Abstract
Proteins perform most cellular functions in macromolecular complexes. The same protein often participates in different complexes to exhibit diverse functionality. Current ensemble approaches of identifying cellular protein interactions cannot reveal physiological permutations of these interactions. Here, we describe a single molecule pull-down (SiMPull) assay that combines the principles of conventional pull-down assay with single molecule fluorescence microscopy and enables direct visualization of individual cellular protein complexes. SiMPull can reveal how many proteins and of which kinds are present in the in vivo complex, as we show using protein kinase A. We then demonstrate a wide applicability to various signaling proteins found in cytosol, membrane, and cellular organelles, and to endogenous protein complexes from animal tissue extracts. The pulled down proteins are functional and are used, without further processing, for single molecule biochemical studies. SiMPull should provide a rapid, sensitive and robust platform for analyzing protein assemblies in biological pathways.
Dynamic interactions between proteins guide almost every aspect of cellular function1. Understanding how the macromolecules in living cells interact, holds the key to deciphering their roles in cellular function and regulation2, 3. Individual proteins are also part of diverse sets of protein networks, making it very challenging to tease apart various permutations of protein-protein interactions occurring in the cellular context4. Currently, the gold standard for determining interactions between proteins is the co-immunoprecipitation assay5–7 which relies on affinity-based co-purification of interacting proteins, followed by identification via Western blot (WB) or mass spectrometry. It is however difficult to determine how many copies of which proteins are present in the physiological complex using the conventional immunoprecipitation. In addition, many hours and multiple steps that often exist between sample preparation and measurements present uncertainties over the extent to which in vivo interactions are preserved prior to analysis.
In situ imaging methods based on resonance energy transfer8, 9, fluorescence correlation spectroscopy10, 11, two-hybrid methods12, 13 and bimolecular fluorescence complementation assay14 are other popular tools for studying pair-wise protein interactions. However, these methods cannot be applied to endogenous proteins and are, in general, blind to heterogeneous interactions between proteins and their stoichiometry.
Here we present a simple, direct and sensitive method to study cellular protein complexes with single complex resolution. We call this method single molecule pull-down or SiMPull because physiological macromolecular complexes are pulled down from cell or tissue extracts directly to the imaging surface of single molecule fluorescence microscopy.
Experimental strategy and YFP pull-down
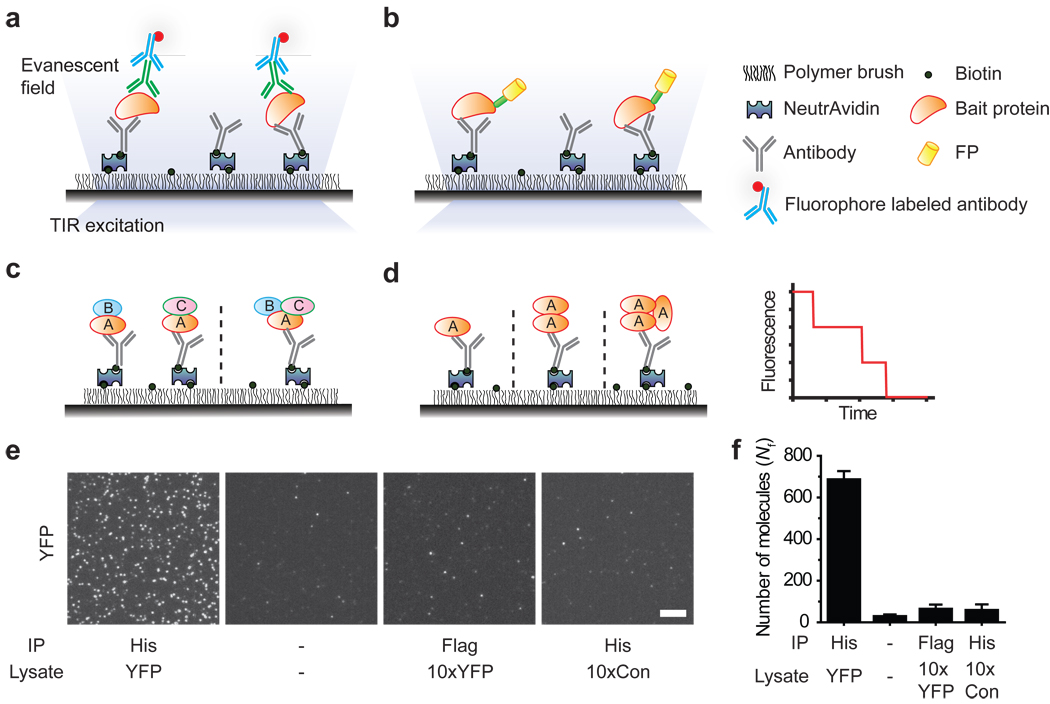
The key requirement for pull-down assays is the selective capture of a protein of interest (bait), which will bring along its binding partners (prey). We constructed a flow chamber using a microscope slide and a cover slip, passivated with mPEG (methoxy polyethylene glycol)15 to prevent non-specific adsorption of cell extracts and antibodies, which should minimize false positives7. The imaging surface was also doped with biotinylated PEG and streptavidin, followed by biotinylated antibodies against the bait protein (Fig. 1a–d, Supplementary Fig. 1). When cell extracts are infused in the flow chamber, the surface tethered antibody captures the bait protein together with any interacting partners. After washing away the unbound cell extract, co-immunoprecipitated prey proteins are visualized either through immunofluorescence labeling (Fig. 1a) or using genetically encoded fluorescent protein tags (Fig. 1b). This approach is extendable to multi-protein complexes via multi-color labeling and has the potential to differentiate between multiple sub-complexes and configurations (Fig. 1c). When proteins are fluorescently labeled with a fixed ratio, photobleaching events yield stoichiometric information16, 17 (Fig. 1d).
Figure 1.
