserted segments are shown in the right, in which conserved residues are underlined and altered sequences are shown in Italic. “No” indicates empty vector. Experiments were performed as described previously (Ohashi et al., 2009).
While hPol κ (and hPol ι, as described below) has a single Rev1-interacting region (RIR), hPol η has two RIRs around FF483–484 and FF531–532 (Ohashi et al, 2009), both of which are embedded in a large unstructured region (see Fig. 3B). Each of the two RIRs independently binds to hRev1-CTD. In the case of hPol κ, FF567–568AA substitution in the RIR sequence completely abolished its hRev1-binding activity and the FF567–568AA mutant could not correct the BPDE-sensitivity of Polk-defective mouse embryonic fibroblast (MEF) cells (Ohashi et al., 2009). In contrast, the mutant of hPol η carrying both FF483–484AA and FF531–532AA substitution that lost most of the hRev1-binding activity could fully complement the UV sensitivity of XP-V cells, as the wild type did (Akagi et al., 2009). The double mutant also suppressed UV-induced mutations in XP-V cells, but it reduced spontaneous mutations only partially. These results imply that the interaction with hRev1 is dispensable for accurate bypass of UV-induced lesions by hPol η, yet it might be necessary for bypass of some DNA lesions, which probably depend on coordinated actions by multiple TLS polymerases (see below for further discussions).
Another interesting feature of hPol η is that it interacts with many other proteins. Watanabe et al. (2004) showed that Rad18 directly interacts with hPol η and help it form nuclear foci in UV-irradiated cells. The C-terminal 158 aa region of hPol η (from 556 to the C-terminus) containing the UBZ domain and the PIP2 site was sufficient for the Rad18-interaction. Furthermore, Kannouche et al. (2002) reported that hPol ι interacted mainly with the 352–595 region (and also weakly with the 595–713 region) of hPol η and concluded that hPol η is required for targeting hPolι to the replication machinery. In these two cases, the interaction sites are not precisely mapped and no mutants specifically affecting such interactions are available. The significance of these interactions will be discussed in a later section.
Pol ι subfamily, enigmatic enzymes inserting G opposite undamaged T
Pol ι subfamily proteins are present only in eukaryotes including mammals, but lacking in S. cerevisiae or S. pombe while present in Drosophila melanogoster and Neurospora crossa (for recent progresses on the in vivo functions of Pol ι, see a review by Vidal & Woodgate, 2009). Human and mouse Pol ι have been thought to contain 715 and 717 residues, respectively; however, another in-frame ATG codon was recently found to exist in the upstream of the presumed start codon in each case. Furthermore, because the extended sequences of Pol ι homologues in mammals are well conserved (Fig. 5), it seems more likely that human and mouse Pol ι proteins are longer than originally thought. Interestingly, there is heterogeneity within the extended coding region in human genomic and cDNA sequences, because of the presence of CGA repeat. The sequence with 4 CGA repeats has an N-terminal extension of 25 amino acids and that with 3 CGA repeats has the extension of 24 amino acids which is completely identical with that of chimpanzee.
Figure 5. Multiple alignment of the N-terminal regions in mammalian Pol.
ι homologues. In the upper amino acid sequence alignments, the Met residues of human and mouse Pol ι that were previously assigned as the first residue are underlined. In the lower sequences, the regions from the newly assigned Met start codon to the previously assigned one in the gene coding for the 740 or 739 aa protein are shown for human Pol ι, and are derived from the NCBI entries NM_007195 and AK301578, respectively. Many cDNA clones with the 5′-end sequence identical to either one of the two entries are found in human EST libraries at almost equal frequencies, implying that the difference is due to heterogeneity, not to sequence error. CGA repeats in the newly identified N-terminal sequences are denoted by a horizontal line with an arrow.
A unique feature of hPol ι is that the enzyme shows extremely high error rates when replicating on template pyrimidines, while showing accurate replication on template purines. For example, hPol ι misinserts G opposite T 3–10 times more frequently than the correct A. Structures of hPol ι have been studied, all as binary complexes with DNA or ternary complexes with DNA and substrate, but none as apoenzyme. Nair et al. (2004 (2005a) reported that in a ternary complex of hPol ι with DNA and an incoming substrate, the template purines (A or G) adopt a syn conformation for Hoogsteen base pairing. It explained the mechanism by which hPol ι could bypass 1,N6-ethenoadenine and N2-ethylguanine by inserting the correct T and C, respectively, because both of the adducts cannot form the normal Watson-Crick base-paring if they keep anti conformation (Nair et al., 2006a; Pence et al., 2009). While the template purines form anti conformation in the binary complex of hPol ι and DNA, the incoming substrate imposes an anti to syn conformational change because hPol ι has a constricted active site (Nair et al., 2006b). In such a constricted active site, Watson-Crick base-paring that requires the C1′-C1′ distance of 10.5 A is disfavored and Hoogsteen base-paring that requires the distance of around 8.5 A is favored. When T is the template base, it remains in an anti conformation and the selection of dGTP over dATP is partly due to the hydrogen bonding between the N2 amino of dGTP and Gln59 (the numbering of residues is followed as the old ones for the total length of 715 residues) in the finger domain of hPol ι (Kirouac & Ling, 2009; Jain et al., 2009).
The results by Kirouac & Ling (2009) explain well why hPol ι has a narrowed active site (Fig. 1G). The loop between β2 and β3 in the finger domain of hPol ι is much shorter, for example when compared with that of Dpo4. The residue Lys60 in the top of the loop interacts with Asp306 in the LF/PAD and Tyr61, together with Ser307 and Arg347 in the LF/PAD, interacts with the template DNA strand, thereby constituting a lid to the template DNA chain. The other end of the active site is defined by the three invariant acidic residues (Asp34, Asp126 and Glu127), which are essential for phosphodiester bond formation. Thus, the narrowed active site limits the C1′-C1′ distance of the replicating base pair to within 9 A in hPol ι. Furthermore, hPol ι has amino acids with relatively large side chains in the finger domain that contact the replicating base pair in the active site. Among them, Gln59, which is conserved among Pol ι subgroup proteins, forms a unique hydrogen bond with the incoming dGTP that maintains an anti form, thereby facilitating the misincorporation of dGTP opposite template T.
Since no apo form of hPol ι has been reported, we may only speculate whether hPol ι is a “convertible” or “preassembled” type, based on the structures of the binary complex with DNA. As described above, the interaction between the finger and LF/PAD domains of hPol ι appears to be very weak, mostly depending on the interaction between Lys60 in the finger domain and Asp306 in the LF/PAD. The interaction is probably facilitated by binding of Ser307 and Arg347 in the LF/PAD to template DNA chain. Since there seems no close contact between the finger domain and LF/PAD (as seen in the case of scPol η, Fig.1F), it is likely that apo form of hPol ι may have a conformation different from the DNA-bound form.
It has been thought that hPol ι does not have an N-terminal extension ahead of the catalytic domain; however, we now know that hPol ι has an N-terminal extension, which has relatively high disorder probabilities peaking around residue 40 (corresponding to the new numbering of full-length 740 residues), as shown in Fig. 6. Thus far, structures of hPol ι have been analyzed with a truncated species comprising residues 26–445 (corresponding to 1~420 of the old numbering system). Such analyses indicate that N-terminal residues 26~50 of hPol ι (1~25 of the old numbering system) are disordered. The N-terminal extension of hPol ι is shorter than the length of the N-clasp in hPol κ (70~100 residues) and very similar in length to the N-digit of hRev1 (around 40~50 residues). Therefore, hPol ι may have an N-terminal domain, similar to the N-digit of hRev1.
Figure 6. Disordered profile plot of hPol ι.
The entire 740 aa hPol ι sequence including the newly identified N-terminal 25 amino acids was analyzed for disorder probability, but the positions of the motifs and domains are presented using the old numbering system (reflecting the N-terminally-truncated 715 amino acid species). The old numbering may be converted to the new full-length sequence by addition of 25.
The PIP-box sequence of hPol ι exists immediately downstream of the catalytic domain. Until recently, the PIP-box sequence of hPol ι has been thought to be 420-KKGLIDYY-427, in which YY supposedly corresponds to FF in the consensus sequence of PIP-box (Vidal et al, 2004; Haracska et al., 2005). However, more detailed in vitro PCNA-binding assays using peptides of altered sequences and structural determination of peptide-bound PCNA revealed that 421-KGLIDYYL-428 corresponds to the PIP-box consensus sequence (Hishiki et al., 2009). The region downstream of the catalytic domain is predicted to be mostly unstructured, except for the two ubiquitin-binding motifs (UBMs). UBZs in hPol η or hPol κ were originally recognized to be zinc-finger motifs and were subsequently found to bind ubiquitin. However, UBMs in hPol ι and Rev1 were originally identified as ubiquitin-binding domains in screens for proteins that interact with ubiquitin (Ub) and a mutant form of Ub in which Ile44 (critical for binding to many proteins) was substituted to Ala (Bienko et al., 2005). UBMs are predicted to comprise two α-helices separated by the central proline residue conserved among them. The entire sequence of hPol ι has only one FF site at 547–548 and therefore the flanking sequence (540-SRGVLSFF-548) was once presumed to be a PCNA-binding site (Haracska et al., 2001). However, FF547–548AA substitution of hPol ι did not affect PCNA-binding (Vidal et al., 2004), yet completely abolished the hRev1-CTD binding (Ohashi et al., 2009) demonstrating that FF 547–548 is a Rev1-interacting region (RIR). Here again, the RIR in hPol ι is located in a region of high disorder probabilities, similar to RIRs in hPol κ and hPol η. The C-terminal 224 residues of hPol ι were shown to be required for the interaction with hPol η (Kannouche et al., 2002), but the precise interaction site has not been identified yet.
Rev1 subfamily, dCMP transferase with non-catalytic function important for in vivo TLS
Rev1 proteins, together with Pol ζ (a complex consisting of the Rev3 catalytic subunit and the Rev7 accessory subunit) and Pol η, have been found in most eukaryotic organisms whose genomes have been sequenced. Rev1 proteins are composed of three portions, the N-terminal portion containing a BRCT domain, the central catalytic domain and the C-terminal portion containing multiple motifs for interactions with other proteins. Yeast Rev1 was the first member of all the Y-family proteins that was found to have a catalytic activity, but it utilized only dCTP as substrate to insert opposite abasic sites, dU and undamaged dG (Nelson et al., 1996b). The inserted dC was extended efficiently by the yeast Pol ζ. Furthermore, a mutant (rev1-1 carrying the G193R substitution in the N-terminal BRCT domain) that retained the dCMP transferase activity was found to be defective for bypass of T-T (6-4)PP but proficient for bypass of T-T CPD (Nelson et al., 2000). Thus, Rev1 was thought to have a “second” role other than dCMP transferase, probably for coordinated action with Pol ζ that is essentially required for bypass of abasic and (6-4)PP lesions.
