Abstract
Although genome-wide association studies (GWAS) are widely used to identify the genetic and environmental etiology of a trait, several key issues related to their statistical power and biological relevance have remained unexplored. Here, we describe a novel statistical approach, called functional GWAS or fGWAS, to analyze the genetic control of traits by integrating biological principles of trait formation into the GWAS framework through mathematical and statistical bridges. fGWAS can address many fundamental questions, such as the patterns of genetic control over development, the duration of genetic effects, as well as what causes developmental trajectories to change or stop changing. In statistics, fGWAS displays increased power for gene detection by capitalizing on cumulative phenotypic variation in a longitudinal trait over time and increased robustness for manipulating sparse longitudinal data.
Introduction
The past 3 years have witnessed a revolution in mapping the distribution of polygenes for human diseases and other complex traits by genome-wide association studies (GWAS) (Altshuler et al. 2008; Ikram et al. 2009; Psychiatric GCCC 2009; Hirschhorn 2009). This revolution has greatly inspired our hope that detailed genetic control mechanisms for complex phenotypes can be understood at the level of individual nucleotides or nucleotide combinations. Up until now, GWAS have reproducibly identified hundreds of loci, many of which affect the outcome of a disease or trait through its biochemical and metabolic pathways (Lettre and Rioux 2008; Mohlke et al. 2008; Hirschhorn and Lettre 2009; Shete et al. 2009; Styrkarsdottir et al. 2009; Turnbull et al. 2010). In the next few years, with ever-improving technologies for genotyping, GWAS methods will play a more important role in identifying genetic associations for complex traits and diseases, which may lead to new physiological or pathological hypotheses.
Despite its potential in genetic studies, there has been recognition of the limitations of using current GWAS approaches to elucidate a comprehensive genetic atlas of complex phenotypes. Some of the limitations include the following: (1) most GWAS have found genes that explain only small proportions of the genetic variance that occurs for many traits; (2) GWAS are based on a simple genotype– phenotype analysis, thus incapable of providing substantial knowledge about the biological and biochemical functions of significant genetic variants required for therapeutic applications; (3) GWAS in which phenotypes are assessed at a single time point cannot capitalize on full information of phenotypic expression, particularly in trials, in which phenotypic data are collected at irregular time intervals. Here, we argue that the power and biological relevance of GWAS can be enhanced by integrating the biological principle of trait formation into a general GWAS framework through mathematical or statistical functions. Such integration, leading to the birth of a new so-called functional GWAS (or fGWAS) approach, is shown to be able to address the limitations of classic GWAS methods.
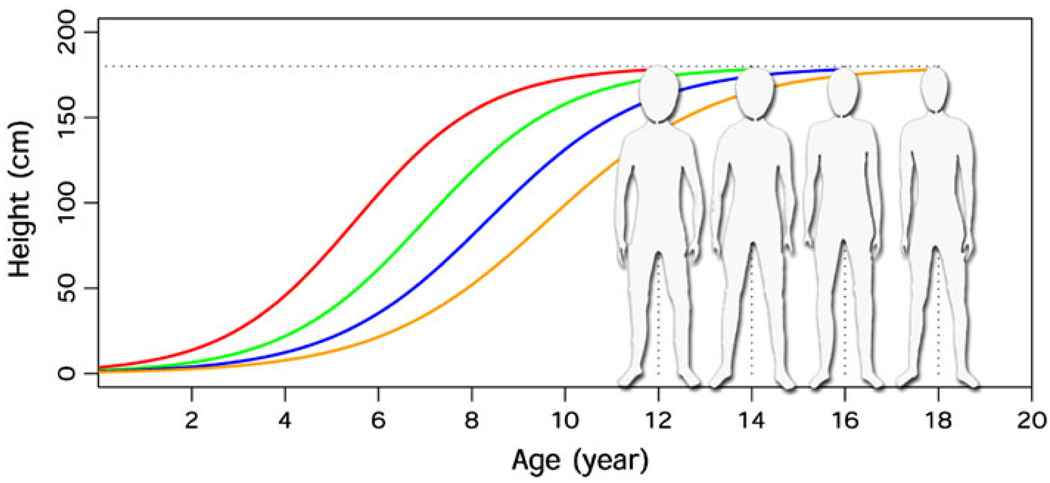
The central tenet of fGWAS is founded on the fact that every trait or disease develops over a period of time. Ignoring this developmental process reduces the power of GWAS. Elaborating on this argument, Fig. 1 provides an example in which four individuals show a similar body height at adult age. Thus, if GWAS are used to detect genes contributing to final height using adult data of these individuals, no genes would be detected with the end point data. However, because these individuals use a different amount of time to reach the same final height, there is considerable inter-individual variation in growth rate. Therefore, compared to the final height at a single adult age, the use of growth trajectories as a phenotype to conduct GWAS will remarkably increase the power for gene detection. In general, human growth includes several distinct phases that children pass through from birth to adult, i.e., infancy, childhood, and puberty (Thompson and Thompson 2009). The difference in overall growth curves should be due to differences in one or more of these phases of development; thus, GWAS of growth curves allows one to identify specific stages in which genes play a central role in governing growth rates. Given that different developmental phases include a particular set of hormonal signals and physiological functions, GWAS are a powerful tool to identify and study genes that control growth-phase specific biochemical pathways.
Fig. 1.
Final heights and growth curves of four subjects
Developmental genetics of complex traits is a science that has intrigued researchers for many decades. Traditional quantitative genetics can be integrated with developmental models to estimate the genetic variation of growth and development (Atchley and Zhu 1997). Meyer (2000) used random regression models to study the ontogenetic control of growth for animal breeding, whereas Kirkpatrick, Pletcher, and colleagues utilized the orthogonal, additive and universally practicable properties of Legendre polynomials to derive a series of genetic models for growth trajectories in the evolutionary context (Kirkpatrick et al. 1994a, b; Pletcher and Geyer 1999). These models have been instrumental in understanding the genetic control of growth by modeling the covariance matrix for growth traits measured at different time points. Ma et al. (2002) implemented a mixture-based method to map quantitative trait loci (QTLs) for developmental processes, greatly facilitating an understanding of the genetic and developmental regulation of trait formation (Cui et al. 2006; Wu and Lin 2006; He et al. 2010).