Schematic for SiMPull assay. Immunoprecipitated protein complexes are visualized using TIRF microscopy via (a) fluorophores-labeled antibody or (b) fluorescent protein tags. (c) Multi-color colocalization can distinguish between subcomplexes (e.g. AB+AC vs. ABC). (d) Photobleaching analysis can provide stoichiometric information. A simulated photobleaching trajectory for a trimeric protein. (e) TIRF images for YFP pulled down from cells expressing (His)6-YFP (YFP) and control cells (Con) using His-tag or a control (Flag-tag) antibody. (−) indicates no antibody or sample. Scale bar is 5 µm. (f) Average number of fluorescent molecules per imaging area, Nf. Error bars denote s. d. (n > 20).
We first validated the SiMPull assay for specific pull-down of yellow fluorescent protein (YFP) from cell extracts. When the crude lysate from cells over-expressing (His)6-tagged YFP was infused into the flow chamber coated with anti-His antibody, we observed single YFP molecules (Fig. 1e, f), similar to the analysis performed using purified protein18 (Supplementary Fig. 2). Binding of YFP to the antibody was stable over two hours (Supplementary Fig. 3). The blank slide surface showed ~30 fluorescent spots per imaging area, 2,500 µm2, possibly due to surface impurities. The number of fluorescent spots per imaging area, Nf, due to specifically pulled down proteins was 10–20 fold over the background; we could maintain > 10 fold signal to noise ratio by controlling the lysate dilution factor. Control experiments with YFP lysate on non-specific antibody and lysate without YFP expression showed the same Nf as blank. Even lysate with 10-fold higher concentration of YFP yielded only ~30 additional spots relative to the blank, implying less than 0.5% contribution from non-specifically adsorbed proteins (Fig. 1e, f). Nf increased monotonically as the cell lysate concentration is increased over three orders of magnitude (Supplementary Fig.4), showing that SiMPull can provide a quantitative estimate of protein concentration in cell lysate.
Single molecule photobleaching analysis performed using the monomeric YFP and tandem dimeric YFP constructs (Supplementary Fig. 5, 6) showed that accurate stoichiometric information can be obtained from the pulled down proteins when we account for the fact that about 75% of YFP is fluorescently active16.
Two-color SiMPull of PKA complex
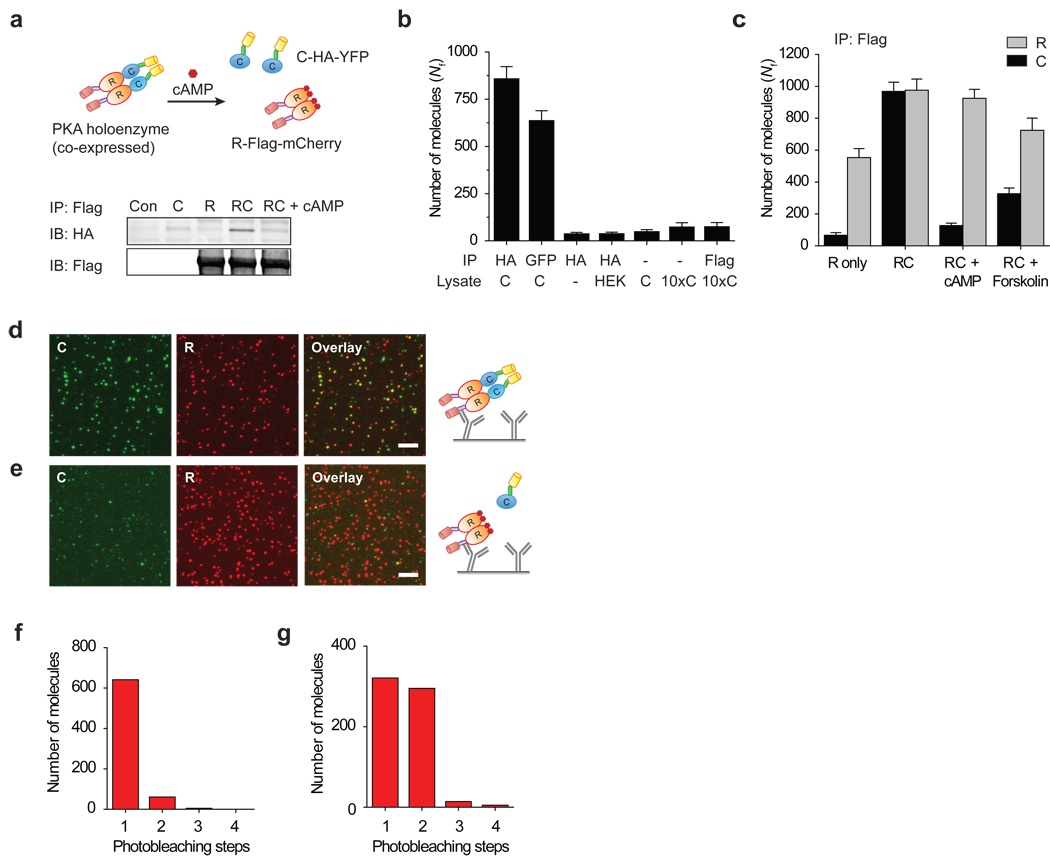
Next, we demonstrate the ability to pull-down single protein complexes from cell extracts using cyclic adenosine monophosphate (cAMP) dependent protein kinase, PKA, as our model system. PKA is a ubiquitous serine/threonine kinase that acts downstream of the G-protein coupled receptor (GPCR) pathway19. In the inactive state, PKA is a tetrameric complex consisting of two regulatory (R) and two catalytic (C) subunits. In the presence of cAMP, the complex dissociates, thereby activating the enzyme. We prepared C-HA-YFP and R-Flag-mCherry constructs (Fig. 2a).
Figure 2.
PKA pull-down. (a) Schematic of PKA construct. In WB, C-HA-YFP is pulled down via R-Flag-mCherry; on adding cAMP, PKA dissociates. (b) Nf for C-HA-YFP (C) as a function of lysates and antibodies demonstrate the specificity of pull-down. (c–e) PKA complex pull-down. (c) Nf for YFP (C) and mCherry (R) spots. (d) Images of single PKA complexes, YFP (left), mCherry (center) and overlay (right). (e) On adding cAMP, YFP spots decrease significantly. Photobleaching step distribution (f) for C-HA-YFP only lysate and (g) for C-HA-YFP pulled down via R-Flag-mCherry. Error bars denote s. d. (n > 20). Scale bar is 5 µm.