While all Y-family DNA polymerases have five common motifs within the catalytic domains, Rev1 subfamily proteins share one additional motif containing the SRLHH sequence at the N-terminus. The additional motif was necessary for the human Rev1 to exhibit dCMP transferase activity in vitro (Masuda et al., 2001). Structural analysis of the yeast Rev1 catalytic core in complex with DNA and incoming dCTP revealed that the template G is evicted from the DNA helix by the Leu residue in the additional motif and the adjacent residue Arg interacts with the incoming dCTP (Nair et al., 2005b). The additional sequence of Rev1 catalytic domain constitutes an extra domain called “N-digit”, which occupies a space between the palm and PAD. The Rev1 PAD has a relatively long loop, designated “G loop”, which accommodates evicted template G in preference to other template bases. Thus, the structure explains well how Rev1 incorporates dCMP opposite abasic sites.
Recently, the structure of human Rev1 catalytic core in complex with DNA and dCTP was solved (Fig. 1H) and was demonstrated to be very similar to that of yeast Rev1, with the exception of two insertions in hRev1 (Swan et al., 2009). One insert (I1, ~40 residues of the 378~417 region) in the palm domain extends away from the active site and the other insert (I2, 54 residues of the 449~504 region) in the finger domain may constitute, together with the G-loop in the PAD, a hydrophobic pocket to accommodate the evicted template G with bulky adducts at N2 position. While some portion of this I2 region is disordered in the complex with non-damaged DNA (Swan et al., 2009), it may form a defined structure when complexed with DNA containing a bulky N2 dG adduct. In fact, hRev1 was shown to bind DNA containing dG with CH2-(6-benzo[a]pyrene) at N2 position 3-fold more tightly than unmodified G-containing DNA (Choi & Guengerich, 2008). In the human Rev1-DNA-dCTP ternary complex, the G-loop in the PAD has multiple contacts with the αE helix in the I2 region of the finger domain, so as to encircle the hole through which the template DNA strand extends its chain without any bending or kink. This implies that the interaction between the finger domain and PAD occurs after DNA binding; otherwise, longer chains of the template DNA cannot penetrate into such a hole. Thus, it is expected that the apo form of hRev1 should have a different conformation from the DNA-bound form.
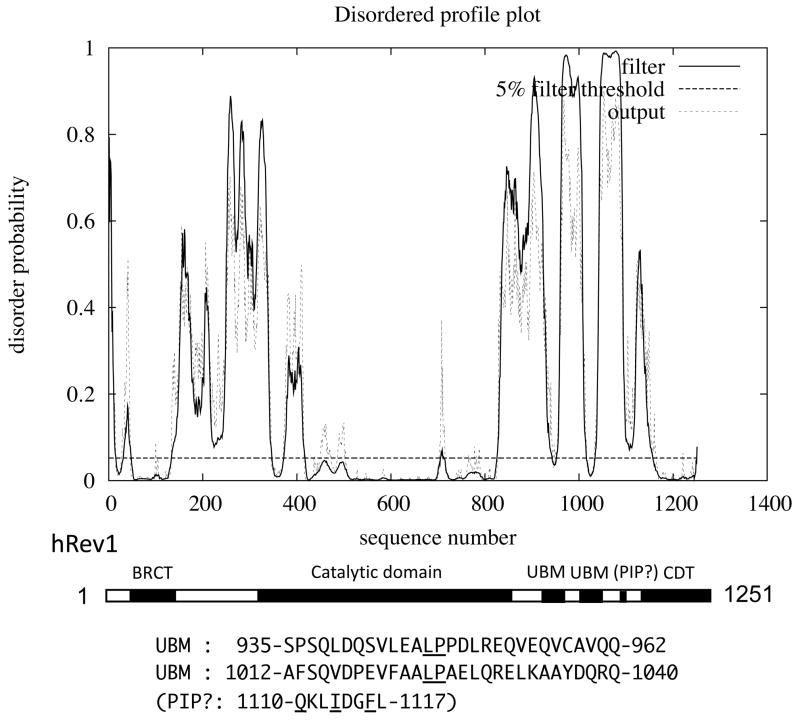
From the disordered profile plot shown in Fig. 7, hRev1 is predicted to have a mosaic structure composed of multiple ordered and disordered regions. The ordered region near the N-terminus (50~130) corresponds to the BRCT domain. The catalytic domain (340~830) is flanked with long disordered regions. The two small peaks around 400 correspond to the I1 region that was disordered in the hRev1-DNA-dCTP ternary complex (Swan et al., 2009).
Figure 7. Disordered profile plot of hRev1.
The location and sequence of a putative PIP-like sequence (1110–1117) in the C-terminal region is indicated by ‘PIP?’. However, as shown in Fig 4A, a region of Rev1 spanning the putative PIP sequence (1102–1124) showed very weak PCNA binding activity in yeast two-hybrid assay.
In the case of hRev1, a PCNA-binding site has not been definitively identified as yet. Ross et al. (2005) localized a PCNA-binding site between 923–1047 of hRev1 using a mammalian two-hybrid system. However, since two UBMs (934~962 and 1012~1040) were later found to exist within the same region, it is plausible that the putative PCNA-binding site in hRev1 might represent one or both of the UBMs. Since these workers could not detect an interaction between PCNA and hRev1 in a yeast two-hybrid assay, they suggested that the interaction might be indirect. Guo et al. (2006a) reported that mouse Rev1 (mRev1) could bind directly to PCNA, in a manner that depends on the functional BRCT domain. According to these authors, “yeast two-hybrid experiments demonstrated that the N-terminal half of mouse Rev1 protein interacts directly with PCNA (data not shown)”. Furthermore, two truncated forms of HA-tagged mRev1 containing the 1–240 or 1–413 region were pulled down with a GST-PCNA fusion protein, but not with GST. However, using yeast two-hybrid assays as shown in Fig. 4, we have not observed differences in PCNA-binding activity between the intact form of hRev1 and a truncated hRev1 lacking the N-terminal BRCT domain (our unpublished observations). As noted in a recent review by Guo et al. (2009), it cannot be excluded that the BRCT-dependent interaction between mouse Rev1 and PCNA was indirectly mediated by other protein(s) bound to the BRCT domain. In this context, it is worthy to note that some BRCT domains are known to bind to DNA rather than to proteins. Kobayashi et al. (2006) showed that the BRCT domain in the N-terminal region of human RFC1 (the largest subunit of replication factor C, a clamp loader) is involved in binding to 5′-phosphorylated double-stranded (ds) DNA. The binding required a region N-terminal to the BRCT domain that was expected to form an α-helix. These workers noted that the N-terminal region of hRev1 aligns well with the BRCT region (BRCT domain plus N-terminal region) of hRFC1 and observed that the hRev1 BRCT region bound dsDNA (cited in the above paper). Clearly, further work is necessary to test whether hRev1 BRCT motifs mediate direct or indirect interactions with PCNA.
The C-terminal region in the downstream of the catalytic domain of hRev1 is predicted to have multiple disordered regions. The two regions with low disorder probabilities located at around 950 and 1020 correspond to two UBMs (934~962 and 1012~1040). Another region with low disorder probabilities is located at 1110, where we found a sequence, 1110-QKLIDGFL-1117, which is similar to the PIP-box consensus sequence. We detected only a very weak signal for PCNA-binding activity of the sequence when the 1102–1124 sequence was examined by yeast two-hybrid assay (Fig. 4A). Moreover, we did not see any significant increase in PCNA-binding activity when we introduced L1117F substitution to change it to QKLIDGFF, a sequence perfectly matching the PIP-box consensus sequence (data not shown). It seems possible that some residues near or within the PIP-box-like sequence may negatively influence PCNA-binding.
About 90 aa near the C-terminus (1160~1251) of hRev1 is predicted to be highly ordered, probably because the region is expected to have secondary structure containing multiple α-helices. The C-terminal region of hRev1 (hRev1-CTD) was first recognized to be important for the interaction with hRev7, the non-catalytic subunit of hPol ζ (Murakumo et al., 2001). The interaction seemed to explain well how Pol ζ might function in conjunction with Rev1 for bypass of various DNA lesions. A stable 1:1 complex of hRev1 and hRev7 proteins, both of which were overproduced in E. coli cells, was purified through gel filtration, but the enzyme activity of hRev1 in the complex showed no significant difference from that of uncomplexed hRev1 (Masuda et al., 2003). Subsequently, the Rev1-CTD was found to interact also with three other Y-family polymerases (Guo et al., 2003; Ohashi et al., 2004, Tissier et al., 2004), thus suggesting that hRev1 plays a central role during in vivo TLS processes. More recently, the hRev1-CTD was found to recognize short (~10 aa) sequences containing FF, which are present in hPol κ, hPol ι and hPol η (Ohashi et al., 2009). However, all the sequences containing FF were not recognized by hRev1-CTD; for example, the C-terminal PIP-box sequences in hPol κ and hPol η did not bind to hRev1-CTD. The role of interactions between hRev1 and other Y family polymerases is discussed in more detail in a later section.
Jansen and colleagues generated mutant mice containing a defined deletion of the BRCT domain in the Rev1 gene or a completely defective Rev1 gene (denoted as Rev1B/B and Rev1−/−, respectively) (Jansen et al., 2005, 2006). While Rev1B/B mice were healthy and displayed normal somatic hypermutation of immunoglobulin (Ig) genes, Rev1−/− mice showed a transient growth retardation and strand-biased defect in C-to-G transversions in Ig genes, which most likely involved the Rev1-mediated dCMP incorporation opposite abasic lesions. Immortalized MEF lines from such mice were established and Rev1B/B cells showed a milder UV-sensitivity than Rev1−/− cells, implicating multiple roles of Rev1 in intracellular TLS processes (Jansen et al., 2009). Rev1-disrupted mutant of the chicken lymphocyte cell line DT40 also showed high sensitivities to UV and cisplatin (Okada et al., 2005). When various constructs carrying the intact or mutant form of the human REV1 gene were expressed in the rev1 mutant of DT40, the full-length form (1–1251), a N-terminal deletion mutant (333–1251) lacking the BRCT domain and a catalytic mutant (D570A/E571A) fully corrected the sensitivities to UV and cisplatin, but a C-terminal deletion mutant (1–827) or a mutant carrying only the C-terminal portion (923–1251) did not (Ross et al., 2005). The results indicated that the C-terminal portion of Rev1 is necessary for effective tolerance of DNA damage in DT40 cells, but requiring some additional function(s) encoded by the central portion (other than the catalytic function), while the N-terminal portion including the BRCT domain was not essential for the tolerance mechanism.