The integration of a biological process into GWAS will provide a useful means for studying developmental genetics by deciphering temporal mechanisms of genetic control of growth and development. Several fundamental questions in genetics can be addressed by fGWAS:
How does a specific gene affect the pattern of development? In other words, what is the temporal pattern of expression and action of genes that control growth and development of dynamic traits?
Are there different temporal patterns of genetic effects during development?
Do genes interact with each other and the environment in a web of interactive network to regulate growth and development?
Can we identify genes that exert pleiotropic effects of multiple traits during development?
fGWAS can capture genotypic differences at the level of phenotypic curves accumulated in the entire process of growth, thereby increasing the statistical power of gene detection. The statistical advantage of fGWAS can be strengthened when longitudinal data are measured irregularly, leading to data sparsity, a common phenomenon in a clinical trial. This type of phenotypic data, shown in Table S1, is characterized by three features: (1) each subject is measured at a limited number of time points, (2) time intervals are unevenly spaced for each subject, and (3) there are different schedules of measurement among subjects. Traditional GWAS have inherent limitations in their ability to handle such sparse longitudinal data. First, because only a small fraction of the subjects has measurements at a given time point, traditional GWAS based on a single time are unable to capitalize on all subjects, thus leading to biased parameter estimation and reduced gene detection ability. Second, individual subjects are measured at a few number of time points, limiting the fit of an informative curve.
In this manuscript, we describe the procedures we have implemented to evaluate the genetic, biological, and statistical merits of fGWAS using a GWAS data set from the Framingham Heart Study (FHS) (Dawber et al. 1951; Jaquish 2007; Fox et al. 2007). fGWAS is used to analyze and model the genetic control of age-specific body mass index (BMI), a highly heritable trait as a typical surrogate of obesity (Frayling 2007; Loos et al. 2008; Frayling et al. 2007; Scuteri et al. 2007). Significant SNPs are detected to affect the rates of change of BMI with ages.
Materials and methods
Study population
The Framingham Heart Study (FHS), a cardiovascular study based in Framingham, Massachusetts, is supported by the National Heart, Lung, and Blood Institute, in collaboration with Boston University. Beginning in 1948 with 5,209 healthy men and women of European descent aged 30–60, the FHS is now on its third generation of participants. This longitudinal project plays a central role in advancing our understanding of the epidemiological cause of hypertensive or arteriosclerotic cardiovascular disease and continues to exert important impacts on the molecular and genetic mechanisms for cardiovascular diseases. All subjects underwent a medical history and physical examination (including body height and weight), laboratory tests, and electrocardiography. Examinations have been repeated every two or more years although different subjects may have different numbers and time intervals of measurements. Recently, 550,000 SNPs have been genotyped for the entire Framingham cohort (Jaquish 2007; Fox et al. 2007), from which we chose 977 subjects for fGWAS analysis and modeling using highly inheritable body mass index (BMI). As it is standard practice, SNPs with rare allele frequency <10% were removed from the fGWAS analysis. The numbers and percentages of non-rare allele SNPs vary among different chromosomes and ranges from 4,417 to 28,771 and from 0.64 to 0.72, respectively. All the data were downloaded, with permission, from the NIH webpage from which ethics approval, and informed consent from all participants can be obtained.
Statistical model
In the fGWAS of clinical data sets, longitudinal traits are measured at irregular and possibly subject-specific time points (see Table S1). Let yi = (yi(ti1),…, yi(tiTi)) denote the vector of trait values for subject i measured at multiple times ti = (ti1,…, tiTi). These denotations capture subject-specific differences in the number and interval of time points. Consider a SNP A with two alleles A and a, generating three genotypes AA (coded by 1) with n1 observations, Aa (coded by 2) with n2 observations, and aa (coded by 3) with n3 observations. The phenotypic value of subject i at time tiτ (τ = 1,…, Ti) at this SNP is expressed as
| (1) |
where μj(tiτ) is the mean value for genotype j at time tiτ, β is a vector of regression coefficients of p covariates at time tiτ, xi is the p × 1 covariate vector for subject i, and ei(tiτ) and εi(tiτ) are the permanent and random errors at time tiτ, respectively, together called the residual errors.
With time-dependent genotypic values, we can estimate the additive (a) and dominant effects (d) of the SNP in a time course, expressed as
| (2) |
| (3) |
If the residual errors of any two subjects are independent, we have the likelihood of yi as
| (4) |
where fj(yi) is a multivariate normal distribution of the trait for subject i who carries SNP genotype j (j = 1, 2, 3), with subject-specific genotypic mean vectors
| (5) |
and subject-specific covariance matrix
| (6) |
In the covariance matrix (Eq. 6), the residual variance is composed of the permanent error variance due to the temporal pattern of longitudinal variables and the random error variance (also called the innovative variance) arising from random independent unpredictable errors. The relative magnitude of the permanent and innovative components is described by parameter ϕ. The covariance matrix (∑iP) due to the permanent errors contains autocorrelation structure that can be modeled, whereas the random errors are often assumed to be independent among different time points so that the random covariance matrix ∑iR is diagonal.
The first task for fGWAS involves modeling the mean vector (Eq. 5) for different SNP genotypes in a biologically and statistically meaningful way and modeling the longitudinal structure of covariance matrix (Eq. 6) in a statistically efficient and robust manner. In analyzing the BMI longitudinal data from the FHS project, we used a nonparametric approach based on Legendre orthogonal polynomials (LOP) to model time-dependent genotypic values.
For the given sparse longitudinal BMI data, we implemented a nonparametric approach based on Legendre orthogonal polynomials (LOP) to model age-specific genotypic values for each SNP genotype. The LOP are solutions to a differential equation, the Legendre equation:
Let Pr(x) denote the Legendre polynomial of order r. The polynomials are either even or odd functions of x for even or odd orders r. The general form of a Legendre polynomial of order k is given by the sum,
where k = r/2 or (r – 1)/2 whichever is an integer. This polynomial is defined over the interval [−1, 1] and is orthogonal in this interval in the sense that when r ≠ s. Therefore, the longitudinal phenotype should be scaled to the range [−1, 1] by
where tmin and tmax are the first and last time points, respectively. The application of Legendre orthogonal polynomials (LOP) in nonparametric regression is not a new idea that we propose. Since these polynomials are orthogonal to each other and integrate to 0 in the interval [−1, 1], they have been applied to nonparametric regression (Meyer 2000), with parameter estimates possessing favorable asymptotic properties (Huskova and Sen 1985; McKay 1997). In practice, the LOP have been used to model time-specific phenotypic or genetic variation for milk production (Meyer 2000) and plant growth traits (Cui et al. 2006; Lin and Wu 2006). An ordinary polynomial regression might also work in this case, but we incorporate LOP in a hope to capitalize its well-established effectiveness in nonparametric regression.