When only C-HA-YFP was expressed in HEK293 cells, we could specifically pull-down the protein from the cell lysate using surface immobilized antibodies against HA- or YFP-epitope (Fig. 2b). As expected, these samples did not show any detectable fluorescence above background in the mCherry detection channel (Supplementary Fig. 7).
When the two subunits, R and C, were co-expressed, WB showed that R and C coimmunoprecipitate20 and they dissociate when cAMP is added to the lysate, confirming that the modified constructs retain the known properties of PKA (Fig. 2a). In a similar fashion, when we pulled down R-Flag-mCherry with anti-Flag antibody, we could detect both YFP and mCherry fluorescence spots. The number of fluorescent spots in mCherry and YFP channels was similar, indicating, on average, a one-to-one association (Fig. 2c, d). 57% of YFP spots colocalized with a corresponding mCherry, as shown by individual images and their overlay (Fig. 2d). Incomplete colocalization may arise from basal tonic activation of PKA, inactive chromophores16, 21 or unbalanced expression of two proteins in individual cells. Adding cAMP analog to the flow channel or pre-incubating the lysate with cAMP resulted in a greatly reduced number of C subunit (YFP spots) without a significant change in R subunit (mCherry spots) (Fig. 2c, e). After the reaction, only 4% of remaining YFP molecules colocalized with a corresponding mCherry.
Intracellular levels of cAMP can be increased by stimulating GPCRs for activation of adenylyl cyclases. When the cells over-expressing the PKA complex were stimulated with forskolin, an agonist for adenylyl cyclase for cAMP production, the amount of active PKA in cells increased as seen via dissociation of PKA in WB (Supplementary Fig. 8). Similarly, in our SiMPull assay, the number of C subunit (YFP molecules) pulled down through R subunit decreased significantly (Fig. 2c).
We explored the stoichiometry of PKA via photobleaching analysis. When C-HA-YFP was expressed alone and pulled down using HA-antibody, 91% of YFP traces exhibited one-step photobleaching indicating a monomeric population (Fig. 2f). When C-HA-YFP and R-Flag-mCherry were co-expressed and pulled down using anti-Flag antibody, 47% of YFP traces displayed two photobleaching steps (Fig. 2g) while 51% molecules bleached in one step. Assuming a 75% active fraction of YFP16, this is consistent with the known stoichiometry of two catalytic subunits in each PKA. We did not perform photobleaching-based stoichiometry analysis for mCherry due to its inferior photophysical properties22.
Applications of SiMPull
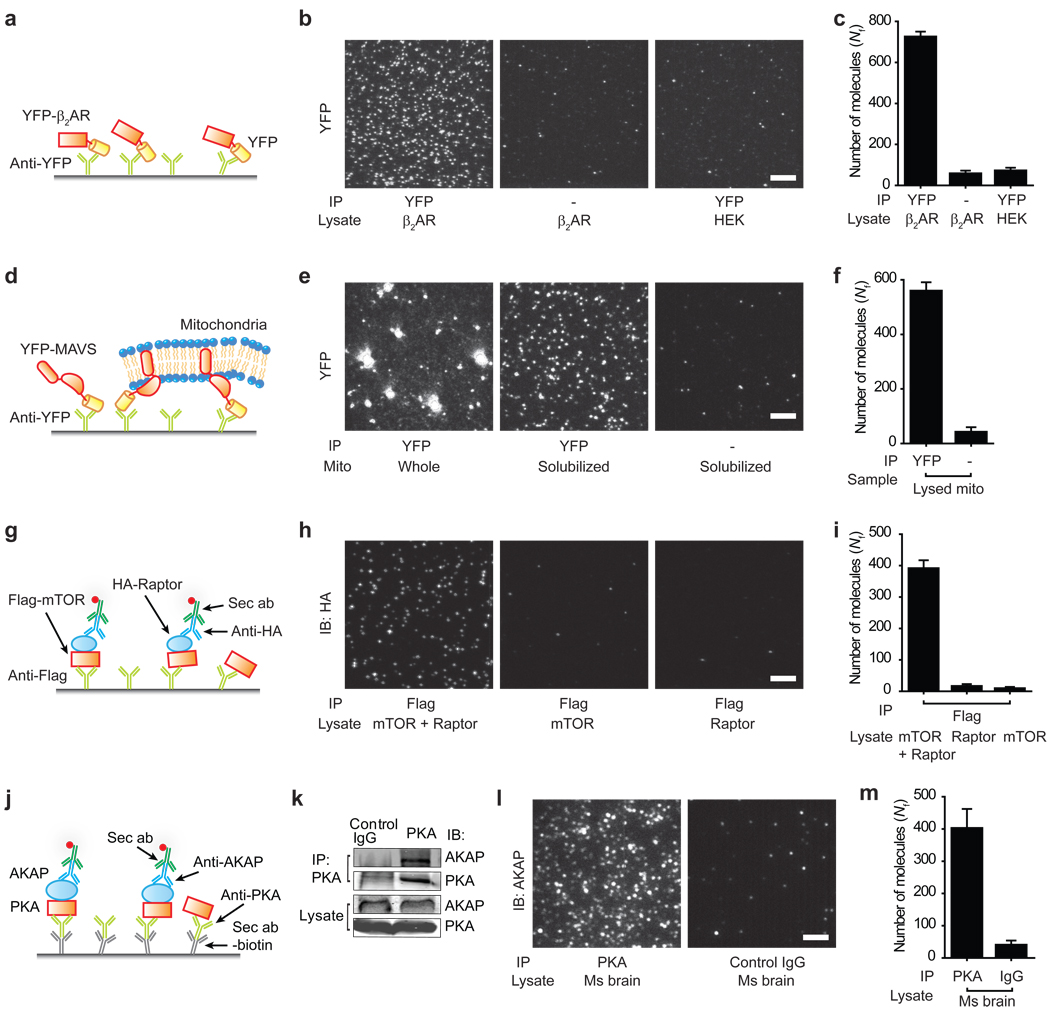
Next, we examined the applicability of SiMPull to protein complexes from various cellular environments using different capture and detection configurations (Fig. 3).
Figure 3.
