Abstract
One of the most studied fish pathogens is Vibrio anguillarum. Development of the genetics and biochemistry of the mechanisms of virulence in this fish pathogen together with clinical and ecologic studies has permitted the intensive development of microbiology in fish diseases. It is the intention of this review to compile the exhaustive knowledge accumulated on this bacterium and its interaction with the host fish by reporting a complete analysis of the V. anguillarum virulence factors and the genetics of their complexity.
Keywords: Vibrio anguillarum, virulence factor, genetics
Vibriosis is one of the most ubiquitous fish diseases caused by bacteria belonging to the genus Vibrio. Vibrio anguillarum is a Gram-negative halophilic bacterial pathogen that causes Vibriosois with lethal hemorrhagic septicimia in cultured and natural fish and shellfish (Actis et al., 2011).
Canestrini (1893) reported epizootics in migrating eels (Anguilla vulgaris) that associated to a bacterium named Bacillus anguillarum. Bergman (1909) first described V. anguillarum as the etiological agent of ‘Red Pest of eels’ in the Baltic Sea. The pathology of the disease as well as the characteristics of the bacterium in these two reports indicated that the etiological agents were the same bacteria. Vibriosis caused by V. anguillarum has been recognized as a major problem for salmonid culture due to the significant economical loses it causes in the fish industry (Schiewe et al., 1977; Trust et al., 1981; Winton et al., 1983; Toranzo et al., 2005; Actis et al., 2011). It was demonstrated that isolates of V. anguillarum displayed an obvious heterogeneity that led to the division of these vibrios into two separate biotypes, 1 and 2 (Harrell et al., 1976; Schiewe et al., 1977). Later, Schiewe et al. (1981) proposed a new species for the V. anguillarum biotype 2 named V. ordalii, based on cultural and biochemical characteristics, as well as DNA homology with biotype 1. There are twenty-three O serotypes that have been reported in V. anguillarum (Sorensen and Larsen, 1986; Grisez and Ollevier, 1995; Pedersen et al., 1999), and serotypes O1, O2 and O3 are mainly causative agents of fish Vibriosis (Toranzo and Barja, 1990; Larsen et al., 1994; Tiainen et al., 1997).
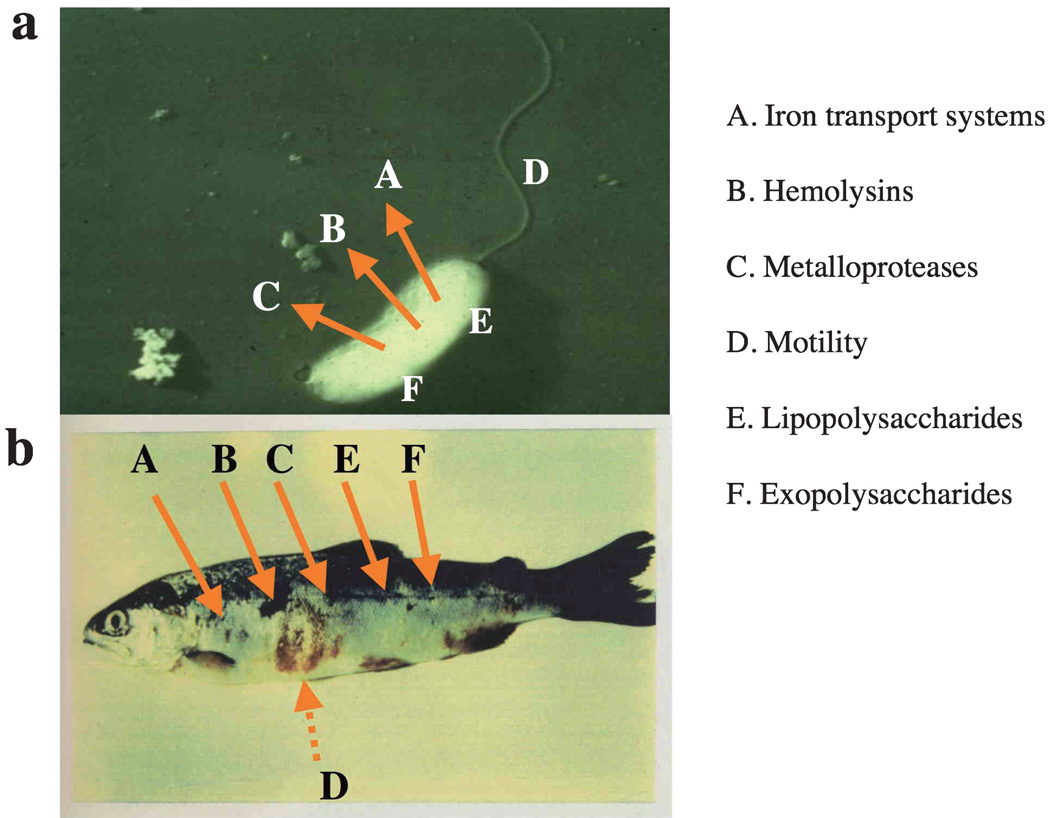
To prevent vibriosis many research groups have attempted to understand the virulence mechanisms of this bacterium. Recently, using molecular genetic approaches have resulted in a large increase in the available information on the virulence of this bacterium. Here we review the main virulence factors identified by molecular genetics as well as biochemical techniques in V. anguillarum. The list of V. anguillarum virulence factors is shown in Table 1, and their action is exemplified in Fig. 1.