Define a family of LOP with a particular order r,
and a vector of genotypic base values
Then time-dependent genotypic values in Eq. 5 can be described as a linear combination of ur weighted by the family of LOP, i.e.,
| (7) |
By assuming different orders of genotypic base vectors, we can find the best order of LOP to fit the longitudinal data set using model selection criteria, such as BIC.
In practice, other nonparametric approaches, such as B-spline, can also be used in fGWAS (Yang et al. 2009). If the traits studied have an explicit mathematical function, such as logistic equations for plant and animal growth (West et al. 2001; von Bertalanffy 1957; Richards 1959; Guiot et al. 2003, 2006), triple-logistic equations for human body growth (Bock and Thissen 1976), then parametric approaches can be used to model the changes of genotypic values over time. Because these mathematical functions are derived from fundamental principles of biology, their incorporation into fGWAS will facilitate the biological interpretation of genetic results (Wu and Lin 2006). If trait formation includes multiple distinct stages, then it is crucial to derive a semiparametric model that combines the precision and biological relevance of parametric approaches and the flexibility of nonparametric approaches because some traits can be modeled parametrically in one phase, but not so in others. Such a semiparametric model was derived in Cui et al. (2006) and can be well implemented in the fGWAS framework.
Robust modeling of longitudinal covariance structure (Eq. 5) is a prerequisite for appropriate statistical inference of genetic effects on longitudinal traits. From longitudinal BMI plots (Fig. S1), we found that interpersonal variation tends to be constant over age. Thus, a stationary autoregressive model was used for the covariance structure by which only two parameters, variance and correlation, are needed to be estimated. If variance and correlation are not stationary, nonstationary approaches, such as the structured antedependence (SAD) model, can be used (Zimmerman and Núñez-Antón 2001; Zhao et al. 2005). More generally, autoregressive moving average models are used [e.g., ARMA(p,q)] because they are capable of handling of complex covariance structure (Li et al. 2001). It is important to determine its optimal order to model covariance structure. A model selection procedure has been established to determine the most parsimonious approach. We have implemented all these approaches into the fGWAS model, allowing geneticists to select an optimal approach for covariance structure for their longitudinal data.
We can also implement nonparametric and semiparametric approaches for longitudinal covariance structure. For sparse longitudinal data, the efficient estimation of covariance structure is a significant concern for detecting the genes that control dynamic traits.
In Table S1’s data structure, subjects are measured at a number of time points (1–3), with intervals and number depending on each subject. But all subjects are projected in a space with a full measure schedule (10). Semiparametric approaches by Fan et al. (2007) and Fan and Wu (2008) will be a better choice for covariance structure modeling in fGWAS.
Hypothesis tests