3) Functional significance of protein-protein interactions involving TLS DNA polymerases
PCNA-binding and formation of nuclear foci by TLS DNA polymerases in genotoxin-treated and –untreated cells
PCNA interacts with a number of proteins involved in replication, repair, cell cycle and other functions (for a recent review, see Moldovan et al., 2007). Most of those proteins have a conserved sequence, PIP-box (Warbrick, 1998), which is often presented as Qxx(M,L,I)xxFF. The p66 subunit of human Pol δ has the sequence 456-QVSITGFFQRK-466 near the C-terminus (K466 is the last residue of the protein). However, neither hPol κ, hPol ι nor hPol η has a sequence completely matching the canonical sequence. Thus the conserved Gln is replaced with Met in hPol η’s PIP2 or Lys in hPol ι and hPol κ’s PIP2. Nevertheless, each of the Y-family polymerase PIP-box sequences gives a clearly positive signal for sequence-specific interactions with PCNA when examined by yeast two-hybrid assay (see Fig. 4A). To evaluate relative affinities of the non-canonical PIP-box sequences for PCNA, synthetic peptides containing each of PIP-box sequences were examined for quantitative measurements of binding to unmodified PCNA by surface plasmon resonance (SPR) (Hishiki et al., 2009). As summarized in Table 1, the results indicate that the PIP-box sequences of hPolη (PIP2) and hPol ι have a similar level of affinity for PCNA with estimated Kd (dissociation constant) values of 0.40 and 0.39 μM, respectively, and that the PIP2 of hPol κ has a much lower affinity. While the wild-type PIP2 of hPol κ showed a clearly positive signal for PCNA interaction by yeast two-hybrid assay (data not shown), significant levels of PCNA binding to the PIP2 of hPol κ by SPR were detected only with an extension of the sequence PLTH at the C-terminus to make the sequence similar to the PIP2 of hPol η. In SPR assays, the variant of hPol η’s PIP2 lacking the C-terminal PLTH sequence did not show any signal for PCNA-binding (Hishiki et al., 2009). However, the data shown in Fig. 4A clearly indicates that the PLTH-deleted sequence still retains PCNA-binding activity, demonstrating that yeast two-hybrid assay is more sensitive for detecting PCNA-interaction than SPR.
Table 1.
Estimated Kd values for binding to PCNA, Ubiquitin and hRev1-CTD
| PCNAa) | Ubiquitin | hRev1-CTDb) | |
|---|---|---|---|
| hPol η | 0.40 (PIP2) | 81 c) | 13 |
| hPol ι | 0.39 | 180 d) | 69 |
| hPol κ | 4.5 (PIP2+PLTH) | 38±2 e) | 7.6 |
(all in μM)
All obtained by SPR (Hishiki et al., 2009).
All obtained by SPR (Ohashi et al., 2009)
Obtained by NMR (Bomar et al., 2007).
Obtained by NMR (Bienko et al., 2005).
Kd value for UBZ of hPol κ is not available. The value obtained for the UBZ of Rad8 by SPR (Crosetto et al., 2008) is shown. The UBZ domain sequence of Rad18 is very similar to that of hPol κ.
Judging from Kd values obtained by SPR assays, the affinities of the PIP-box sequences for PCNA seem to be much stronger than those of ubiquitin-binding domains (UBZ in hPol η and hPol κ or UBM in hPol ι and Rev1) for ubiquitin (see Table 1). For example, the Kd value in the interaction between free ubiquitin and the UBZ of hPol η was estimated to be around 81 μM by isothermal titration calorimetry (ITC) (Bomar et al., 2007). Therefore, it seems likely that the PIP2 of hPolη contributes to the interactions between hPol η and mono-ubiquitinated PCNA (mUb-PCNA) more significantly than does the UBZ, thereby determining the binding specificity. For evaluation of the affinities for homotrimeric PCNA, it is desirable to re-estimate Kd values by methods other than SPR, for example, ITC. Structural analyses of the PCNA bound to the peptide containing each of the PIP-box sequences provided reasonable explanations about why the non-canonical PIP-box sequences have lower affinities than that of the canonical PIP-box sequence (Hishiki et al., 2009).
Much of the information on in vivo functions of PIP-box and UBZ or UBM domains has been obtained from nuclear focus formation assays in genotoxin-treated cultured cells. Kannouche et al. (2001) described that GFP-fused form of hPol η (GFP-hPolη) formed nuclear foci in an approximately 10~15% of MRC5 cells without any DNA-damaging treatment and that the frequency of the cells with such foci increased up to 80% after UV-irradiation. Because the proportion of the cells with GFP-hPol η foci in non-damaged cells corresponded to that of the cells in S-phase and also because such foci co-localized with PCNA, they suggested that hPol η is associated with replication factories during S-phase in non-damaged cells. Subsequently, Kannouche et al. (2004) found that PCNA becomes mono-ubiquitinated in a variety of human cells following UV irradiation, similar to previous findings in yeast (Hoedge et al., 2002), and described that hPol η interacted specifically with mUb-PCNA, but not unmodified PCNA in vivo. Furthermore, Bienko et al. (2005) showed that both UBZ and PIP motifs of hPol η were essential for the nuclear foci formation in UV-irradiated cells. However, it remains unanswered how hPol η formed nuclear foci co-localizing with PCNA (presumably unmodified) in non-damaged cells. A similar study for hPol κ showed that the frequency of the cells with nuclear foci of GFP-hPol κ was around 5% of MRC5 cells expressing the fusion proteins under non-damaged conditions and it increased up to at most 23 % of the cells (Ogi et al., 2005). As the C-terminal 119 residues of hPol η containing UBZ, NLS (nuclear localization signal) and PIP were required for nuclear foci formation (Kannouche et al., 2001), the C-terminal 97 residues of hPol κ containing one of the two UBZs, NLS and PIP were necessary and sufficient for foci formation induced by DNA damages. Similarly, nuclear foci formation of hPol ι in DNA-damaged cells required the intact PIP (Vidal et al, 2004) and at least one of the two UBMs (Bienko et al., 2005). These results are consistent with the notion that formation of hPol η, hPol κ or hPol ι nuclear foci in DNA-damaged cells depends on both PIP and UBZ or UBM, probably for their stable binding to mUb-PCNA.
However, results of nuclear focus formation assays should be interpreted with caution, since the nature of nuclear foci is poorly understood. For instance, we do not know how many molecules of a protein in question must accumulate to be detected as foci or how many other different proteins are present in the same foci. Even if two proteins (e.g., hPol η and hPol ι) co-localize in foci, the two proteins are not necessarily interacting directly with each other in such foci. Potentially, co-localization may occur through binding to a common binding partner (e.g., PCNA). The results obtained by Gueranger et al. (2008) suggested that TLS by hPol η might occur without the accumulation of microscopically visible foci. These workers isolated a mutant of the Burkitt’s lymphoma cell line BL2 by inactivating the gene coding for hPol η and observed that the hPol η-deficient mutant exhibited a UV-sensitive phenotype. The UV-sensitive phenotype was fully restored by transfection of an EGFP expression vector carrying the wild-type hPol η, but also by expressing a mutant of hPol η with the alteration of the C-terminal PIP-box sequence (701-MQTLESFF-708) to MATAESAA that did not form detectable nuclear foci. The result implied that the functional complementation and accumulation at nuclear foci are separable, but it does not rule out a possibility that hPol η has another PIP-box in addition to the C-terminal one. Acharya et al. (2008) noted the presence of an additional PIP-like sequence 437-STDITSFL-444 (named PIP1, and the C-terminal one was named PIP2), just C-terminal to the PAD (at a very similar position to that of the PIP-box in hPol ι). Their results showed that the PIP1 mutant (F443A, L444A) or the PIP2 mutant (F707A, F708A) conferred intermediate levels of UV-resistance to XP-V cells and the PIP1 PIP2 double mutant was completely defective in imparting UV-resistance to XP-V cells. They concluded that hPol η has two PCNA-binding PIP domains that can functionally substitute for one another. We are interested in comparing relative strength in PCNA-binding activity between the PIP1 and PIP2 of hPol η and examined both sequences for PCNA-binding by yeast two-hybrid and SPR assays. As shown in Fig. 4A, we could not detect any signal of PIP1 for PCNA interaction in yeast two-hybrid assay while we could observed a very strong signal for PCNA interaction with PIP2. It seems that PCNA-binding activity of the PIP1 sequence is much weaker than that of the PIP2 sequence, while a possibility that the PIP1 requires some additional sequence for exhibiting its PCNA-binding activity cannot be ruled out.
Acharya et al. (2008) examined nuclear foci formation by co-expressing GFP-PCNA and FLAG-hPol η in MRC5 cells and observed that the D652A mutation in the UBZ domain, which inactivated ubiquitin-binding of hPol η (Bienko et al., 2005), still retained the ability to form nuclear foci that co-localized with PCNA. However, Sabbioneda et al. (2009) argued against the interpretation, by showing that the eGFP-hPol η carrying the D652A mutation failed to form foci without over-expression of PCNA. Thus, requirements for nuclear foci formation are variable depending on experimental conditions examined.
The question of what elements are required for nuclear foci formation of hRev1 is also under debates. Tissier et al. (2004) reported that when expressed as a fusion with YFP, each of N-terminal half (1–730) and C-terminal half (730–1251) of hRev1 formed nuclear foci in UV-irradiated cells, whereas Murakumo et al. (2006) reported that nuclear foci formation of GFP-hRev1 in UV-irradiated was observed with the C-terminal region (e.g., 826–1251), but not with the N-terminal region (e.g., 1–825). Furthermore, Guo et al. (2006a) reported that the intact BRCT domain was required for foci formation of GFP-mRev1 in non-damaged cells, but not UV-irradiated cells. They also showed that UBMs are required for increased level of foci formation of GFP-mRev1 in UV-irradiated cells (Guo et al., 2006b). If we assume that the C-terminal region of hRev1 has a weak PCNA-binding site as well as two UBMs, we can interpret the above results as indicating that requirements for nuclear foci formation of the C-terminal region of hRev1 in DNA-damaged cells are similar to those of hPol η, hPol κ and hPol ι.