Table 1.
List of V. anguillarum genes associated with virulence
| Category | Genes name | Functions or products |
|---|---|---|
| Iron transport systems | angA–E, G, H, M, N, R, T and U | Anguibactin biosynthesis |
| vabA–E | Anguibactin biosynthesis | |
| fatA–D | Anguibactin transport | |
| ttpC, tonB2, exbB2 and exbD2 | Energy transducer for anguibactin and heme transport | |
| huvA–D, S, X and Z | Heme transport | |
| tonB1, exbB1 and exbD1 | Energy transducer for heme transport | |
| Hemolysins | rtxA | Hemolytic and cytotoxic activity |
| vah2–5 | Hemolytic activity | |
| mltD | Hemolytic, phospholipase, gelatinase and diastase activity | |
| Metalloproteases | empA | Hemolytic and cytotoxic activity |
| prtV | Gelatinase, protease, and glycosidase activity | |
| Motility | flaA–E | Flagellin |
| flgI | Flagellar P-ring protein | |
| flgB | Flagellar rod protein | |
| flgL | Flagellar hook-associated protein HAP3 | |
| flhA | Flagellum export protein | |
| rpoN | Sigma-54 subunit that affects expression of flagellin subunits | |
| cheR | chemotaxis | |
| motY | Stator for the sodium-driven motor of the polar flagella | |
| Lipopolysaccharide | rmlA–D | O1 side chain biosynthesis |
| virA (wbhS), virB (wbhR) and virC | O-antigen biosynthesis | |
| Exopolysaccharide | wza–c, wbfB–D, orf1 | Exopolysaccharide biosynthesis |
Fig. 1.
Production of V. anguillarum virulence factors and action on the host fish. (a) Virulence factors of V. anguillarum. Arrows indicate that virulence factors are secreted outside of bacterial cell. (b) Location where virulence factors affect the host fish during infection. Arrows indicate that virulence factors aid in the attachment or to go through the fish integument and/or inside the fish, while the dotted arrow indicates that the virulence factor only attaches to the fish integument.
Iron transport systems
Iron is an essential metal for nearly all living organisms since it is indispensable for their metabolism. However the available free iron in the host environment is extremely limited because the majority of the iron inside the host is strongly bound to host factors such as transferrin and lactofrerrin thus pathogenic bacteria need to harbor high affinity systems to acquire iron from those complexes to survive and multiply inside the host and cause disease.
V. anguillarum virulence is associated with the presence of a plasmid-mediated iron uptake system (Crosa, 1980). The 65-kilobasepairs (kb) pJM1 plasmid that presents in the serotype O1 strain 775 encodes a very efficient iron uptake system mediated by the siderophore anguibactin. This is the most well studied and important virulence factor of V. anguillarum. The siderophore anguibactin is a small molecular weight peptide (348 Da) and it steals iron from the transferrin-iron complex. Many O1 strains of V. anguillarum carry the 65kb virulence plasmid pJM1 or pJM1-like plasmids. The pJM1 plasmid cured strain shows dramatically lower virulence than the wild type strain (the difference of LD50 is > 104) thus the pJM1 plasmid is an essential virulence factor for V. anguillarum to cause diseases. The pJM1 plasmid encodes the majority of the anguibactin biosynthesis as well as transport genes. In the pJM1 plasmid, angB, angC, angD, angE, angG, angH, angM, angN, angR, angT and angU are involved in anguibactin biosynthesis (Crosa, 1980; Tolmasky et al., 1988; Wertheimer et al., 1999; Welch et al., 2000; Crosa and Walsh, 2002; Di Lorenzo et al., 2003; Di Lorenzo et al., 2004; Liu et al., 2004; Alice et al., 2005; Liu et al., 2005; Di Lorenzo et al., 2008; Naka et al., 2008). The fatA gene encodes the cognate receptor for ferric anguibactin, and fatB, fatC and fatD encode an iron ABC transporter that is essential to lead the ferric siderophore from the periplasmic space to the cytosol (Actis et al., 1985; Actis et al., 1988; Koster et al., 1991; Actis et al., 1995; Lopez and Crosa, 2007; Lopez et al., 2007; Naka et al., 2010). In addition to the pJM1 plasmid, some of the anguibactin biosynthesis genes such as vabA, vabB, vabC, vabD and vabE are located in the chromosome, and angC and vabC, angD and vabD, and angE and vabE have been shown to be functional homologues (Alice et al., 2005; Naka et al., 2008). It has been shown that the plasmid pJM1-cured strain of V anguillarum can neither transport the ferric anguibactin complex nor does it colonize the skin (or colonized only 5–10% of the fish) although it colonizes the intestine, and shows a decrease in motility through the mucus as compared with the wild type strain (Weber et al., 2010). The authors also found that this plasmidless strain is more sensitive to lysozyme than the wild type strain.