The significance test of the genetic effect of a SNP is a key for detecting significant genetic variants. This can be done by formulating the hypotheses as follows:
| (8) |
The likelihoods under the null and alternative hypotheses are calculated, from which the log-likelihood ratio (LR) is computed. The LR value is supposed to be asymptotically χ2-distributed with the degree of freedom equal to the difference in the numbers of unknown parameters under the H1 and H0. The significance of individual SNPs will be adjusted for multiple comparisons with a standard approach such as the false discovery rate (FDR).
We can also test the additive (H0: a(tiτ) = 0) and dominant effects (H0: a(tiτ) = 0) of a SNP after it is detected to be significant. Similarly, the LR values are calculated separately for each test and compared with critical values determined from the χ2-distribution.
Results and discussion
Gene detection
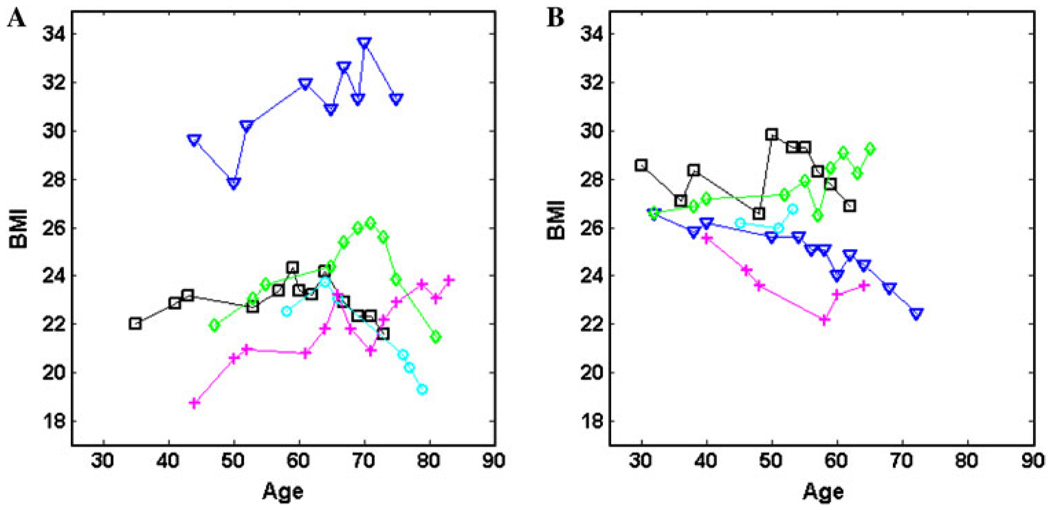
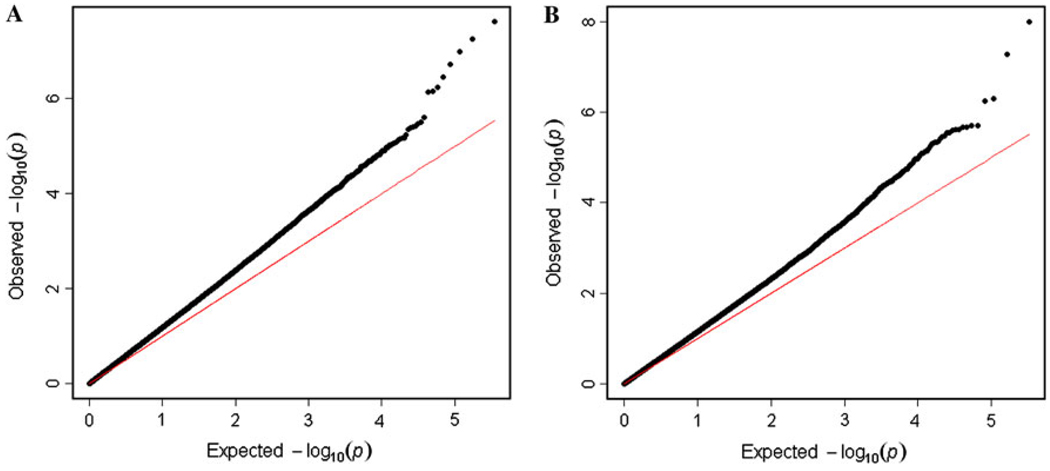
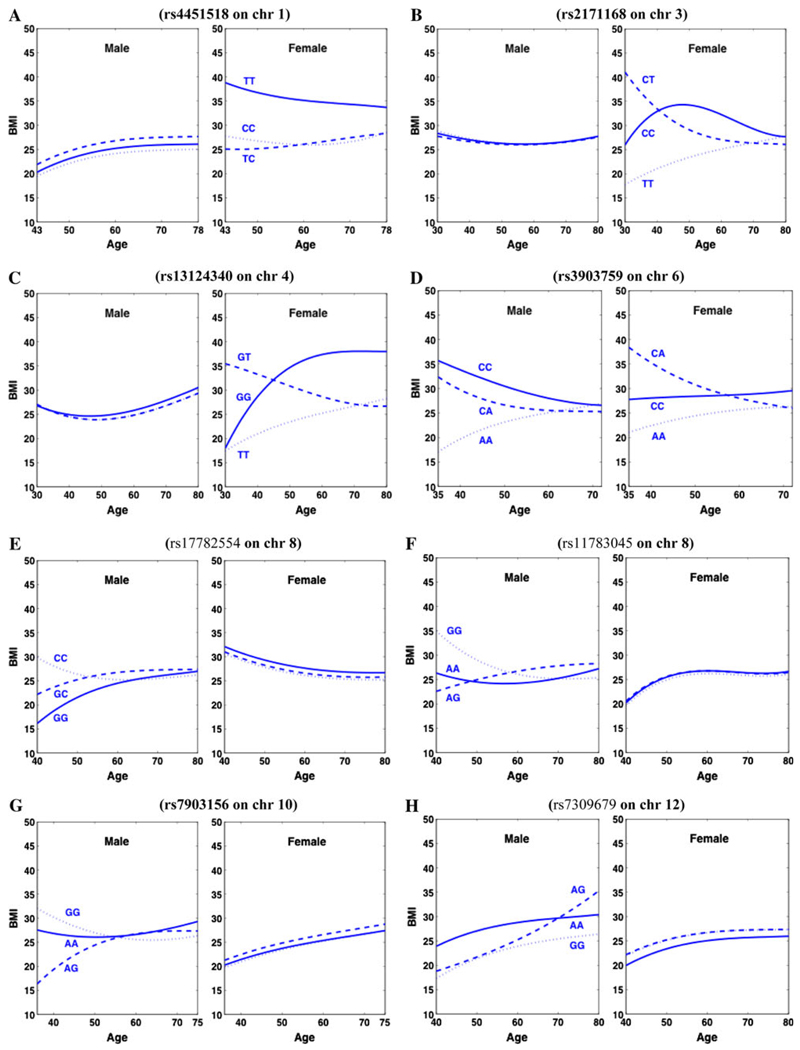
The phenotypic measurements of BMI for the FHS are collected on an irregular schedule. Although individual subjects have a few repeated measurements (1–19), collectively they display many time points (75). Figure 2 gives age-specific trajectories of BMI for five males and five females randomly drawn from the FHS data. We divided the population according to sex group and used Legendre orthogonal polynomials (LOP)-based nonparametric approach (Meyer 2000; Lin and Wu 2006) to model age-specific changes of BMI. For this given data set, in which variance tends to be constant over age (Fig. S1), a simple first-order autoregressive model was used for the longitudinal covariance structure within the fGWAS framework. It turns out that longitudinal curves were fitted most parsimoniously by the LOP of order 3 for both sexes (see Fig. S2 for the BIC plot). SNP loci with a significant effect on age-specific changes in BMI were detected on different chromosomes for the two sexes (Fig. S3). Since we have millions of SNPs to be tested, multiple testing procedures need to be applied. We avoid using Bonferroni’s adjustment since it is too conservative; we focus to control false discovery rate (FDR) by using Benjamini-Hochberg (1995) algorithm. After adjusting for multiple comparisons with FDR, 8 and 4 SNPs were significant at p < 10−6 for males and females, respectively. Table 1 provides detailed information about the names of these significant SNPs and their chromosomal locations, alleles, and significance levels. Figure 3a, b shows the observed (black) versus expected (red) p values in −log scale with base 10 for male and female population, respectively. For both the populations, most of the observed p values are above the red line (expected p values). We observe 8 extreme points for male and 4 for female which are far from the other points. These extremes are eight SNPs we detect as significant for male and four SNPs significant for female. The trajectory curves of BMI for different genotypes at each significant SNP are illustrated in Fig. 4.
Fig. 2.
Plots of BMI measured at different ages for five males (a) and five females (b) randomly sampled from the Framingham Heart Study. The sparsity and irregularity of longitudinal data for BMI are reflected by subject-specific uneven-spaced age intervals
Table 1.