Applications of SiMPull assay. (a–c) β2AR-YFP pull-down. (d–f) MAVS pull-down. Mitochondrial fraction from cells over-expressing YFP-MAVS was added either directly or after detergent solubilization (g–i) mTORC1 pull-down. Lysate from cells expressing Flag-mTOR, HA-Raptor or both was applied on chambers with Flag antibody, and probed through primary antibody against HA and labeled secondary antibody. (j–m) Endogenous PKA-AKAP complex pull-down from mouse brain extract. (k) WB shows AKAP immunoprecipitation with PKA antibody. (l) Immunofluorescence images of AKAP150 pulled down through PKA antibody. (c, f, i, m) show Nf. Scale bars are 5 µm. Error bars denote s. d. (n > 20).
Receptor pull-down
Membrane protein complexes are particularly difficult to analyze using conventional methods, thus motivating new approaches23, 24. Their stoichiometry cannot be determined via photobleaching in the cell unless the areal density of protein complexes is low enough for single molecule detection16, 25, 26. SiMPull should be able to detect individual complexes if membrane patches containing one complex can be isolated. As a test, we applied SiMPull on β2 adrenergic receptor (β2AR), a prototypical GPCR. HEK293 cells were transfected with Flag-YFP-β2AR and membrane proteins were solubilized. We could specifically pull-down the receptor using antibodies against YFP or Flag (Fig. 3a–c). 29% of the traces displayed two distinct bleaching steps (Supplementary Fig. 9a), indicating a ~51% dimer population, assuming 75% of active fluorophores. Less than 3% of the traces showed 3 or more photobleaching steps. Our observation of β2AR homo-dimerization is consistent with previous studies27, 28. To test if β1AR may form heterooligomers with β2AR28, we co-expressed mCherry-β1AR and YFP-β2AR. Using antibody against mCherry-β1AR, we could pull-down YFP-β2AR and vice versa (Supplementary Fig. 9b–e). The two fluorophores colocalized with ~42% overlap, consistent with heterooligomer formation.
Mitochondrial protein pull-down
MAVS (mitochondrial antiviral signaling) is a mitochondrial outer membrane protein involved in innate immune response29. When isolated mitochondrial fraction from cells over-expressing YFP-MAVS30 was applied to a surface with anti-YFP, we observed bright fluorescent structures (Fig. 3e, left), indicating the presence of multiple YFP molecules. Upon pre-solubilizing the mitochondrial preparation using mild detergent, we observed isolated single YFP spots (Fig. 3e, center), 86% of which displayed single photobleaching steps (Supplementary Fig. 10), supporting monomeric state of solubilized MAVS. This observation suggests that the bright fluorescent structures observed in unsolubilized preparations are due to immunoprecipitated mitochondrial membrane patches or whole mitochondria. It may be possible to specifically immobilize cellular organelles or their components using antibodies against suitable marker proteins and perform single molecule measurements in a physiologically relevant context.
Immunofluorescence detection of single complexes
We extended the assay to detection via antibodies using mammalian target of rapamycin complex 1 (mTORC1) as a model system. mTORC1 is a key signaling complex that regulates cell growth and metabolism in response to nutrient availability to the cells31, 32. In addition to mTOR (mammalian target of rapamycin), a defining component of mTORC1 is Raptor (regulatory associated protein of mTOR), which associates with mTOR at an equimolar ratio31, 33. We expressed Flag-mTOR and HA-Raptor in HEK293 cells. Flag-mTOR was pulled down using biotinylated Flag-antibody; Raptor was detected using HA-antibody followed by fluorescently labeled secondary antibody (Fig. 3g). When both Flag-mTOR and HA-Raptor were co-expressed, we observed the detection antibody binding as fluorescent spots whereas background level of fluorescence was detected when only one of the two proteins was expressed (Fig. 3h, i), demonstrating antibody-based detection in SiMPull.
Pull-down of endogenous complexes in native tissues
Exogenous expression may lead to non-physiological associations between proteins. Pull-down of endogenously expressed proteins, though desirable, is challenging due to low abundance, high background interaction with other cellular proteins, and general lack of high affinity antibodies. We tested if SiMPull can be used to detect interactions between endogenous proteins. A kinase anchoring proteins (AKAPs) bind to PKA and confine it to discrete locations in the cell34. Fig. 3k shows AKAP150 can be co-immunoprecipitated with PKA from mouse brain extract.
Primary antibodies against proteins are often expensive and difficult to label with biotin or fluorophores. Thus, to keep our approach general, we used biotin labeled secondary antibody to immobilize the antibody against the bait (PKA), and applied mouse brain extract. On probing for the prey protein (AKAP150) using its primary antibody and fluorescently labeled secondary antibody, we observed 10-fold more fluorescent spots in the channel with PKA antibody as compared to the control channel (Fig. 3l, m). SiMPull required a 20-fold lower sample volume as compared to the corresponding WB. This sensitivity allowed detection of PKA-AKAP binding from mouse heart tissue, which was below the detection limit of the conventional WB under the same conditions (Supplementary Fig. 11).