Also, it should be noted that all the above results were obtained with ectopically expressed human or mouse Rev1, but not with the endogenous level of the Rev1 proteins. Akagi et al. (2009) studied accumulation of the endogenous hRev1 into locally UV-irradiated areas of nuclei, using antibody with high affinity for hRev1, because nuclear foci of the endogenous hRev1 were not clearly observed even after UV-irradiation. Importantly, they observed the accumulations of the endogenous hRev1 into locally UV-irradiated areas of nuclei in the cells expressing hPol η, but not in hPolη-deficient XP-V cells. This makes a sharp contrast to the observation by Tissier et al. (2004) that GFP-hRev1 formed nuclear foci in UV-irradiated XP-V cells. Furthermore, when XP-V cells were reconstituted with wild-type hPol η or its mutant defective for the hRev1-interaction, UV-sensitivity of the XP-V cells was corrected by either the wild type or the mutant, yet the accumulation of the endogenous hRev1 into UV-irradiated areas was observed with the wild type, but not with the mutant. These results indicate that accumulation of the endogenous hRev1 to UV-irradiated areas depends on the interaction with hPol η, while nuclear foci formation of ectopically (over)expressed hRev1 occurred independently of hPol η. Thus, there are two different pathways for targeting hRev1 to sites of DNA damage, hPol η-dependent and -independent ones. The latter becomes more easily detectable when ectopically (over)expressed.
Mechanism and biological significance of Rev1-TLS polymerase interactions
Sequences as short as 10 aa residues in hPol κ, hPol ι and hPol η are sufficient for mediating interaction with hRev1-CTD (Ohashi et al., 2009). Thus far, only four such sequences have been identified, all of which contain two consecutive phenylalanines (FF) as critical residues. As the PIP-box consensus sequence is often denoted as Qxx(I,L,M)xxFF, many PIP-box sequences contain FF. However, none of the four RIR sequences binds to PCNA and the PIP-box of hPol ι did not bind to hRev1-CTD (Fig. 4B). The PIP-box sequence containing FF at the C-terminus of hPol η or hPol κ did not bind to hRev1-CTD (Ohashi et al., 2009). In PIP-box sequences, several residues in the N-terminal side of FF are required for interaction with multiple residues in PCNA and adopting a 310 helical structure, while residues C-terminal to FF are not essential although they contribute to stabilizing the interaction with PCNA. By contrast, in RIR sequences, no conserved amino acid is present in either N-terminus or C-terminus to FF; however, the presence of several sequences in the C-terminal side of FF is essential for binding to hRev1-CTD. Proline substitution of the four residues C-terminal to FF abrogated the hRev1-CTD interaction, while alanine substitution did not. Thus, the consensus sequence for RIR is denoted as xxxFFyyyy. For evaluating relative affinities of the RIRs in hPol η, hPol ι and hPol κ, synthetic peptides carrying each of the RIR sequences were examined for binding to His-hRev1(1130–1251) by SPR. As summarized in Table 1, the RIR of hPol κ showed higher affinity than that of hPol η or hPol ι. The hRev1-CTD is known to bind to hRev7, which contains no FF sequence in the entire sequence of 211 residues. Moreover, a longer region (>150 residues) of hRev7 is required for binding to hRev1-CTD (Murakumo et al., 2001), while hPol η, hPol ι and hPol κ interact with hRev1-CTD via short RIR sequences. It therefore seems likely that hRev1-CTD has two different interfaces for interactions with other proteins, one recognizing short RIR sequence in hPol η, hPol ι and hPol κ and the other recognizing conformation of hRev7. How hRev1-CTD recognizes the RIR sequences is an intriguing question, but has not been solved as yet because of the difficulty in purifying the hRev1-CTD at large scales for structural analysis.
The Saccharomyces cerevisiae (sc) proteins involved in TLS are known to show physical interactions, while some differences exist in such interactions among yeast and human proteins. The scRev7 protein interacts mainly with the PAD domain of scRev1 (Acharya et al., 2005), also interacting weakly with the CTD and BRCT domains (D’Souza & Walker, 2006). The PAD of scRev1 interacts with a C-terminal region of scPol η (Acharya et al., 2007) and the scRev1-CTD interacted with a central region of scREV3 (Acharya et al., 2006). More recently, interactions among the Y-family proteins in the fruit fly Drosophila melanogaster (dm) were reported (Kosarek et al., 2008). The C-terminal 117 amino acids of dmRev1 were necessary and sufficient for an interaction with dmPol η, but a region adjacent to the C-terminus of dmRev1 was required for its interaction with dmPol ι. Interestingly, dmPol η, but not dmPol ι, interacted with the C-terminal region (~100 residues) of mRev1. Since the C-terminal sequences of the human and mouse Rev1 proteins are well conserved with 95% identity (Masuda et al., 2002), the mRev1-CTD is expected to recognize the same RIR sequences as the hRev1-CTD. While dmPol ι has no FF in the entire sequence, dmPol η has 5 sites containing FF (26–27, 500–501, 564–565, 786–787, 880–881). A mutant of dmPol η with FF26–27AA and FF564–565AA substitutions was found to have lost most of the dmRev1-interacting activity (J. Tomida and T. Todo, personal communication). This suggests that the dmRev1-CTD, which is distantly related to the hRev1- and mRev1-CTDs (24% identity), recognizes sequences containing FF, while at the present it is not explainable why three other sites containing FF do not interact with dmRev1-CTD. In any case, interactions of Rev1 with other TLS polymerases appear to be conserved in eukaryotes with some variations.
The most important unanswered question about Rev1 concerns its central role during the intracellular TLS processes. As described above, a hPol κ mutant defective for interaction with hRev1 could not correct the BPDE-sensitivity of Polk-defective mouse embryonic fibroblast (MEF) cells (Ohashi et al., 2009), implying that the hRev1-interaction is essential for hPol κ to execute its function in vivo. In contrast, a similar mutant of hPol η defective for hRev1-interaction corrected the two phenotypes of XP-V cells, namely, UV-sensitivity and elevated mutation rates by UV-irradiation as efficiently as the wild type (Akagi et al., 2009). Nevertheless, the hPol η mutant suppressed only partially another phenotype of XP-V cells, that is, a higher incidence of spontaneous mutations, while the wild type suppressed the phenotype completely. This suggested that, while the Rev1 interaction is dispensable for hPol η to carry out accurate TLS of UV-induced lesions such as CPDs, it contributes to suppression of spontaneous mutations, probably by promoting accurate TLS past endogenous DNA lesions such as those generated by oxidative stress. While no data are available at the present to infer whether or not the Rev1-interaction is essential for hPol ι function, hPol ι is known to interact with hPol η and re-localization of GFP-hPol ι in foci after UV-irradiation was shown to depend on hPol η (Kannouche et al., 2002). It is perhaps surprising that Pol ι recruitment is Pol η-dependent since hPol ι has a PIP-box with affinity for PCNA similar to that of hPol η’s PIP2 (Hishiki et al., 2009) and two UBMs for binding to mUb-PCNA (Bienko et al., 2005). These results cannot be explained by a model which presumes that TLS polymerases are recruited to DNA-damaged sites randomly on a simple “try and error” basis.
We therefore proposed a “sequential recruitment” model to explain the mechanism by which TLS polymerases are recruited to stalled replication forks at sites of DNA damage (Barkley et al., 2007). When the replication fork encounters a site of DNA damage, uncoupling between the preceding DNA helicase and the stalled replicative DNA polymerase generates single-stranded (ss) regions on the template DNA, to which RPA (a heterotrimeric protein with ssDNA-binding activity) binds. The RPA-coated ssDNA region then recruits the Rad6 (an E2 ubiquitin-conjugating enzyme)-Rad18 (an E3 ubiquitin ligase) complex via the interaction between RPA and Rad18 (Davies et al., 2008; Tsuji et al., 2008) to mediate mono-ubiquitination of PCNA in the stalled replication fork, which facilitates transfer of hPol η from the Rad6-Rad18 complex to mUb-PCNA. Preferential recruitment of hPol η to all stalled forks could be advantageous for cells because hPol η is a versatile enzyme capable of bypassing many different DNA lesions correctly in many cases (Masutani et al, 2000). However, hPol η forms nuclear foci in response to various DNA-damaging treatments, including BPDE that mainly generates N2-BPDE-dG adducts which the enzyme cannot bypass (Bi et al., 2005). Therefore, the hPol η recruited in response to BPDE lesions needs to be replaced with hPol κ that is able to bypass N2-BPDE-dG adducts correctly (Ogi et al., 2002). Such a polymerase switching might be dependent on hRev1, which interacts with both hPol η and hPol κ. A similar scenario can be envisaged for other lesions to explain the exchange between two TLS polymerases, one for inserting a nucleotide opposite a lesion and the other for extending further. For example, at the site of T-T (6-4)PP, hPolη inserts one of the 4XMPs opposite 3′-T, but does not extend further (Masutani et al., 1999a). If hPol η is replaced with hPol ι before inserting any nucleotide opposite the 3′-T (either via direct hPolη-hPolι interactions, or through interactions of hPolι and hPolη with hRev1), hPol ι can insert the correct A opposite the 3′-T without extending further. In both cases, hPol ζ is believed to carry out the extension reaction, after being recruited most probably through the interaction between hRev1 and hRev7, the non-catalytic subunit of hPol ζ.