Associations of SNPs with age-specific changes of BMI in different sexes
| Chr | Most significant SNP |
Position | Alleles | MAF | p value |
|---|---|---|---|---|---|
| Male-specific | |||||
| 8 | rs17782554 | 11059516 | G/C | 0.3984(C) | 7.079 × 10−7 |
| 8 | rs11783045 | 11093585 | A/G | 0.3843(G) | 1.862 × 10−7 |
| 10 | rs7903156 | 133498454 | A/G | 0.1197(G) | 5.888 × 10−7 |
| 12 | rs7309679 | 96235821 | A/G | 0.1402(G) | 3.548 × 10−7 |
| 17 | rs9915696 | 2688526 | G/C | 0.2596(C) | 2.340 × 10−8 |
| 18 | rs948716 | 73293243 | T/C | 0.1435(C) | 7.413 × 10−7 |
| 20 | rs747911 | 22580931 | C/T | 0.2505(T) | 1.047 × 10−7 |
| Female-specific | |||||
| 1 | rs4451518 | 180384945 | T/C | 0.1907(C) | 5.623 × 10−7 |
| 3 | rs2171168 | 122054294 | C/T | 0.2728(T) | 5.128 × 10−7 |
| 4 | rs13124340 | 125989067 | G/T | 0.1306(T) | 5.370 × 10−7 |
| Males/Females | |||||
| 6 | rs3903759 | 131562687 | C/A | 0.1645(A) | 5.620 × 10−8 |
The chromosomal positions, alleles, minor allele (in parentheses) frequencies (MAF), and significance levels of these SNPs are given
Fig. 3.
a p plot (expected vs. observed p values) for male population in Framingham Heart Study. b p plot (expected vs. observed p values) for female population in Framingham Heart Study
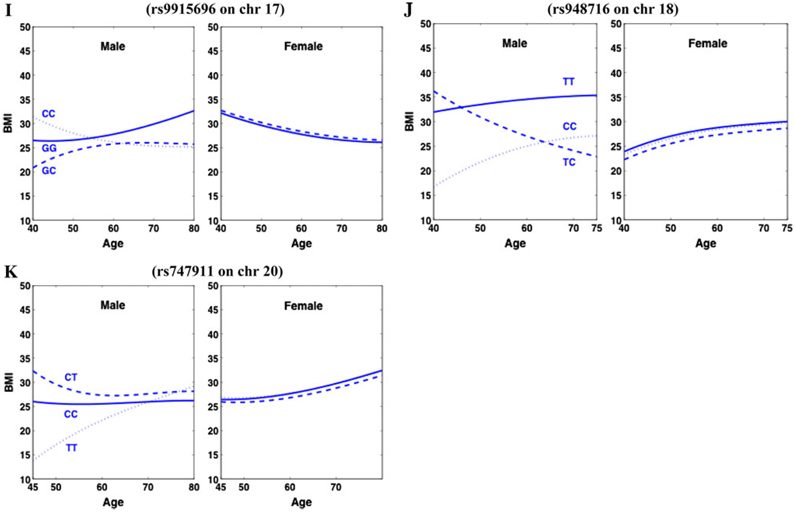
Fig. 4.
Age-specific trajectories of BMI in different sexes for three genotypes at each significant SNP detected from various chromosomes
Of all these significant SNPs detected, only one was found to affect BMI in both sexes, while the expression of all the others was sex-specific. Three genotypes at SNP rs4451518 on chromosome 1, rs2171168 on chromosome 3, and rs13124340 on chromosome 4 display strikingly different shapes of BMI curves in females, whereas genotypic curves at these SNPs are similar or overlap in males. There are notable discrepancies in BMI trajectories among three genotypes at SNP rs17782554 and rs11783045 on chromosome 8, rs7903156 on chromosome 10, rs7309679 on chromosome 12, rs9915696 on chromosome 17, rs948716 on chromosome 18, and rs747911 on chromosome 20 in males, but no differences were found in females. Such sex-specific expression should be one of the major causes of genotype × sex interactions for BMI trajectories. Unlike these SNPs, rs3903759 on chromosome 6 is expressed in both sexes but to different extents. The pattern of this sex-biased effect also contributes to genotype × sex interactions (Anholt and Mackay 2004).
Male-specific significant SNP, rs948716, is located on a similar region of the MC4R (melanocortin-4 receptor) gene on chromosome 18. Previous GWAS work using adult and child BMI data (also of European descent) detected a common SNP, rs17782313, mapped 188 kb downstream from MC4R (Loos et al. 2008). The results of our fGWAS analysis are in agreement with previous reports about the presence of common variants near MC4R which influences fat mass, weight and obesity risk. In a recent review on gene identification for type 2 diabetes, nine regions have been confirmed to harbor significant signals with this disease (Frayling 2007). These regions include two on chromosomes 3 and 10, respectively, and one on each chromosome 4, 6, 9, 12, and 16. We have noticed that a high proportion of SNP associations detected with fGWAS overlap with those detected on chromosomes 3, 4, 6, 10 and 12 by previous studies targeting type 2 diabetes. This is not surprising because BMI and type 2 diabetes are highly correlated (Frayling 2007). This observation also confirms the effectiveness of fGWAS.
In other GWAS for BMI (Frayling et al. 2007; Scuteri et al. 2007), significant noncoding SNPs were detected in an intron of the FTO (fat mass and obesity associated) gene on chromosome 16. Although fGWAS did not find highly significant SNPs on this chromosome, it did detect some SNPs with borderline significance (p < 10−4) in both sexes. fGWAS has identified several SNPs associated with BMI (Table 1) which have not been detected in previous studies, possibly showing unique power of this new approach. To show the biological relevance of this gained power, we searched for the presence of candidate genes within the 500 kb region in which SNPs associated with related traits have been reported by other groups (Table S2). The detailed biochemical functions of these genes are given in Table S3, many of which are related to energy intake, obesity, and cardiovascular diseases (http://www.ncbi.nlm.nih.gov/gene).
Temporal pattern of genetic control
Apart from increased power for gene detection, fGWAS can produce more biologically relevant results by studying the interplay between gene actions and trait progression. To show how the significant SNPs affect the change of BMI with age, we drew age-specific BMI curves for different genotypes at each significant SNP (Fig. 4). SNP rs3903759 on chromosome 6 triggers a pronounced effect on BMI in mid-age males and females, with the magnitude of the effect reducing with age and tending to be zero when entering old ages (Fig. 4d). Some SNPs, such as rs4451518 on chromosome 1 (Fig. 4a), rs13124340 on chromosome 4 (Fig. 4c), and rs948716 on chromosome 18 (Fig. 4j), actively affect BMI changes in middle and old ages.