SiMPull as a preparatory tool
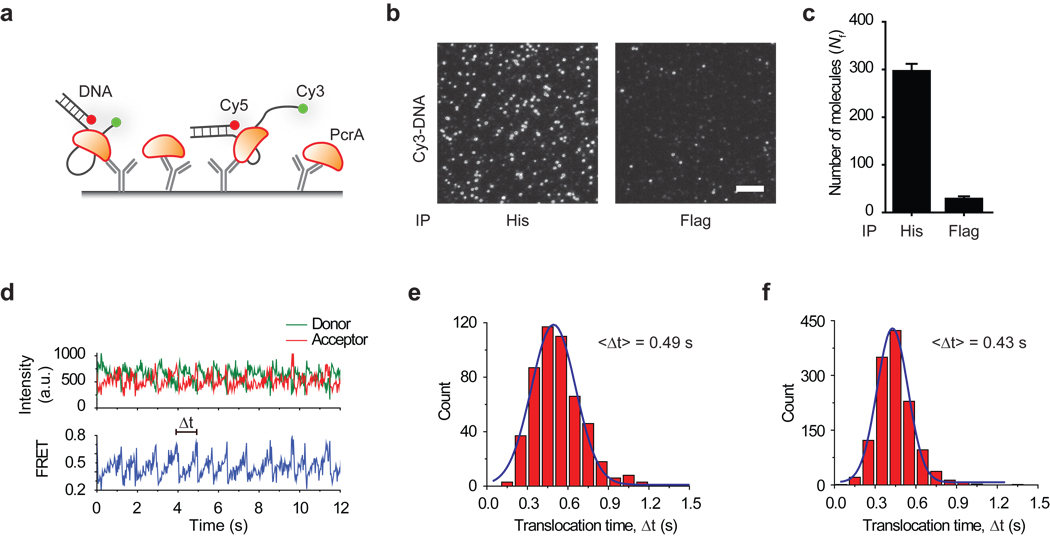
A key advantage of SiMPull is that protein complexes can be directly observed from a fresh cell lysate, bypassing purification procedures. We tested if SiMPull can be used for functional analysis of pulled down proteins. PcrA, a superfamily 1 helicase, is an ATP driven motor protein that binds and translocates on single stranded DNA (ssDNA)35. (His)6-tagged PcrA was pulled down from bacterial lysate using anti-His antibody and fluorescently labeled DNA molecules were added to the flow channel (Fig. 4a). Fluorescent spots due to labeled DNA binding appeared in the flow channel with pulled down PcrA, while the control channel showed minimal DNA binding (Fig. 4b, c).
Figure 4.
PcrA pull-down and activity test (a) schematic, (b, c) labeled DNA binding to immunoprecipitated PcrA. Scale bar is 5 µm. Error bars represent s. d. (n > 20). (d) A typical time trace of repetitive reeling-in activity of PcrA monitored by FRET. The distribution of translocation times (Δt) and its mean, <Δt>, (e) for purified PcrA and (f) for PcrA pulled down from cell extracts, at 1 mM ATP concentration.
When PcrA binds to a partial duplex DNA with a 5’ overhang, it anchors itself to the junction and repetitively reels-in the ssDNA35. By labeling the DNA with a donor at the tail end and an acceptor at the junction (Fig. 4a), we could observe the reeling-in activity as a gradual increase in fluorescence resonance energy transfer (FRET) (Fig. 4d). Once PcrA reaches the end of ssDNA, it runs off the ssDNA track and repeats the process from the junction over and over, resulting in cyclic increases and decreases in FRET. 86% (161 of 188) of bound FRET-labeled DNA molecules exhibited repetitive cycling. On increasing the ATP concentration, translocation became faster (Supplementary Fig. 12a), and in the absence of ATP, DNA remained bound but no reeling-in activity was observed (Supplementary Fig. 12b). The mean translocation time matched well with the data obtained with purified protein (Fig. 4e, f). Thus, SiMPull can pull-down functional macromolecules directly from cell extracts for subsequent single molecule biochemistry in situ.
Discussion
We have established a single molecule platform for analyzing cellular association of macromolecules. SiMPull can be used as an extension of commonly used WB analysis without requiring additional sample preparation (Supplementary Fig. 13) and confers several key advantages. 1) It can provide quantitative data on sub-populations of different association states. 2) It provides information on complex stoichiometry if the proteins can be stoichiometrically labeled. 3) The high sensitivity allows the studies of complexes of low abundance and a suitable calibration (Supplementary Fig. 4) should make it possible to determine the expression level of protein complexes in cell lysate. 4) The whole assay took about 30 minutes, considerably shorter than conventional WB. In a pilot experiment, we could dilute the cell lysate, pull-down and quantify YFP in 2.5 min (Supplementary Fig. 14). Therefore, it should be possible to analyze even relatively weak protein complexes as long as the dissociation rate constant koff is equal to or smaller than 0.01 s−1. Combining this with microfluidics platform, cross-linking methods or zero mode waveguide36 may extend the method to complexes with even higher koff.
Cellular processes are under a tight spatio-temporal regulation. In order to overcome ensemble averaging over heterogeneous cell populations, SiMPull may be combined with fluorescence aided cell sorting to selectively analyze a subpopulation or may even be pushed to the single cell level. In a preliminary experiment, we could pull-down and quantify proteins from 10 cells obtained through cell sorting (Supplementary Fig. 15) compared to ~5000 cells usually required for a WB37. Single cell SiMPull may enhance the recently demonstrated ability to quantify proteins and RNA numbers in single cells38. As in conventional WB, the sensitivity and specificity of SiMPull are determined by the quality of capture and detection antibodies. The assay may be combined with recent developments in labeling strategies39 for further improvement in sensitivity and labeling efficiency.
Post-translational modifications play an important role in cellular processes but are difficult to reproduce in recombinant proteins. In addition, the necessary co-factors or ligands for a protein of interest are often unknown. SiMPull as a preparatory tool provides a possibility to study these modified proteins or protein complexes that cannot be purified using conventional methods.
Methods summary
Flow chambers were prepared on mPEG passivated quartz slides doped with biotin PEG15. Biotinylated antibodies were immobilized by incubating ~10 nM of antibody for 10 min on NeutrAvidin (Thermo) coated flow chambers. Prism type total internal reflection fluorescence (TIRF) microscope was used to acquire the single molecule data40. Samples with fluorescent protein tag were serially diluted to obtain well-isolated spots on the surface upon 20 min of incubation over immobilized antibody surface. All dilutions were made immediately before addition to the flow chamber in 10 mM Tris-HCl pH 8.0, 50 mM NaCl buffer with 0.1 mg/ml bovine serum albumin (New England Biolabs), unless specified. Unbound antibodies and sample were removed from the channel by washing with buffer twice between successive additions. For immunofluorescence detection, immunoprecipitated complexes were incubated with a different antibody against prey protein (~10 nM) for 20 min and fluorescent-dye-labeled secondary antibody (2–5 nM) for 5 min before imaging. Single molecule analysis was performed using scripts written in Matlab.