Another model was proposed, which postulates that TLS enzymes might be recruited by two different mechanisms, depending on the coding capacity of the damaged nucleotide at the template strand (Jansen et al., 2007). First, damaged nucleotides retaining good coding capacity (e.g., CPD) may be readily bypassed by hPolη, without conferring a significant replication arrest. Second, forms of damage causing severe distortion of the DNA structure and/or having poor coding capacity [e.g., (6-4)PP] activates a more elaborate pathway that requires functioning of multiple TLS polymerases for the insertion and extension steps separately, re-priming of replication downstream of the lesion and activation of DNA damage signaling which involves ATR and an alternative clamp, the Rad9-Rad1-Hus1 (9-1-1) complex. Further according to this model, the 9-1-1 clamp binds to the re-primed 5′ terminus and recruits the hRev1/hPol ζ complex, which then moves up to the stalled 3′ terminus by the unique activity of hRev1 to bind to stretches of ssDNA and then translocate to 3′ primer-template junctions (Masuda and Kamiya, 2006). The authors also consider that hPol η is the default primary TLS polymerase recruited to a stalled fork and that hRev1 may dislodge the defunct hPol η from the stalled 3′ terminus. It may be also possible that hRev1 is recruited to the re-primed 5′ terminus, either as the hRev1/Pol ζ complex via a presumed interaction between hRev7 and the 9-1-1 complex or by itself through binding of its N-terminal BRCT domain to 5′ recessed end of dsDNA (as discussed earlier).
Thus far, there are two reports on physical interactions between TLS proteins and the 9-1-1 complex, both from studies on yeasts; in S. cerevisiae, Rev7 binds to the Hus1 and Rad9 orthologs (Sabbioneda et al., 2005) and in S. pombe, DinB/Pol κ interacts with the 9-1-1 complex, especially with Hus1 (Kai & Wang, 2003). However, at the moment, it is not known whether such interactions occur among the mammalian counterparts. Very recently, the structure of the human 9-1-1 complex has been determined (Dore et al., 2009; Xu et al., 2009; Sohn & Cho, 2009), which revealed that the 9-1-1 complex has a structure very similar to that of PCNA. It should be very intriguing to examine whether PIP-box of any human TLS polymerase (and hREV7) can bind to the human 9-1-1 complex or its subunit and if it does bind, how strongly it binds in comparison with PCNA-binding.
As pertains to the two different pathways for recruitment of TLS proteins to sites of DNA damage, one may argue that the replication block should be more severe when the replication fork encounters a blocking DNA damage present on the leading strand template than on the lagging strand template, because re-priming occurs constantly for the lagging strand synthesis of a new Okazaki fragment, even in the absence of DNA damage. Therefore, it may be plausible that different pathways for TLS may become activated, depending on which strand a blocking lesion is present. In contrast to the models described above, Edmunds et al. (2008) proposed a different model in which Rev1 functions for TLS at a stalled replication fork independently of PCNA ubiquitination and that Rad18-dependent PCNA ubiquitination controls TLS at a post-replicative gap filling. These authors proposed that Rev1 might be recruited to the stalled fork via interaction with the replicative DNA polymerase and/or accessory proteins and then recruits another TLS polymerase, such as Pol η. In this connection, it is noteworthy that scRev1 protein interacts via its PAD with the Pol32 protein, which is the non-essential subunit of scPol δ involved in mutagenesis (Acharya et al., 2009). Previous site-specific mutagenesis experiments using plasmid DNA containing an abasic site, T-T CPD or T-T (6-4)PP demonstrated that Pol32 is required for the bypass of abasic sites and T-T (6-4)PP in a manner that is dependent on scRev1 and scPol ζ, but not for the bypass of T-T CPD which is scPol η–dependent (Gibbs et al., 2005). In contrast, the rev6-1 mutation corresponding to the G178S substitution in PCNA abolishes the bypass of all three lesions (Zhang et al., 2006). Because Pol32 binds to the scRev1-scPol ζ complex via its central region, binding to the Pol31 subunit of scPol δ via its N-terminal region and to PCNA via the C-terminal PIP-box (Acharya et al., 2009), it follows that the scRev1-scPol ζ complex can bind to the scPol δ bound to PCNA in the replisome. However, even if such an interaction between Pol δ and the Rev1/Pol ζ occurs in human cells, it does not explain why re-localization of the endogenous hRev1 after DNA damaging is dependent on hPol η.
Interactions with other proteins and post-translational modifications
There are many reports describing that the activities of hPol η and other TLS polymerases are stimulated by the addition of various proteins such as Msh2 (Wilson et al. 2005), or WRN (Werner syndrome protein, Kamath-Loeb et al., 2007) and Ctf18-Replication factor C complex (Shiomi et al., 2007). However, since no mutant specifically affecting such interactions is available at the present, we cannot speculate on the biological relevance of such interactions and we must wait for more detailed results of further experimentations. Rather, we’d like to point out that disordered regions of a TLS protein or any given protein have much more potentials for interaction with other proteins than ordered regions, in which most of the sequences in ordered regions are employed for the maintenance of specific secondary structures and only limited sequences of them are available for interactions with other proteins. Interactions of short motif sequences in disordered regions with other proteins may be mostly weak and transient ones, involving dynamic exchanges of binding partners. Such interactions should be important in vivo during TLS because TLS is an inherently transient process, acting only during the period when replicative DNA polymerases are stalled. It is also known that unstructured regions are susceptible to various post-translational modifications, such as phosphorylaiton (Fink, 2005). A recent paper suggested that hPolη might be phosphorylated at S587 and T617, both of which are located in the C-terminal unstructured region (Chen et al., 2008). It is likely that future studies will identify additional post-translational modifications of Y-family polymerases as important regulatory mechanisms for TLS.
Acknowledgments
We thank Katsuhiko Sasaki for his excellent technical assistance and also Junya Tomida and Takeshi Todo for permitting us to cite their unpublished results. Our works described and cited here were supported by grants-in-aids (17013041 to H. O.) from the Ministry of Education, Sports, Science and technology of Japan and by grants (ES09558 to C. V.) from the National Institutes of Health, USA.
References
- Acharya N, Haracska L, Johnson RE, Unk I, Prakash S, Prakash LL. Complex formation of yeast Rev1 and Rev7 proteins: a novel role for the polymerase-associated domain. Mol Cell Biol. 2005;25:9734–9740. doi: 10.1128/MCB.25.21.9734-9740.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya N, Johnson RE, Prakash S, Prakash LL. Complex formation with Rev1 enhances the proficiency of Saccharomyces cerevisiae DNA polymerase ζ for mismatch extension and for extension opposite from DNA lesions. Mol Cell Biol. 2006;26:9555–9563. doi: 10.1128/MCB.01671-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya N, Haracska L, Prakash S, Prakash LL. Complex formation of yeast Rev1 with DNA polymerase η. Mol Cell Biol. 2007;27:8401–8408. doi: 10.1128/MCB.01478-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya N, Yoon JH, Gali H, Unk I, Haracska L, Johnson RE, Hurwitz J, Prakash L, Prakash S. Roles of PCNA-binding and ubiquitin-binding domains in human DNA polymerase η in translesion DNA synthesis. Proc Natl Acad Sci USA. 2008;105:17724–17729. doi: 10.1073/pnas.0809844105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya N, Johnson RE, Pages V, Prakash S, Prakash LL. Yeast Rev1 protein promotes complex formation of DNA polymerase ζ with P32 subunit of DNA polymerase δ. Proc Natl Acad Sci USA. 2009;106:9631–9636. doi: 10.1073/pnas.0902175106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagi J, Masutani C, Kataoka Y, Kan T, Ohashi E, Mori T, Ohmori H, Hanaoka FF. Interaction with DNA polymerase η is required for nuclear accumulation of REV1 and suppression of spontaneous mutations in human cells. DNA Repair. 2009;8:585–599. doi: 10.1016/j.dnarep.2008.12.006. [DOI] [PubMed] [Google Scholar]
- Alt A, Lammens K, Chiocchini C, Lammens A, Pieck JC, Kuch D, Hopner KP, Carell T. Bypass of DNA lesions generated during anticancer treatment with cisplatin by DNA polymerase η. Science. 2007;318:967–970. doi: 10.1126/science.1148242. [DOI] [PubMed] [Google Scholar]
- Auffret van der Kemp P, de Padula M, Burguiere-Slezak G, Ulrich HD, Boiteux S. PCNA monoubiquitylation and DNA polymerase η ubiquitin-binding domain are required to prevent 8-oxoguanine-induced mutagenesis in Saccharomyces cerevisiae. Nucleic Acids Res. 2009;37:2549–2559. doi: 10.1093/nar/gkp105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkley LR, Ohmori H, Vaziri C. Integrating S-phase checkpoint signaling with trans-lesion synthesis of bulky DNA adducts. Cell Biochem Biophys. 2007;47:392–408. doi: 10.1007/s12013-007-0032-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X, Slater DM, Ohmori H, Vaziri C. DNA polymerase κ is specifically required for recovery from the benzo[a]pyrene-dihydrodiol epoxide (BPDE)-induced S-phase checkpoint. J Biol Chem. 2005;280:22343–22355. doi: 10.1074/jbc.M501562200. [DOI] [PubMed] [Google Scholar]
- Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, Hofmann K, Dikic I. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- Bomar MG, Pai MT, Tzeng SR, Li SSC, Zhou P. Structure of the ubiquitin-binding zinc finger domain of human DNA Y-polymerase η. EMBO Reports. 2007;8:247–251. doi: 10.1038/sj.embor.7400901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq F, Kokoska RJ, Plosky BS, Vaisman A, Ling H, Kunkel TA, Yang W, Woodgate R. Investigating the role of the little finger domain of Y-family DNA polymerases in low fidelity synthesis and translesion replication. J Biol Chem. 2004;279:32932–32940. doi: 10.1074/jbc.M405249200. [DOI] [PubMed] [Google Scholar]
- Carpio RVD, Silverstein TD, Lone S, Swan MK, Choudhury JR, Johnson RE, Prakash S, Prakash L, Aggarwal AK. Structure of human DNA polymerase κ inserting dATP opposite an 8-oxoG DNA lesion. PLoS One. 2009;4:e5766. doi: 10.1371/journal.pone.0005766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-w, Cleaver JE, Hatahet Z, Honkamen RE, Chang JY, Yen Y, Chou K-m. Human DNA polymerase h activity and translocation is regulated by phosphorylaiton. Proc Natl Acad Sci USA. 2008;105:16578–16583. doi: 10.1073/pnas.0808589105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JY, Angel KC, Guengerich FP. Translesion synthesis across bulky N2-alkyl guanine DNA adducts by human DNA polymerase κ. J Biol Chem. 2006;281:21062–21072. doi: 10.1074/jbc.M602246200. [DOI] [PubMed] [Google Scholar]
- Choi JY, Guengerich FP. Kinetic analysis of translesion synthesis opposite bulky N2- and O6-alkylguanine DNA adducts by human DNA polymerase REV1. J Biol Chem. 2008;283:23645–23655. doi: 10.1074/jbc.M801686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosetto N, Bienko M, Hibbert RG, Perica T, Ambrogio C, Kensche T, Hofmann K, Sixma TK, Dikic I. Human Wrnip1 is localized in replication factories in a ubiquitin-binding zinc finger dependent manner. J Biol Chem. 2008;283:35173–35185. doi: 10.1074/jbc.M803219200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies AA, Huttner D, Daigaku Y, Chen S, Ulrich HD. Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein A. Mol Cell. 2008;29:625–636. doi: 10.1016/j.molcel.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Padula M, Slezak G, Auffret van Der Kemp P, Boiteux S. The post-replication repair RAD18 and RAD6 genes are involved in the prevention of spontaneous mutations caused by 7,8-dihydro-8-oxoguanine in Saccharomyces cerevisiae. Nucleic Acids Res. 2004;23:5003–5010. doi: 10.1093/nar/gkh831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dore AS, Kilkenny M, Rzechorzek NJ, Pearl LH. Crystal structure of the human Rad9-Rad1-Hus1 DNA damage checkpoint complex-Implications for clamp loading and regulation. Mol Cell. 2009;34:735–745. doi: 10.1016/j.molcel.2009.04.027. [DOI] [PubMed] [Google Scholar]
- D’Souza S, Walker GC. Novel role for the C terminus of Saccharomyces cerevisiae mediating protein-protein interactions. Mol Cell Biol. 2006;26:8173–8182. doi: 10.1128/MCB.00202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr Opi Struc Biol. 2008;18:756–764. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Edmunds CE, Simpson LJ, Sale JE. PCNA ubiquitination and REV1 define temporally distinct mechanisms for controlling translesion synthesis in the avian cell line DT40. Mol Cell. 2008;30:519–529. doi: 10.1016/j.molcel.2008.03.024. [DOI] [PubMed] [Google Scholar]
- Fink AL. Natively unfolded proteins. Curr Opi Struc Bol. 2005;15:35–41. doi: 10.1016/j.sbi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Shultz RA, Ellenberger T. DNA repair and mutagenesis. 2. ASM Press; Washington, D. C.: 2006. [Google Scholar]
- Gibbs PEM, Wang XD, Li Z, McManus TP, McGregor WG, Lawrence CW, Maher VM. The function of the human homolog of Saccharomyces cerevisiae REV1 is required for mutagenesis induced by UV light. Proc Natl Acad Sci USA. 2000;97:4186–4191. doi: 10.1073/pnas.97.8.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs PEM, McDonald J, Woodgate R, Lawrence CW. The relative roles in vivo of Saccharomyces cerevisiae Pol η, Pol ζ, Rev1 protein and Pol32 in the bypass and mutation induction of an abasic site, T-T (6–4) photoadduct and T-T cis-syn cyclobutane dimer. Genetics. 2005;169:575–582. doi: 10.1534/genetics.104.034611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueranger Q, Stary A, Aoufouchi S, Fali A, Sarasin A, Reynaud CA, Weill JC. Role of DNA polymerases η, ι and ζ in UV resistance and UV-induced mutagenesis in a human cell line. DNA Repair. 2008;7:1551–1562. doi: 10.1016/j.dnarep.2008.05.012. [DOI] [PubMed] [Google Scholar]
- Guo C, Fischhaber PL, Luk-Paszyc MJ, Masuda Y, Zhou J, Kamiya K, Kisker C, Friedberg EC. Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. EMBO J. 2003;22:6621–6630. doi: 10.1093/emboj/cdg626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Sonoda E, Tang TS, Parker JL, Bielen AB, Takeda S, Ulrich HD, Friedberg EC. REV1 protein interacts with PCNA: significance of the REV1 BRCT domain in vitro and in vivo. Mol Cell. 2006a;23:265–271. doi: 10.1016/j.molcel.2006.05.038. [DOI] [PubMed] [Google Scholar]
- Guo C, Tang TS, Bienko M, Parker JL, Bielen AB, Sonoda E, Takeda S, Ulrich HD, Dikic I, Friedberg EC. Ubiquitin-binding motifs in REV1 protein are required for its role in the tolerance of DNA damage. Mol Cell Biol. 2006b;26:8892–8900. doi: 10.1128/MCB.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Kosarek-Stancel JN, Tang TS, Friedberg EC. Y-family DNA polymerases in mammalian cells. Cell Mol Life Sci. 2009;66:2363–2381. doi: 10.1007/s00018-009-0024-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Yu SL, Johnson RE, Prakash L, Prakash S. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase η. Nat Genet. 2000;25:458–461. doi: 10.1038/78169. [DOI] [PubMed] [Google Scholar]
- Haracska L, Johnson RE, Unk I, Phillips BB, Hurwitz J, Prakash L, Prakash S. Targeting of human DNA polymerase ι to the replication machinery via interaction with PCNA. Proc Natl Acad Sci USA. 2001;98:14256–14261. doi: 10.1073/pnas.261560798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Prakash S, Prakash L. Role of human DNA polymerase κ as an extender in translesion synthesis. Proc Natl Acad Sci USA. 2002;99:16000–16005. doi: 10.1073/pnas.252524999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Acharya N, Unk I, Johnson RE, Hurwitz J, Prakash L, Prakash S. A single domain in human DNA polymerase ι mediates interaction with PCNA: implications for translesion DNA synthesis. Mol Cell Biol. 2005;25:1183–1190. doi: 10.1128/MCB.25.3.1183-1190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hishiki M, Hashimoto H, Hanafusa T, Kamei K, Ohashi E, Shimizu T, Ohmori H, Sato M. Structural basis for novel interactions between human translesion synthesis polymerases and proliferating cell nuclear antigen. J Bio Chem. 2009;284:110552–10560. doi: 10.1074/jbc.M809745200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldova GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- Irimia A, Eoff RL, Guengerich FP, Egli M. Structural and functional elucidation of the mechanism promoting error-prone synthesis by human DNA polymerase κ opposite 7,8-dihydro-8-oxo-2′-deoxyguanosine adduct. J Bio Chem. 2009 doi: 10.1074/jbc.M109.003905. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R, Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Replication across template T/U by human DNA polymerase-ι. Structure. 2009;17:974–980. doi: 10.1016/j.str.2009.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaloszynski P, Ohashi E, Ohmori H, Nishimura S. Error-prone and inefficient replication across 8-hydroxyguanine (8-oxoguanine) in human and mouse ras gene fragments by DNA polymerase κ. Genes Cells. 2005;10:543–550. doi: 10.1111/j.1365-2443.2005.00858.x. [DOI] [PubMed] [Google Scholar]
- Jansen JG, Tsaalbi-Shtylik A, Langerak P, Calleja F, Meijers CM, Jacobs H, de Wind N. The BRCT domain of mammalian Rev1 is involved in regulating DNA translesion synthesis. Nucleic Acids Res. 2005;33:356–365. doi: 10.1093/nar/gki189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen JG, Langerak P, Tsaalbi-Shtylik A, van den Berk P, Jacobs H, de Wind N. Strand-biased defect in C/G transversions in hypermutating immunoglobulin genes in Rev1-deficient mice. J Exp Med. 2006;203:319–323. doi: 10.1084/jem.20052227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen JG, Fousteri MI, de Wind N. Send in the clamps: control of DNA translesion synthesis in eukaryotes. Mol Cell. 2007;28:522–529. doi: 10.1016/j.molcel.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Jansen JG, Tsaalbi-Shtylik A, Hendriks G, Gali H, Hendel A, Johansson F, Erixon K, Livneh Z, Mullenders LH, Haracska L, de Wind N. Separate domains of Rev1 mediate two modes of DNA damage bypass in mammalian cells. Mol Cell Biol. 2009;29:3113–3123. doi: 10.1128/MCB.00071-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosz DF, Godoy VG, Delaney JC, Essigmann JM, Walker GC. A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature. 2006;439:225–228. doi: 10.1038/nature04318. [DOI] [PubMed] [Google Scholar]