pJM1-less O1 strains as well as O2 serotype strains produce a chromosome mediated siderophore vanchrobactin (Lemos et al., 1988; Soengas et al., 2008). The chromosomal locus encoding vanchrobactin biosynthesis (vabA, vabB, vabC, vabD, vabE, vabF and vabG), transport (fvtA), secreting (vabS) genes have been identified and characterized (Balado et al., 2006; Balado et al., 2008; Naka et al., 2008; Balado et al., 2009). The vanchrobactin system is important to grow in iron limiting conditions although it has not been tested whether vanchrobactin is important for virulence of V. anguillarum (Balado et al., 2006; Balado et al., 2008). Furthermore, this vanchrobactin system has also been found in the serotype O1 anguibactin producer harboring pJM1, but in this case one of the vanchrobactin biosyntheis gene vabF was interrupted by a transposon originated from pJM1 (Naka et al., 2008). It has been proposed that an anguibactin producer was generated evolutionary because the anguibactin system is better fit to deal with the ability to scavenge iron as compared to the vanchrobactin system (Naka et al., 2008).
Furthermore, tonB2 mediated energy transport systems consisting of ttpC, tonB2, exbB2 and exbD2 that are required for ferric-anguibactin and exogenous siderophore uptake are also essential virulence factors for V. anguillarum (Stork et al., 2004; Stork et al., 2007; Kuehl and Crosa, 2009; Kuehl and Crosa, 2010).
In addition to siderophore mediated iron transport system V. anguillarum also harbors another iron transport system, the heme system by which V. anguillarum can possibly acquire iron from heme-containing proteins that exist in host. The outer membrane heme receptor huvA gene has been identified, and an huvA mutant has growth defects under iron limiting condition in the presence of hemoglobin and shows lower virulence than the wild type strain in turbot pretreated with hemin (Mazoy et al., 2003). Furthermore, it was found that the huvA gene exists as linked to the tonB1 cluster consisting of huvA, huvZ, huvX, tonB1, exbB1, exbD1, huvB, huvC and huvD, and the deletion mutant of huvA, huvZ, huvX, huvB, huvC and huvD showed hemin transport defect (Mourino et al., 2004). It has been demonstrated that V. anguillarum can use tonB1 or tonB2 systems for heme transport (Stork et al., 2004). In addition to huvA there is another heme receptor gene, huvS, identified in V. anguillarum, and it has been shown that the existence of huvA and/or huvS depend on the strains examined (Mourino et al., 2005).
Hemolysins
A hemolysin gene from V. anguillarum, vah1, a homologue of V. cholerae EI Tor hemolysin was first reported by Hirono et al. (1996). The product of this gene showed strong hemolytic activity to erythrocytes from fish such as carp and rainbow trout albeit lower activity to mammalian erythrocytes such as sheep, rabbit, bovine and horse. Rock and Nelson (2006) found that the mutation of plp, a phospholipase gene located downstream of vah1 increased two- to threefold the hemolytic activity of V. anguillarum suggesting that the plp gene product must repress or destabilize the vah1 transcripts. Furthermore, the upstream gene of vah1 was annotated as a putative lactonizing lipase gene, llpA. Mutation of vah1 or llpA in the plp mutant background decreased the hemolytic activity of the plp mutant to the wild type level. From those results they concluded that at least these three genes, vah1, plp and llpA, are involved in one of the hemolytic activities of V. anguillarum. Moreover, they also found that the highest hemolytic activity was detected in the exponential growth phase where the highest expression of vah1 and plp also occurred. An insertion mutant obtained by integrating a suicide vector in vah1, attenuated V. anguillarum virulence for juvenile Atlantic salmon, even though the mutant still showed hemolytic activity similar to the wild type. Furthermore, insertion mutations in llpA or plp did not affect the virulence of V. anguillarum. Later on, the same group demonstrated that the vah1 deletion mutant, that was constructed using the same parent strain and carrying an insert of the kanamycin resistance gene, did not show a reduction of the V. anguillarum virulence in the same animal model (Li et al., 2008). This could be due to the fact that the vah1 insertion mutant that Rock and Nelson (2006) constructed caused a polar effect, or there was a secondary mutation. Since the deletion mutant would also affect downstream genes in the same operon albeit with a lower effect it seems that the vah1 single mutation does not directly affect V. anguillarum virulence. Li et al. (2008) found that the residual hemolytic activity of the vah1 mutant is due to the existence of another hemolysin gene, rtxA which is highly related