Methods
Overview
Flow chambers were prepared on mPEG passivated quartz slides doped with biotin PEG15. Biotinylated antibodies were immobilized by incubating ~10 nM of antibody for 10 min on NeutrAvidin (Thermo) coated flow chambers. Prism type total internal reflection fluorescence (TIRF) microscope was used to acquire the single molecule data40. Samples with fluorescent protein tag were serially diluted to obtain well-isolated spots on the surface upon 20 min of incubation over immobilized antibody surface. All dilutions were made immediately before addition to the flow chamber in 10 mM Tris-HCl pH 8.0, 50 mM NaCl buffer with 0.1 mg/ml bovine serum albumin (New England Biolabs), unless specified. Unbound antibodies and sample were removed from the channel by washing with buffer twice between successive additions. For immunofluorescence detection, immunoprecipitated complexes were incubated with a different antibody against prey protein (~10 nM) for 20 min and fluorescent-dye-labeled secondary antibody (2–5 nM) for 5 min before imaging. Single molecule analysis was performed using scripts written in Matlab.
Single molecule imaging and spot counting
Prism type TIRF microscope was used to acquire single molecule data40. YFP was excited at 488 nm; mCherry was excited at 532 or 568 nm. Narrow band-pass filters were used to avoid cross-talk between channels (HQ 535/30 from Chroma Technology for YFP and BL 607/36 from Semrock Inc. for mCherry). All experiments were performed at room temperature (22 – 25 °C) unless specified. Single molecule analysis was performed as described earlier15. Mean spot count per image (imaging area 2500 µm2) and standard deviation were calculated from images taken from 20 or more different regions.
Photobleaching analysis
Single molecule fluorescence time traces of surface immobilized YFP-tagged proteins were manually scored for the number of bleaching steps16. To avoid false colocalization, samples were immobilized at an optimal surface density (~300 molecules in 2500 µm2 imaging area). The number of photobleaching steps (single frame intensity drops of equal size) in each trace was manually determined, following published procedures16. Fluorescence trace of each molecule was classified as having 1–4 bleaching steps or was discarded if no clean bleaching steps could be identified (Supplementary Fig. 5). Some fluorescent protein molecules exhibited blinking but under most circumstances, distinct fluorescence intensity levels could still be readily determined in spite of blinking (Supplementary Fig. 5a, b). Separate counts were maintained for each case. At least 500 molecules were analyzed for each sample. The probability of missed bleaching events, due to simultaneous bleaching of both FPs within the same imaging window is ~5%. The population distribution of observed bleaching events and discarded traces is reported in Supplementary Table 1. For future extensions to complexes with many more copies of the same protein, automated algorithms for scoring photobleaching steps would be required41.
Single molecule co-localization
Co-localization between YFP and mCherry was performed using a method similar to Ulbrich et al.42. Briefly, we took two separate movies of the same region using YFP and mCherry excitation. The fluorescent spots in both images were fit with Gaussian profiles to determine the center positions of molecules to half-pixel accuracy. Next, for each molecule in YFP image, we determined the mCherry molecules with center within 2 pixel (~300 nm) distance. The number of molecules where this co-localization occurred divided by the total number of YFP molecules was presented as overlap %.
YFP constructs and pull-down
As YFP has been shown to dimerize, monomeric YFP was generated through site directed mutagenesis of alanine 207 to lysine using pEYFP-C1 as DNA template (Clontech). For bacterial expression, monomeric YFP was cloned into Sal I and Xho I sites of pET-28b(+) vector. BL21 DE3 cells were transformed with YFP construct and induced by 0.2 mM IPTG for protein expression. Cells were resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole pH 8.0) and sonicated. The lysate was centrifuged at 15,000g for 20 min to collect supernatant used for SiMPull.
For expression in mammalian cells, YFP-(His)6 was generated through addition of a 6xHis-tag to the C-terminal of YFP and subcloned into Xho I and Xba I sites of pCDNA3.1(+). A second YFP was subcloned into the Hind III and EcoR I sites of pCDNA3.1-YFP-(His)6 to make a tandem dimeric YFP construct. Monomeric and dimeric YFP constructs were transiently expressed in HEK293 cells and purified using standard Ni-NTA chromatography. Proteins were detected by WB using GFP antibody (Clontech) or Penta-His antibody (Qiagen). For single molecule analysis, samples were immobilized on biotinylated anti-Penta-His antibody (Qiagen) or on biotinylated polyclonal anti-GFP antibody (Rockland Immunochemicals, Inc.).
Protein kinase A (PKA) constructs and pull-down
HEK293 cells were transfected with R-Flag-mCherry and C-HA-YFP constructs. The regulatory subunit used was PKA RIIβ, and the catalytic subunit is the Cα isoform. After 24 hr expression, cells were harvested into lysis buffer (10 mM Tris pH 7.5, 1% NP-40, 150 mM NaCl, 1 mM EDTA, 1 mM benzamidine, 10 µg/ml leupeptin, 1 mM NaF, 1 mM Na3VO4). This lysate was centrifuged at 14,000g for 20 min and used for SiMPull. For bulk immunoprecipitation, anti-Flag M2 beads were added to the lysate for 3 hr at 4 °C. Proteins were separated by SDS-PAGE and transferred onto nitrocellulose membranes for blot with anti-HA antibody and anti-mCherry antibody.
cAMP treatment: A non-hydrolysable analog (8-Br-cAMP, Sigma), was used to activate PKA. For in vivo stimulation in cells, R-Flag-mCherry and C-HA-YFP were transiently expressed in HEK293 cells for 24 hr. Cells were pre-treated for 10 min with 10 µM 3-Isobutyl-1-methlxanthine (IBMX, Sigma) and 5 min stimulation with 10 µM Forskolin (Sigma). Cells were immediately washed with cold PBS and lysed as described earlier.
Adrenergic receptor constructs and pull-down
Flag-YFP-β2AR, HA-YFP-β2AR, Flag-mCherry-β1AR and HA-mCherry-β1AR were transiently expressed in HEK293 cells for 24 hr. Cells were harvested into hypotonic lysis buffer (10 mM Tris pH 7.4, 1 mM EDTA, 1 mM benzamidine, 10 µg/ml leupeptin, 0.3% DDM (n-dodecyl-beta-D-maltoside), and incubated for 30 min before centrifugation at 600g for 10 min. Supernatants were collected and used for SiMPull with antibodies as indicated.