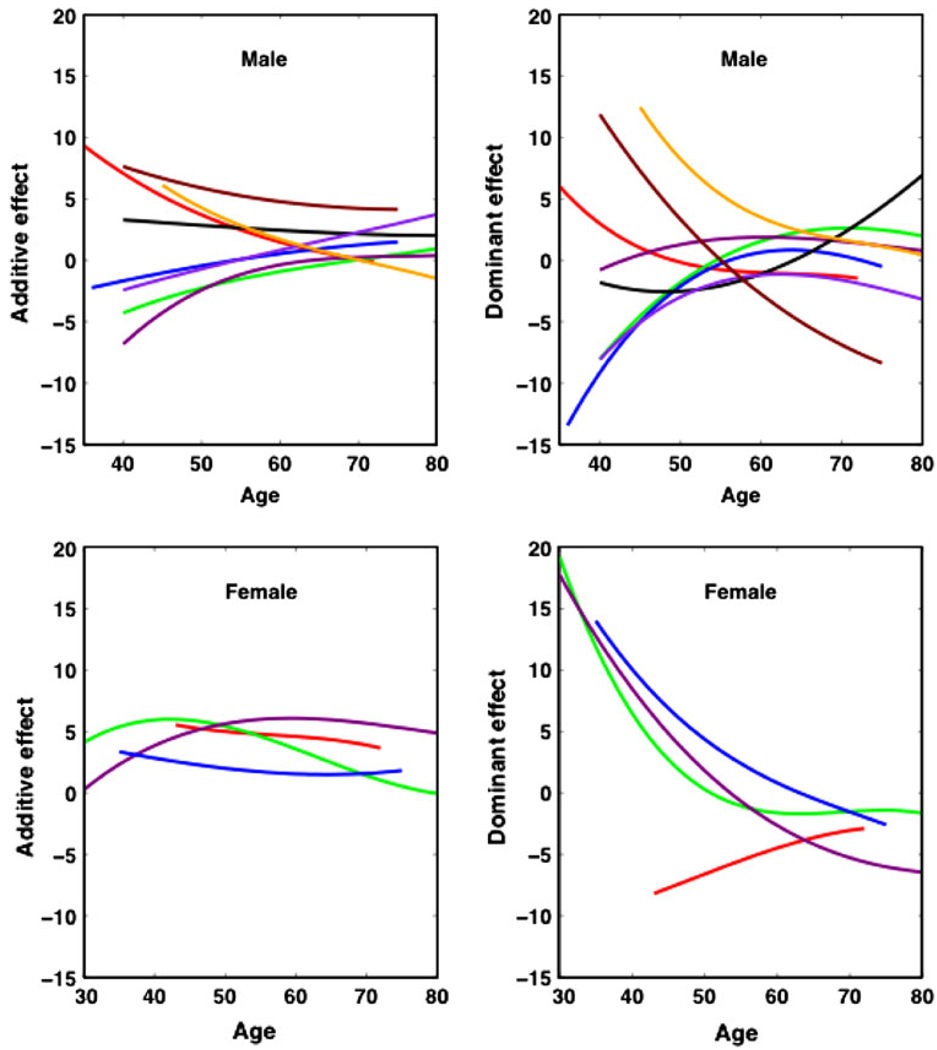
Based on three genotypes at each SNP, we drew the curves of additive and dominant genetic effects for age-specific changes of BMI (Fig. 5). The additive effect is defined as the homozygote for the common allele minus the homozygote for the minor allele. The dominance effect reflects the interaction between the common and minor alleles at the same SNP. In general, SNPs control age-specific changes of BMI in different patterns. In males, additive effects of all SNPs on BMI tends to reduce with age, but the age-specific change of dominant effects show a complicated pattern, with some SNPs altering the direction of their effects. In females, there are more remarkable dominant effects compared with additive effects, especially at middle ages. Overdominant effects were detected at middle ages. A quantitative genetic theory has been established to explain the molecular basis of dominance and overdominance in terms of metabolic pathways (Kacser and Burns 1981; Keightley and Kacser 1987).
Fig. 5.
Age-specific change of the additive and dominant genetic effects on BMI in different sexes for each significant SNP detected from various chromosomes. The additive effect is defined as the homozygote for the common allele minus the homozygote for the minor allele. The additive and dominant effects near zero suggest their absence
Power and FPR analysis
We performed a cross-validation analysis to test the reproducibility of results obtained from fGWAS. By randomly splitting all individuals in each sex into two groups of roughly the same size and analyzing each group with fGWAS, we obtained the same set of significant SNPs from each of these two groups and complete samples, suggesting an excellent reproducibility of our results. We performed simulation studies to examine the power of fGWAS to detect significant genes for longitudinal traits and the false-positive rates of the new approach. The data were simulated by assuming a segregating SNP with different minor allele frequencies (MAF) which are associated with a longitudinal trait. The trait was assumed to follow a sparse structure characterized by the FHS.
As shown in Table S4, fGWAS provides good estimates of the parameters that model genotypic curves using Legendre orthogonal polynomials and the autoregressive covariance structure. In general, a sample size of 1,000 is adequate for precise estimates of parameters for all genotypic curves when the MAF is 0.3 or larger. For those with MAF < 0.3, a larger sample size, say 2,000, is required. The power of gene detection estimated from the simulated data is about 0.8 or greater (Table 2), whereas the false-positive rates of fGWAS are less than 0.10 for a sample size of 1,000. Because our simulation was conducted by mimicking the FHS, it is likely that our power and FPR analyses reflect an actual case.
Table 2.
Theoretical power and false-positive rates (FPR) of fGWAS
| Sample size | MAF | Power | FPR |
|---|---|---|---|
| 1,000 | 0.1 | 0.79 | 0.094 |
| 0.2 | 0.78 | 0.098 | |
| 0.3 | 0.83 | 0.056 | |
| 0.4 | 0.80 | 0.056 | |
| 0.5 | 0.81 | 0.061 | |
| 2,000 | 0.1 | 0.84 | 0.083 |
| 0.2 | 0.83 | 0.088 | |
| 0.3 | 0.84 | 0.049 | |
| 0.4 | 0.81 | 0.055 | |
| 0.5 | 0.82 | 0.052 |
Conclusions
Genome-wide association studies with single nucleotide polymorphisms have proven to be a powerful tool for elucidating the role genetics plays in human health and disease. By analyzing hundreds of thousands of genetic variants in a particular population, this approach can identify the chromosomal distribution and function of multiple genetic changes that are associated with polygenic traits and diseases. Indeed, in the last 3 years, we have seen successful applications of GWAS in the study of complex traits and diseases of major medical importance such as human height, obesity, diabetes, coronary artery disease, and cancer (Altshuler et al. 2008; Ikram et al. 2009; Psychiatric GCCC 2009; Hirschhorn 2009; Lettre and Rioux 2008; Mohlke et al. 2008; Hirschhorn and Lettre 2009; Shete et al. 2009; Styrkarsdottir et al. 2009; Turnbull et al. 2010).