- Jia L, Geacintov NE, Broyde S. The N-claps of human DNA polymerase κ promotes blockage or error-free bypass of adenine- or guanine-benzo[a]pyrenyl lesions. Nucleic Acids Res. 2008;36:6571–6584. doi: 10.1093/nar/gkn719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Pol η. Science. 1999a;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Kondratick CM, Prakash S, Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999b;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- Kai M, Wang TS. Checkpoint activation regulates mutagenic translesion synthesis. Genes Dev. 2003;17:64–76. doi: 10.1101/gad.1043203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath-Loeb AS, Lan Li, Nakajima S, Yasui A, Loeb LA. Werner syndrome protein interacts functionally with translesion DNA polymerases. Proc Natl Acad Sci USA. 2007;104:10394–10399. doi: 10.1073/pnas.0702513104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannouche P, Broughton BC, Volker M, Hanaoka F, Mullenders LH, Lehmann AR. Domain structure, localization, and function of DNA polymerase η, defective in xeroderma pigmentosum variant cells. Genes Dev. 2001;15:158–172. doi: 10.1101/gad.187501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannouche P, Fernandez de Henestrosa AR, Coull B, Vidal A, Gray C, Zicha D, Woodgate R, Lehmann AR. Localization of DNA polymerase η and ι to the replication machinery is tightly coordinated in human cells. EMBO J. 2002;21:6246–6256. doi: 10.1093/emboj/cdf618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannouche P, Wing J, Lehmann AR. Interaction of human DNA polymerase η with monoubiquitinated PCNA; A possible mechanism for the polymerase switch defective in response to DNA damage. Mol Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- Kim SR, Maenhaut-Michel G, Yamada M, Yamamoto Y, Matsui K, Sofuni T, Nohmi N, Ohmori HH. Multiple pathways for SOS-induced mutagenesis in Escherichia coli: an overproduction of dinB/dinP results in strongly enhancing mutagenesis in the absence of any exogenous treatment to damage DNA. Proc Natl Acad Sci USA. 1997;94:13792–13797. doi: 10.1073/pnas.94.25.13792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirouac KN, Ling H. Structural basis of error-prone replication and stalling at a thymine base by human DNA polymerase ι. EMBO J. 2009;28:1644–1654. doi: 10.1038/emboj.2009.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Figaroa F, Meeuwenoord N, Jansen LE, Siegal G. Characterization of the DNA binding and structural properties of the BRCT region of human replication factor C p140 subunit. J Biol Chem. 2006;281:4308–4317. doi: 10.1074/jbc.M511090200. [DOI] [PubMed] [Google Scholar]
- Kornberg A, Baker T. DNA Replication. 2. W. H. Freeman and Company; New York: 1991. [Google Scholar]
- Kosarek JN, Woodruff RV, Rivera-Begeman A, Guo C, D’Souza S, Koonin EV, Walker GC, Friedberg EC. Comparative analysis of in vivo interactions between Rev1 protein and other Y-family DNA polymerases in animals and yeasts. DNA Repair. 2008;7:439–451. doi: 10.1016/j.dnarep.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulaeva OI, Koonin EV, McDonald JP, Randall SK, Rabinovich N, Connaughton JF, Levine AS, Woodgate R. Identification of a DinB/UmuC homolog in the archeon Sulfolobus solfataricus. Mutat Res. 1996;357:245–253. doi: 10.1016/0027-5107(96)00164-9. [DOI] [PubMed] [Google Scholar]
- Lawrence CW. Cellular Functions of DNA polymerase ζ and Rev1 protein. Adv Prot Chem. 2004;69:167–203. doi: 10.1016/S0065-3233(04)69006-1. [DOI] [PubMed] [Google Scholar]
- Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- Lone S, Townson SA, Uljoin SN, Johnson RE, Brahma A, Nair DT, Prakash S, Prakash L, Aggarwal AK. Human DNA polymerase κ encircles DNA: implications for mismatch extension and lesion bypass. Mol Cell. 2007;25:601–614. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- Masuda Y, Takahashi M, Tsunekuni N, Minami T, Sumii M, Miyagawa K, Kamiya K. Deoxycytidyl transferase activity of the human REV1 protein is closely associated with the conserved polymerase domain. J Biol Chem. 2001;276:15051–15058. doi: 10.1074/jbc.M008082200. [DOI] [PubMed] [Google Scholar]
- Masuda Y, Takahashi M, Fukuda S, Sumii M, Kamiya K. Mechanism of dCMP transferase reactions catalyzed of mouse Rev1 protein. J Biol Chem. 2002;277:3040–3046. doi: 10.1074/jbc.M110149200. [DOI] [PubMed] [Google Scholar]
- Masuda Y, Ohmae M, Masuda K, Kamiya KK. Structure and enzymatic properties of a stable complex of the human REV1 and REV7 proteins. J Biol Chem. 2003;278:12356–12360. doi: 10.1074/jbc.M211765200. [DOI] [PubMed] [Google Scholar]
- Masuda Y, Kamiya K. Role of single-stranded DNA in targeting REV1 to primer termini. J Biol Chem. 2006;281:24313–24321. doi: 10.1074/jbc.M602967200. [DOI] [PubMed] [Google Scholar]
- Masutani C, Araki M, Yamada A, Kusumoto R, Nogimori T, Maekawa T, Iwai S, Hanaoka F. Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J. 1999a;18:3491–3501. doi: 10.1093/emboj/18.12.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (Xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature. 1999b;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- Masutani C, Kusumoto R, Iwai S, Hanaoka F. Mechanisms of accurate translesion synthesis by human DNA polymerase η. EMBO J. 2000;19:3100–3109. doi: 10.1093/emboj/19.12.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch SD, Kokoska RJ, Masutani C, Iwai S, Hanaoka F, Kunkel TA. Preferential cis-syn thymine dimer bypass by DNA polymerase η occurs with biased fidelity. Nature. 2004;428:97–100. doi: 10.1038/nature02352. [DOI] [PubMed] [Google Scholar]
- McDonald JP, Levine AS, Woodgate R. The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics. 1997;147:1557–1568. doi: 10.1093/genetics/147.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcllwraith MJ, Vaisman A, Liu Y, Fanning E, Woodgate R, West SC. Human DNA polymerase η promotes DNA synthesis from strand invasion intermediates of homologous recombination. Mol Cell. 2005;20:783–792. doi: 10.1016/j.molcel.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Murakumo Y, Ogura Y, Ishii H, Numata S, Ichihara M, Croce CM, Fishel R, Takahashi M. Interactions in the error-prone postreplication repair proteins hREV1, hREV3, and hREV7. J Biol Chem. 2001;276:35644–35651. doi: 10.1074/jbc.M102051200. [DOI] [PubMed] [Google Scholar]
- Murakumo Y, Mizutani S, Yamaguchi M, Ichihara M, Takahashi M. Analyses of ultraviolet-induced focus formation of hREV1 protein. Genes Cells. 2006;11:193–205. doi: 10.1111/j.1365-2443.2006.00938.x. [DOI] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash S, Prakash L, Aggarwal AK. Replication by human DNA polymerase-ι occurs by Hoogsteen base-pairing. Nature. 2004;430:377–380. doi: 10.1038/nature02692. [DOI] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Human DNA polymerase ι incorporates dCTP opposite template G via a G.C + Hoogsteen base pair. Structure. 2005a;13:1569–1577. doi: 10.1016/j.str.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Nair DT, Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science. 2005b;309:2219–2222. doi: 10.1126/science.1116336. [DOI] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Hoogsteen base pair formation promotes synthesis opposite the 1,N6-ethenodeoxyadenosine lesion by human DNA polymerase ι. Nat Struct Mol Biol. 2006a;13:619–625. doi: 10.1038/nsmb1118. [DOI] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. An incoming nucleotide imposes an anti to syn conformational change on the templating purine in the human DNA polymerase-ι active site. Structure. 2006b;14:749–755. doi: 10.1016/j.str.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Nelson JR, Lawrence CW, Hinkle DC. Thymine-thymine dimer bypass by yeast DNA Polymerase ζ. Science. 1996a;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- Nelson JR, Lawrence CW, Hinkle DC. Deoxycytidyl transferase activity of yeast REV1 protein. Nature. 1996b;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- Nelson JR, Gibbs PE, Nowicka AM, Hinkle DC, Lawrence CW. Evidence for a second function for Saccharomyces cerevisiae Rev1p. Mol Microbiol. 2000;37:549–554. doi: 10.1046/j.1365-2958.2000.01997.x. [DOI] [PubMed] [Google Scholar]
- Ogi T, Shinkai Y, Tanaka K, Ohmori H. Polκ protects mammalian cells against the lethal and mutagenic effects of benzo[a]pyrene. Proc Natl Acad Sci USA. 2002;99:15548–15553. doi: 10.1073/pnas.222377899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogi T, Kannouche P, Lehmann AR. Localization of human Y-family DNA polymerase κ: relationship to PCNA. J Cell Sci. 2005;118:129–136. doi: 10.1242/jcs.01603. [DOI] [PubMed] [Google Scholar]
- Ohashi E, Ogi T, Kusumoto R, Iwai S, Masutani C, Hanaoka F, Ohmori H. Error-prone bypass of certain DNA lesions by the human DNA polymerase κ. Genes Dev. 2000;14:1589–1594. [PMC free article] [PubMed] [Google Scholar]
- Ohashi E, Murakumo Y, Kanjo N, Akagi J, Masutani C, Hanaoka F, Ohmori H. Interaction of hREV1 with three human Y-family DNA polymerases. Genes Cells. 2004;9:523–531. doi: 10.1111/j.1356-9597.2004.00747.x. [DOI] [PubMed] [Google Scholar]
- Ohashi E, Hanafusa T, Kamei K, Song I, Tomida J, Hashimoto H, Vaziri C, Ohmori H. Identification of a novel REV1-interacting motif necessary for DNA polymerase κ function. Genes Cells. 2009;14:101–111. doi: 10.1111/j.1365-2443.2008.01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori H, Hatada E, Qiao Y, Tsuji M, Fukuda R. dinP, a new gene in Escherichia coli, whose product shows similarity to UmuC and its homologues. Mutat Res. 1995;347:1–7. doi: 10.1016/0165-7992(95)90024-1. [DOI] [PubMed] [Google Scholar]
- Ohmori H, Friedberg EC, Fuchs RPP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, Prakash L, Prakash S, Todo T, Walker GC, Wang Z, Woodgate R. The Y-family of DNA polymerases. Mol Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- Ohmori H, Ohashi E, Ogi T. Mammalian Pol κ: regulation of its expression and lesion substrates. Adv Protein Chem. 2004;69:265–278. doi: 10.1016/S0065-3233(04)69009-7. [DOI] [PubMed] [Google Scholar]
- Okada T, Sonoda E, Yoshimura M, Kawano Y, Saya H, Kohzaki M, Takeda S. Multiple roles of vertebrate REV genes in DNA repair and recombination. Mol Cell Biol. 2005;25:6103–6111. doi: 10.1128/MCB.25.14.6103-6111.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pence GM, Blans P, Zink CN, Holls T, Fishbein JC, Perrino FW. Lesion bypass of N2-ethylguanine by human DNA polymerase ι. J Biol Chem. 2009;284:1732–1740. doi: 10.1074/jbc.M807296200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potapova O, Grindley NF, Joyce CM. The mutational specificity of he Dbh lesion bypass polymerase and its implications. J Biol Chem. 2002;277:28157–28166. doi: 10.1074/jbc.M202607200. [DOI] [PubMed] [Google Scholar]