Mitochondria preparation and MAVS pull-down
HEK293 cells were transiently transfected with YFP-MAVS30. Intact mitochondria were isolated using MITOISO2 kit (Sigma) and diluted in the storage buffer supplied with the kit. Mitochondrial preparation was immobilized on slides either directly or after solubilization by adding 1% DDM to the storage buffer. We obtained similar results using whole cell lysates prepared using several different lysing solutions. YFP-MAVS or mitochondria were immunoprecipitated using biotinylated antibody against GFP.
mTORC1 construct and pull-down
Flag-mTOR was stably transfected in HEK293 cells to obtain near endogenous expression levels of mTOR. For mTORC1 pull-down, Flag-mTOR stable cell lines were transiently transfected with HA-Raptor. HA-Raptor only lysate was obtained by transiently transfecting HEK293 cells with HA-Raptor. Cells were lysed using CHAPS detergent buffer (40 mM HEPES pH 7.5, 0.3% CHAPS, 150 mM NaCl, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM EDTA), with protease inhibitor cocktail. Lysate was diluted in the buffer without CHAPS for SiMPull with biotinylated anti-Flag antibody (Sigma). Co-immunoprecipitated HA-Raptor was detected using Goat-anti-HA antibody (Genscript) and Donkey-anti-Goat secondary antibody (Rockland Immunochemicals, Inc.) labeled with Cy3.
Endogenous protein pull-down
Mouse brain and heart extracts were prepared from two-week-old FVB mice after anesthetization. Both sample were homogenized in lysis buffer (20 mM HEPES pH 7.4, 0.5% Triton X-100, 150 mM NaCl, 10% glycerol, 5 mM EDTA, 5 µg/ml pepstatin, 1 mM PMSF, 1 mM NaF, 1 mM Na3VO4), incubated at 4 °C for one hr followed by centrifugation at 16,000g for 10 min to collect the supernatant. These samples were directly used for SiMPull. For bulk immunoprecipitation, protein-A beads were added to pre-clean the lysate (2 hr incubation at 4 °C). PKARII antibody was then added to the lysate for overnight immunoprecipitation followed by 1 hr incubation with protein-A beads. Control rabbit IgG was added at final concentration 2 µg/ml. The immunoprecipitated proteins were separated by SDS-PAGE and transferred onto nitrocellulose membranes for immunoblot analysis with AKAP150 and PKARII antibodies.
PcrA pull-down and functional assay
(His)6-tagged PcrA purified protein and cell lysate were prepared as previously described35. The protein was immobilized on slides via antibody against polyhistidine tag. A Cy3 and Cy5 dual-labeled partial duplex DNA (Integrated DNA Technologies, Inc.) with a 5’ tail was added to immobilized protein. The sequence of DNA used was: 5’Cy3-(dT)40-GCC TCG CTG CCG TCG CCA-3’ + 5’- TGG CGA CGG CAG CGA GGC-3’-Cy5.
Supplementary Material
Acknowledgements
We thank Sua Myong, Prakrit Jena, Sinan Arslan and Reza Vafabakhsh for helpful discussions. The expression vector encoding YFP-MAVS gene was a gift from Daniel Lamarre (Université de Montréal). This work was funded by NIH grants (AI083025, GM065367 to T.H.; HL082846 to Y.K.X.; AR048914 to J.C.). Additional support was provided by NSF grants (0646550, 0822613 to T.H.). T.H. is an investigator with the Howard Hughes Medical Institute.
Footnotes
Author contributions
A.J., K.Y.X., and T.H. designed research. A.J., R.L. and Y.I. conducted experiments, R.L., B.R., E.A. and J.P. provided samples, K.R. and Y.I. contributed important ideas to the experiments, A.J. and R.L. analyzed the data and A.J., K.Y.X. and T.H. wrote the paper with inputs from other authors.
References
- 1.Alberts B. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell. 1998;92:291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- 2.Papin JA, Hunter T, Palsson BO, Subramaniam S. Reconstruction of cellular signalling networks and analysis of their properties. Nat Rev Mol Cell Biol. 2005;6:99–111. doi: 10.1038/nrm1570. [DOI] [PubMed] [Google Scholar]
- 3.Gavin AC, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 4.Yamada T, Bork P. Evolution of biomolecular networks: lessons from metabolic and protein interactions. Nat Rev Mol Cell Biol. 2009;10:791–803. doi: 10.1038/nrm2787. [DOI] [PubMed] [Google Scholar]
- 5.Barrios-Rodiles M, et al. High-throughput mapping of a dynamic signaling network in mammalian cells. Science. 2005;307:1621–1625. doi: 10.1126/science.1105776. [DOI] [PubMed] [Google Scholar]
- 6.Puig O, et al. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 7.Gingras AC, Gstaiger M, Raught B, Aebersold R. Analysis of protein complexes using mass spectrometry. Nat Rev Mol Cell Biol. 2007;8:645–654. doi: 10.1038/nrm2208. [DOI] [PubMed] [Google Scholar]
- 8.Wallrabe H, Periasamy A. Imaging protein molecules using FRET and FLIM microscopy. Curr Opin Biotechnol. 2005;16:19–27. doi: 10.1016/j.copbio.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 9.Carriba P, et al. Detection of heteromerization of more than two proteins by sequential BRET-FRET. Nat Methods. 2008;5:727–733. doi: 10.1038/nmeth.1229. [DOI] [PubMed] [Google Scholar]
- 10.Slaughter BD, Schwartz JW, Li R. Mapping dynamic protein interactions in MAP kinase signaling using live-cell fluorescence fluctuation spectroscopy and imaging. Proc Natl Acad Sci U S A. 2007;104:20320–20325. doi: 10.1073/pnas.0710336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zamir E, Lommerse PH, Kinkhabwala A, Grecco HE, Bastiaens PI. Fluorescence fluctuations of quantum-dot sensors capture intracellular protein interaction dynamics. Nat Methods. 7:295–298. doi: 10.1038/nmeth.1441. [DOI] [PubMed] [Google Scholar]
- 12.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 340;1989:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 13.Eyckerman S, et al. Design and application of a cytokine-receptor-based interaction trap. Nat Cell Biol. 2001;3:1114–1119. doi: 10.1038/ncb1201-1114. [DOI] [PubMed] [Google Scholar]