The successes and potential of GWAS have not been explored when complex phenotypes arise as a function of time. In any regard, a time function is more informative than a point in describing the biological or clinical feature of a trait. By integrating GWAS and functional aspects of dynamic traits, a new analytical model, called functional GWAS (fGWAS), can be naturally derived, which provides an unprecedented opportunity to study the genetic control of developmental traits. fGWAS is not only able to identify genes that determine the final form of the trait but also displays power to study the temporal pattern of genetic control in a time course. From a statistical standpoint, fGWAS capitalizes on the full information provided by growth and development of complex traits over time, increasing the power of gene identification. In particular, fGWAS is robust for handling longitudinal sparse data in which no single time point has the phenotypic data for all subjects facilitating the application of GWAS to study the genetic architecture of hard-to-measure traits.
With the completion of the Human Genome Project, it has been possible to draw a comprehensive picture of genetic control mechanisms of complex traits and processes and, ultimately, integrate genetic information into routine clinical therapies for disease treatment and prevention. To achieve this goal, there is a pressing need to develop powerful statistical and computational algorithms for detecting genes that determine dynamic traits. Unlike static traits, dynamic traits are described by a series of developmental processes composed of a large number of variables. In this article, we describe fGWAS, derived by integrating mathematical models for the molecular mechanisms and functions of biological processes into a likelihood framework. Next, we will need to extend fGWAS to consider the impact of genetic imprinting (Luedi et al. 2005) and copy number variants (McCarroll et al. 2008) to recover so-called missing heritability in GWAS (Bogardus 2009; Manolio et al. 2009). By then, we will be in an excellent position to ask and address any hypothesis tests at the interplay between genetics and developmental disorders.
Supplementary Material
Acknowledgments
This work is partially supported by grant DMS/NIGMS-0540745 to RW and NIDA, NIH grants R21 DA024260 and R21 DA024266 to RL. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIDA or the NIH.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00439-011-0960-6) contains supplementary material, which is available to authorized users.
Contributor Information
Kiranmoy Das, Department of Statistics, The Pennsylvania State University, University Park, PA, USA.
Jiahan Li, Department of Statistics, The Pennsylvania State University, University Park, PA, USA.
Zhong Wang, Department of Public Health Sciences, Pennsylvania State College of Medicine, Hershey, PA, USA.
Chunfa Tong, Department of Public Health Sciences, Pennsylvania State College of Medicine, Hershey, PA, USA.
Guifang Fu, Department of Statistics, The Pennsylvania State University, University Park, PA, USA.
Yao Li, Department of Statistics, West Virginia University, Morgantown, WV, USA.
Meng Xu, Department of Public Health Sciences, Pennsylvania State College of Medicine, Hershey, PA, USA.
Kwangmi Ahn, Department of Public Health Sciences, Pennsylvania State College of Medicine, Hershey, PA, USA.
David Mauger, Department of Public Health Sciences, Pennsylvania State College of Medicine, Hershey, PA, USA.
Runze Li, Department of Statistics, The Pennsylvania State University, University Park, PA, USA; Department of Public Health Sciences, Pennsylvania State College of Medicine, Hershey, PA, USA.
Rongling Wu, Email: rwu@hes.hmc.psu.edu, Department of Statistics, The Pennsylvania State University, University Park, PA, USA; Department of Public Health Sciences, Pennsylvania State College of Medicine, Hershey, PA, USA.
References
- Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anholt RRH, Mackay TFC. Genetic analysis of complex behaviors in Drosophila. Nat Rev Genet. 2004;5:838–849. doi: 10.1038/nrg1472. [DOI] [PubMed] [Google Scholar]
- Atchley WR, Zhu J. Developmental quantitative genetics, conditional epigenetic variability and growth in mice. Genetics. 1997;147:765–776. doi: 10.1093/genetics/147.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock RD, Thissen D. Fitting multi-component models for growth in stature; Proceedings of the 9th international biometrics conference; 1976. pp. 431–442. [Google Scholar]
- Bogardus C. Missing heritability and GWAS utility. Obesity. 2009;17:209–210. doi: 10.1038/oby.2008.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Zhu J, Wu RL. Functional mapping for genetic control of programmed cell death. Physiol Genomics. 2006;25:458–469. doi: 10.1152/physiolgenomics.00181.2005. [DOI] [PubMed] [Google Scholar]
- Dawber TR, Meadors GF, Moore FE., Jr Epidemiological approaches to heart disease: the Framingham Study. Am J Public Health Nations Health. 1951;41:279–286. doi: 10.2105/ajph.41.3.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Wu Y. Semiparametric estimation of covariance matrixes for longitudinal data. J Am Stat Assoc. 2008;103:1520–1533. doi: 10.1198/016214508000000742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Huang T, Li R. Analysis of longitudinal data with semiparametric estimation of covariance function. J Am Stat Assoc. 2007;102:632–641. doi: 10.1198/016214507000000095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox CS, Heard-Costa N, Cupples LA, Dupuis J, Vasan RS, et al. Genome-wide association to body mass index and waist circumference: the Framingham Heart Study 100K project. BMC Med Genet. 2007;8 Suppl 1:S18. doi: 10.1186/1471-2350-8-S1-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frayling TM. Genome-wide association studies provide new insights into type 2 diabetes aetiology. Nat Rev Genet. 2007;8:657–662. doi: 10.1038/nrg2178. [DOI] [PubMed] [Google Scholar]
- Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiot C, Degiorgis PG, Delsanto PP, Gabriele P, Deisboeck TS. Does tumor growth follow a “universal law”? J Theor Biol. 2003;225:147–151. doi: 10.1016/s0022-5193(03)00221-2. [DOI] [PubMed] [Google Scholar]