- Prakash S, Johnson RE, Prakash S. Eukaryotic translesion synthesis DNA polymerases: Specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- Radman M. Mutation: Enzymes of evolutionary change. Nature. 1999;401:866–869. doi: 10.1038/44738. [DOI] [PubMed] [Google Scholar]
- Rechkoblit O, Zhang Y, Guo D, Wang Z, Amin S, Krzeminsky J, Louneva N, Geacintov NE. Translesion synthesis past bulky benzo[a]pyrene diol epoxide N2-dG and N6-dA lesions catalyzed by DNA bypass polymerases. J Biol Chem. 2002;277:30488–30494. doi: 10.1074/jbc.M201167200. [DOI] [PubMed] [Google Scholar]
- Rechkoblit O, Mainina L, Cheng Y, Kuryavyi V, Broyde S, Geacintov NE, Patel DJ. Stepwise translocation of Dpo4 polymerase during error-free bypass of an 8-oxoG lesion. PLoS Biol. 2006;4:e11. doi: 10.1371/journal.pbio.0040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuven NB, Arad G, Maor-Shoshan A, Livneh Z. The mutagenesis protein UmuC is a DNA polymerase activated by the UmuD′, RecA, and SSB and is specialized for translesion replication. J Biol Chem. 1999;274:31763–31766. doi: 10.1074/jbc.274.45.31763. [DOI] [PubMed] [Google Scholar]
- Ross AL, Simpson LJ, Sale JE. Vertebrate DNA damage tolerance requires the C-terminus but not BRCT or transferase domains of REV1. Nucleic Acids Res. 2005;33:1280–1289. doi: 10.1093/nar/gki279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roush AA, Squarez M, Friedberg EC, Radman M, Siede W. Deletion of the Saccharomyces cerevisiae gene RAD30 encoding an Escherichia coli DinB homolog confers UV radiation sensitivity and altered mutability. Mol Gen Genet. 1998;257:686–692. doi: 10.1007/s004380050698. [DOI] [PubMed] [Google Scholar]
- Sabbioneda S, Minesinger BK, Giannattasio M, Plevani P, Muzi-Falconi M, Jinks-Robertson S. The 9-1-1 checkpoint clamp physically interacts with polζ and is partially required for spontaneous polζ-dependent mutagenesis in Saccharomyces cerevisiae. J Biol Chem. 2005;280:38657–38665. doi: 10.1074/jbc.M507638200. [DOI] [PubMed] [Google Scholar]
- Sabbioneda S, Green CM, Bienko M, Kannouche P, Dikic I, Lehmann AR. Ubiquitin-binding motif in human DNA polymerase η is required for correct localization. Proc Natl Acad Sci USA. 2009;106:E20. doi: 10.1073/pnas.0812744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- Shiomi Y, Masutani C, Hanaoka F, Kimura H, Tsurimoto T. A second proliferating cell nuclear antigen loader complex, Ctf18-Replication Factor C, stimulates DNA polymerase η activity. J Biol Chem. 2007;282:20906–20914. doi: 10.1074/jbc.M610102200. [DOI] [PubMed] [Google Scholar]
- Silvian LF, Toth EA, Pham P, Goodman MF, Ellenberger T. Crystal structure of a DinB family error-prone DNA polymerase from Sulfolobus solfataricus. Nat Struct Biol. 2001;8:984–989. doi: 10.1038/nsb1101-984. [DOI] [PubMed] [Google Scholar]
- Sohn SY, Cho Y. Crystal structure of the human Rad9-Hus1-Rad1 clamp. J Mol Biol. 2009;390:490–502. doi: 10.1016/j.jmb.2009.05.028. [DOI] [PubMed] [Google Scholar]
- Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Ohashi E, Kolbanovskiy A, Geacintov NE, Grollman AP, Ohmori H, Shibutani S. Translesion synthesis by human DNA polymerase κ on a DNA template containing a single stereoisomer of dG-(+)- or dG-(−)-anti-N2-BPDE (7,8-dihydroxy-anti-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene) Biochemistry. 2002;41:6100–6106. doi: 10.1021/bi020049c. [DOI] [PubMed] [Google Scholar]
- Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structure of the human Rev1-DNA-dNTP ternary complex. J Mo Biol. 2009;390:699–709. doi: 10.1016/j.jmb.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Shen X, Frank EG, O’Donnell M, Woodgate R, Goodman MF. UmuD′2C is an error-prone DNA polymerase, Escherichia coli pol V. Proc Natl Acad Sci USA. 1999;96:8919–8924. doi: 10.1073/pnas.96.16.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissier A, McDonald JP, Frank EG, Woodgate R. polι, a remarkably error-prone human DNA polymerase. Genes Dev. 2000;14:1642–1650. [PMC free article] [PubMed] [Google Scholar]
- Tissier A, Kannouche P, Reck MP, Lehmann AR, Fuchs RP, Cordonnier A. Co-localization in replication foci and interaction of human Y-family members, DNA polymerase pol η and REVl protein. DNA Repair (Amst) 2004;3:1503–1514. doi: 10.1016/j.dnarep.2004.06.015. [DOI] [PubMed] [Google Scholar]
- Trincao J, Johnson RE, Escalante CR, Prakash S, Prakash L, Aggarwal AK. Structure of the catalytic core of S. cerevisiae DNA polymerase η: Implications for translesion DNA synthesis. Mol Cell. 2001;8:417–426. doi: 10.1016/s1097-2765(01)00306-9. [DOI] [PubMed] [Google Scholar]
- Tsuji Y, Watanabe K, Araki K, Shinohara M, Yamagata Y, Tsurimoto T, Hanaoka F, Yamamura K, Yamaizumi M, Tateishi S. Recognition of forked and single-stranded DNA structures by human RAD18 complexed with RAD6B protein triggers its recruitment to stalled replication forks. Genes Cells. 2008;13:343–354. doi: 10.1111/j.1365-2443.2008.01176.x. [DOI] [PubMed] [Google Scholar]
- Uljoin SN, Johnson RE, Edwards TA, Prakash S, Prakash L, Aggarwal AK. Crystal structure of the catalytic core of human DNA polymerase κ. Structure. 2004;12:1395–1404. doi: 10.1016/j.str.2004.05.011. [DOI] [PubMed] [Google Scholar]
- Vasiman A, Lehmann AR, Woodgate R. DNA polymerases η and ι. Adv Protein Chem. 2004;69:205–228. doi: 10.1016/S0065-3233(04)69007-3. [DOI] [PubMed] [Google Scholar]
- Vidal AE, Kannouche P, Podust VN, Yang W, Lehmann AR, Woodgate R. Proliferating cell nuclear antigen-dependent coordination of the biological functions of human DNA polymerase ι. J Biol Chem. 2004;279:48360–48368. doi: 10.1074/jbc.M406511200. [DOI] [PubMed] [Google Scholar]
- Vidal AE, Woodgate R. Insights into the cellular role of enigmatic DNA polymerase ι. DNA Repair. 2009;8:420–423. doi: 10.1016/j.dnarep.2008.12.007. [DOI] [PubMed] [Google Scholar]
- Wagner J, Gruz P, Kim SR, Yamada M, Matsui K, Fuchs RP, Nohmi T. The dinB gene encodes a novel E. coli DNA polymerase, DNA PolIV, involved in mutagenesis. Mol Cell. 1999;4:281–286. doi: 10.1016/s1097-2765(00)80376-7. [DOI] [PubMed] [Google Scholar]
- Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Warbrick E. PCNA binding through a conserved motif. Bioessays. 1998;20:195–199. doi: 10.1002/(SICI)1521-1878(199803)20:3<195::AID-BIES2>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Tateishi S, Kawasuji M, Tsurimoto T, Inoue H, Yamaizumi M. Rad18 guides polη to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 2004;23:3886–3896. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weill JC, Reynaud CA. DNA polymerase in adaptive immunity. Nature Reviews Immunology. 2008;8:302–312. doi: 10.1038/nri2281. [DOI] [PubMed] [Google Scholar]
- Wilson RC, Pata JD. Structural insights into the generation of single-base deletions by the Y-family DNA polymerase Dbh. Mol Cell. 2008;29:767–779. doi: 10.1016/j.molcel.2008.01.014. [DOI] [PubMed] [Google Scholar]
- Wilson TM, Vaisman A, Martomo SA, Sullivan P, Lan L, Hanaoka F, Yasui A, Woodgate R, Gearhart PJ. MSH2-MSH6 stimulates DNA polymerase η, suggesting a role for A:T mutations in antibody genes. J Exp Med. 2005;201:637–645. doi: 10.1084/jem.20042066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JH, Fiala KA, Suo Z, Ling H. Snapshots of a Y-family DNA polymerase in replication: substrate-induced conformational transitions and implications for fidelity of Dpo4. J Mol Biol. 2008;379:317–330. doi: 10.1016/j.jmb.2008.03.038. [DOI] [PubMed] [Google Scholar]
- Xing G, Kirouac K, Shin YJ, Bell SD, Ling H. Structural insight into recruitment of translesion DNA polymerase Dpo4 to sliding clamp PCNA. Mol Microbiol. 2009;71:678–691. doi: 10.1111/j.1365-2958.2008.06553.x. [DOI] [PubMed] [Google Scholar]
- Xu M, Bai L, Gong Y, Xie W, Hang H, Jiang T. Structure and functional implications of the human Rad9-Hus1-Rad1 cell cycle checkpoint complex. J Biol Chem. 2009;284:20457–20461. doi: 10.1074/jbc.C109.022384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Woodgate R. What a difference a decade makes: Insights into translesion synthesis. Proc Natl Acad Sci USA. 2007;104:174–183. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa MS, Masutani C, Hirano A, Cohn MA, Yamaizumi M, Nakatani Y, Hanaoka F. A human DNA polymerase η complex containing Rad18, Rad6 and Rev1; proteomic analysis and targeting of the complex to the chromatin-bound fraction of cells undergoing replication fork arrest. Genes Cells. 2006;11:731–744. doi: 10.1111/j.1365-2443.2006.00974.x. [DOI] [PubMed] [Google Scholar]
- Zang H, Irimia A, Choi JY, Angel KC, Loukachevitch LV, Egli M, Guengerich FP. Efficient and high fidelity incorporation of dCTP opposite 7,8-dihydro-8-oxodeoxyguanosine by Sulfolobus solfataricus DNA polymerase Dpo4. J Biol Chem. 2006;281:2358–2372. doi: 10.1074/jbc.M510889200. [DOI] [PubMed] [Google Scholar]
- Zhang H, Gibbs PEM, Lawrence CW. The Saccharomyces cerevisiae rev6–1 mutation, which inhibits both the lesion bypass and the recombination mode of DNA damage tolerance, is an allele of POL30, encoding proliferating cell nuclear antigen. Genetics. 2006;173:1983–1989. doi: 10.1534/genetics.106.058545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yuan F, Wu X, Wang M, Rechkoblit O, Taylor JS, Geacintov NE, Wang Z. Error-free and error-prone lesion bypass by human DNA polymerase κ. Nucleic Acids Res. 2000;28:4138–4146. doi: 10.1093/nar/28.21.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BL, Pata JD, Steitz TA. Crystal structure of a DinB lesion bypass DNA polymerase catalytic fragment reveals a classic polymerase catalytic domain. Mol Cell. 2001;8:427–437. doi: 10.1016/s1097-2765(01)00310-0. [DOI] [PubMed] [Google Scholar]