- 14.Kerppola TK. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu Rev Biophys. 2008;37:465–487. doi: 10.1146/annurev.biophys.37.032807.125842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ulbrich MH, Isacoff EY. Subunit counting in membrane-bound proteins. Nat Methods. 2007;4:319–321. doi: 10.1038/NMETH1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reyes-Lamothe Rodrigo SD, J, Leake MC. Stoichiometry and Architecture of Active DNA Replication Machinery in Escherichia coli. Science. 2010;328:498–501. doi: 10.1126/science.1185757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mashanov GI, Tacon D, Knight AE, Peckham M, Molloy JE. Visualizing single molecules inside living cells using total internal reflection fluorescence microscopy. Methods. 2003;29:142–152. doi: 10.1016/s1046-2023(02)00305-5. [DOI] [PubMed] [Google Scholar]
- 19.Collins S, Caron MG, Lefkowitz RJ. Regulation of adrenergic receptor responsiveness through modulation of receptor gene expression. Annu Rev Physiol. 1991;53:497–508. doi: 10.1146/annurev.ph.53.030191.002433. [DOI] [PubMed] [Google Scholar]
- 20.Taylor SS, et al. PKA: a portrait of protein kinase dynamics. Biochim Biophys Acta. 2004;1697:259–269. doi: 10.1016/j.bbapap.2003.11.029. [DOI] [PubMed] [Google Scholar]
- 21.Maeder CI, et al. Spatial regulation of Fus3 MAP kinase activity through a reaction-diffusion mechanism in yeast pheromone signalling. Nat Cell Biol. 2007;9:1319–1326. doi: 10.1038/ncb1652. [DOI] [PubMed] [Google Scholar]
- 22.Yu Y, et al. Structural and molecular basis of the assembly of the TRPP2/PKD1 complex. Proc Natl Acad Sci U S A. 2009;106:11558–11563. doi: 10.1073/pnas.0903684106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopez-Gimenez JF, Canals M, Pediani JD, Milligan G. The alpha1-badrenoceptor exists as a higher-order oligomer: effective oligomerization is required for receptor maturation, surface delivery, and function. Mol Pharmacol. 2007;71:1015–1029. doi: 10.1124/mol.106.033035. [DOI] [PubMed] [Google Scholar]
- 24.Schwarzenbacher M, et al. Micropatterning for quantitative analysis of proteinprotein interactions in living cells. Nat Methods. 2008;5:1053–1060. doi: 10.1038/nmeth.1268. [DOI] [PubMed] [Google Scholar]
- 25.Yu J, Xiao J, Ren X, Lao K, Xie XS. Probing gene expression in live cells, one protein molecule at a time. Science. 2006;311:1600–1603. doi: 10.1126/science.1119623. [DOI] [PubMed] [Google Scholar]
- 26.Leake MC, et al. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature. 2006;443:355–358. doi: 10.1038/nature05135. [DOI] [PubMed] [Google Scholar]
- 27.Angers S, et al. Detection of beta 2-adrenergic receptor dimerization in living cells using bioluminescence resonance energy transfer (BRET) Proc Natl Acad Sci U S A. 2000;97:3684–3689. doi: 10.1073/pnas.060590697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mercier JF, Salahpour A, Angers S, Breit A, Bouvier M. Quantitative assessment of beta 1- and beta 2-adrenergic receptor homo- and heterodimerization by bioluminescence resonance energy transfer. J Biol Chem. 2002;277:44925–44931. doi: 10.1074/jbc.M205767200. [DOI] [PubMed] [Google Scholar]
- 29.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 30.Baril M, Racine ME, Penin F, Lamarre D. MAVS dimer is a crucial signaling component of innate immunity and the target of hepatitis C virus NS3/4A protease. J Virol. 2009;83:1299–1311. doi: 10.1128/JVI.01659-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim DH, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 32.Sabatini DM. mTOR and cancer: insights into a complex relationship. Nat Rev Cancer. 2006;6:729–734. doi: 10.1038/nrc1974. [DOI] [PubMed] [Google Scholar]
- 33.Yip CK, Murata K, Walz T, Sabatini DM, Kang SA. Structure of the human mTOR complex I and its implications for rapamycin inhibition. Mol Cell. 38:768–774. doi: 10.1016/j.molcel.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tunquist BJ, et al. Loss of AKAP150 perturbs distinct neuronal processes in mice. Proc Natl Acad Sci U S A. 2008;105:12557–12562. doi: 10.1073/pnas.0805922105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park J, et al. PcrA helicase dismantles RecA filaments by reeling in DNA in uniform steps. Cell. 142:544–555. doi: 10.1016/j.cell.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levene MJ, et al. Zero-mode waveguides for single-molecule analysis at high concentrations. Science. 2003;299:682–686. doi: 10.1126/science.1079700. [DOI] [PubMed] [Google Scholar]
- 37.Schulte R, Talamas J, Doucet C, Hetzer MW. Single bead affinity detection (SINBAD) for the analysis of protein-protein interactions. PLoS One. 2008;3:e2061. doi: 10.1371/journal.pone.0002061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taniguchi Y, Quantifying E, et al. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen I, Ting AY. Site-specific labeling of proteins with small molecules in live cells. Curr Opin Biotechnol. 2005;16:35–40. doi: 10.1016/j.copbio.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 40.Myong S, Rasnik I, Joo C, Lohman TM, Ha T. Repetitive shuttling of a motor protein on DNA. Nature. 2005;437:1321–1325. doi: 10.1038/nature04049. [DOI] [PubMed] [Google Scholar]
- 41.Leake MC, et al. Variable stoichiometry of the TatA component of the twin-arginine protein transport system observed by in vivo single-molecule imaging. Proc Natl Acad Sci U S A. 2008;105:15376–15381. doi: 10.1073/pnas.0806338105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ulbrich MH, Isacoff EY. Rules of engagement for NMDA receptor subunits. Proc Natl Acad Sci U S A. 2008;105:14163–14168. doi: 10.1073/pnas.0802075105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.