- Guiot C, Delsanto PP, Carpinteri A, Pugno N, Mansury Y, Deisboeck TS. The dynamic evolution of the power exponent in a universal growth model of tumors. J Theor Biol. 2006;240:459–463. doi: 10.1016/j.jtbi.2005.10.006. [DOI] [PubMed] [Google Scholar]
- He QL, Berg A, Li Y, Vallejos CE, Wu RL. Modeling genes for plant structure, development and evolution: functional mapping meets plant ontology. Trends Genet. 2010;26:39–46. doi: 10.1016/j.tig.2009.11.004. [DOI] [PubMed] [Google Scholar]
- Hirschhorn JN. Genomewide association studies—illuminating biologic pathways. New Engl J Med. 2009;360:1699–1701. doi: 10.1056/NEJMp0808934. [DOI] [PubMed] [Google Scholar]
- Hirschhorn JN, Lettre G. Progress in genome-wide association studies of human height. Horm Res. 2009;71:5–13. doi: 10.1159/000192430. [DOI] [PubMed] [Google Scholar]
- Huskova M, Sen PK. On sequentially adaptive asymptotically efficient rank statistics. Seq Anal. 1985;4:125–151. [Google Scholar]
- Ikram MA, Seshadri S, Bis JC, Fornage M, DeStefano AL, et al. Genomewide association studies of stroke. New Engl J Med. 2009;360:1718–1728. doi: 10.1056/NEJMoa0900094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaquish C. The Framingham Heart Study, on its way to becoming the gold standard for cardiovascular genetic epidemiology? BMC Med Genet. 2007;8:63. doi: 10.1186/1471-2350-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacser H, Burns JA. The molecular basis of dominance. Genetics. 1981;97:639–666. doi: 10.1093/genetics/97.3-4.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley PD, Kacser H. Dominance, pleiotropy and metabolic structure. Genetics. 1987;117:319–329. doi: 10.1093/genetics/117.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M, Hill W, Thompson R. Estimating the covariance structure of traits during growth and ageing, illustrated with lactation in dairy cattle. Genet Res. 1994a;64:57–69. doi: 10.1017/s0016672300032559. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M, Lofsvold D, Bulmer M. Analysis of the inheritance, selection and evolution of growth trajectories. Genetics. 1994b;124:979–993. doi: 10.1093/genetics/124.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettre G, Rioux JD. Autoimmune diseases: insights from genome-wide association studies. Hum Mol Genet. 2008;17:R116–R121. doi: 10.1093/hmg/ddn246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, McMurry T, Berg A, Wang Z, Berceli SA, Wu RL. Functional clustering of periodic transcriptional profiles through ARMA(p,q) PLoS One. 2001;5(4):e9894. doi: 10.1371/journal.pone.0009894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M, Wu RL. A joint model for nonparametric functional mapping of longitudinal trajectories and time-to-events. BMC Bioinformatics. 2006;7(1):138. doi: 10.1186/1471-2105-7-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos RJ, Lindgren CM, Li S, Wheeler E, Zhao JH, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedi PP, Hartemink AJ, Jirtle RL. Genome-wide prediction of imprinted murine genes. Genome Res. 2005;15:875–884. doi: 10.1101/gr.3303505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma CX, Casella G, Wu RL. Functional mapping of quantitative trait loci underlying the character process: a theoretical framework. Genetics. 2002;161:1751–1762. doi: 10.1093/genetics/161.4.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll SA, Kuruvilla FG, Korn JM, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- Mckay MD. Non-parametric variance based methods for assessing uncertainty importance. Reliab Eng Syst Saf. 1997;57:267–279. [Google Scholar]
- Meyer K. Random regressions to model phenotypic variation in monthly weights of Australian beef cows. Livest Prod Sci. 2000;65:19–38. [Google Scholar]
- Mohlke KL, Boehnke M, Abecasis GR. Metabolic and cardiovascular traits: an abundance of recently identified common genetic variants. Hum Mol Genet. 2008;17:R102–R108. doi: 10.1093/hmg/ddn275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletcher SD, Geyer CJ. The genetic analysis of age-dependent traits: modeling the character process. Genetics. 1999;151:825–835. doi: 10.1093/genetics/153.2.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psychiatric GCCC. Genomewide association studies: history, rationale, and prospects for psychiatric disorders. Am J Psychiatry. 2009;166:540–556. doi: 10.1176/appi.ajp.2008.08091354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards FJ. A flexible growth function for empirical use. J Exp Bot. 1959;10:290–300. [Google Scholar]
- Scuteri A, Sanna S, Chen WM, Uda M, Albai G, et al. Genomewide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3(7):e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shete S, Hosking FJ, Robertson LB, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009;41:899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styrkarsdottir U, Halldorsson BV, Gretarsdottir S, Gudbjartsson DF, Walters GB, et al. New sequence variants associated with bone mineral density. Nat Genet. 2009;41:15–17. doi: 10.1038/ng.284. [DOI] [PubMed] [Google Scholar]
- Thompson P, Thompson PJL. Introduction to coaching theory. UK: Meyer & Meyer Sport; 2009. [Google Scholar]
- Turnbull C, Ahmed S, Morrison J, Pernet D, Renwick A, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42:504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bertalanffy L. Quantitative laws for metabolism and growth. Q Rev Biol. 1957;32:217–231. doi: 10.1086/401873. [DOI] [PubMed] [Google Scholar]
- West GB, Brown JH, Enquist BJ. A general model for ontogenetic growth. Nature. 2001;413:628–631. doi: 10.1038/35098076. [DOI] [PubMed] [Google Scholar]
- Wu RL, Lin M. Functional mapping—how to study the genetic architecture of dynamic complex traits. Nat Rev Genet. 2006;7:229–237. doi: 10.1038/nrg1804. [DOI] [PubMed] [Google Scholar]
- Yang J, Wu RL, Casella G. Nonparametric functional mapping of quantitative trait loci. Biometrics. 2009;65:30–39. doi: 10.1111/j.1541-0420.2008.01063.x. [DOI] [PubMed] [Google Scholar]
- Zhao W, Chen YQ, Casella G, Cheverud JM, Wu R. A nonstationary model for functional mapping of complex traits. Bioinformatics. 2005;21:2469–2477. doi: 10.1093/bioinformatics/bti382. [DOI] [PubMed] [Google Scholar]
- Zimmerman D, Núñez-Antón V. Parametric modelling of growth curve data: an overview (with discussions) Test. 2001;10:1–73. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.