to the repeat in toxin genes that are virulence factors of different Vibrio species such as V. cholerae and V. vulnificus (Lee et al., 2007; Liu et al., 2007; Olivier et al., 2009). The rtxA single insertion mutant still shows hemolytic activity whereas no hemolytic activity was observed in the double vah1 and rtxA mutant. Furthermore, they discovered that both vah1 and rtxA participate in the cytotoxicity to Atlantic salmon kidney (ASK) cells. They determined that the RtxA toxin is necessary to cause cell rounding of the ASK cells, while Vah1 causes their vacuolation. Moreover, they demonstrated that RtxA is a major virulence factor of V. anguillarum as determined using intraperitoneal injection in juvenile Atlantic salmon. In addition to Vah1, four additional hemolysins (Vah 2–5) have been reported (Rodkhum et al., 2005). It was of interest that E. coli expressing each hemolysin gene showed hemolytic activity on rainbow trout blood agar indicating that those hemolysins can be exported outside of the E. coli cells. Furthermore, the purified hemolysin proteins showed hemolytic activities not only against fish erythrocytes, but also for sheep and rabbit red cells. Each deletion mutant of vah 2–5 showed a reduction of virulence against rainbow trout as compared with the parent strain (low virulence strain cured of the pJM1 plasmid) indicating that each of the hemolysins contributes to the virulence of V. anguillarum. It is also clear from these studies that Vha4 is the most important for the virulence of V. anguillarum. However, all of the single mutants still show hemolytic activity, even though the activity is less than the wild type strain. Xu et al. (2010) identified the mltD gene, a homologue of E. coli mltD encoding a membrane-bound lytic murein transglycosylase D. The mltD mutant showed reduction in extracellular protease as well as gelatinase activity, and a total loss of hemolytic activity. They also found that the mutation in mltD resulted in better growth at higher concentration of sodium chloride and an increase antibiotic resistance i.e. to aminoglycosides, cephalosporines and penicillins as compared with the wild type strain. The purified MltD showed hemolytic, phospholipase, gelatinase and diastase activities. The mltD mutant showed higher virulence to zebrafish than the wild type strain indicating that mltD somehow affects the virulence of V. anguillarum.
Metalloproteases
Norqvist et al. (1990) found that a spontaneous rifampicin resistance mutant of V. anguillarum showed lower virulence, to rainbow trout as compared with the wild type. Comparison of the rifampicin mutant with the wild type revealed that the mutant also shows much lower protease activity than the wild type. The purified protease was 36 kDa and required Zn2+ for its activity and Ca2+ for its stability. Farrell and Crosa (1991) also identified and purified a 38 kDa protease from V. anguillarum. The protease was proposed to be a metalloprotease since the protease activity was inhibited by EDTA and 1,10-phenathroline, while classical inhibitors of serine, cysteine, and acid proteases did not inhibit the protease activity.
The empA gene encoding the metalloprotease was cloned and sequenced from V. anguillarum NB10 (Milton et al., 1992). EmpA consists of 611 amino acids with a molecular weight of approximately 66,700. This 611-amino-acid polypeptide may contain a putative signal sequence, a putative leader peptide and a mature protein. The signal peptide cleavage site was predicted in the Ala-25-Ala-26. The mature protein consists of 411 amino acids with a calculated molecular weight of 44,600. The purified protein was previously demonstrated to have a molecular mass of approximately 36 kDa, thus the mature protein is most likely processed a third time. The V. anguillarum metalloprotease EmpA was 69.3% and 47.3% identical to the V. cholerae HA/protease and the Pseudomonas aeruginosa elastase, respectively. Putative zinc-binding as well as other active sites were well conserved (greater than 30%) as compared with numerous other bacterial metalloproteases. The empA mutant constructed by integrating a suicide vector in the empA gene showed much lower proteolytic activity on 2% gelatin agar indicating that EmpA is responsible for the V. anguillarum protease activity. Varina et al. (2008) demonstrated that protease Epp processes a 46 kDa pro-EmpA to a 36 kDa mature EmpA by removing a peptide of about 10 kDa. The epp gene is located in the locus that includes the vah1 gene coding for the Vah1 hemolysin and this cluster is considered to be a pathogenicity island of V. anguillarum. They also showed that Epp can be secreted independently of EmpA, and secreted Epp and EmpA interact in the extracellular environment.
Yang et al. (2007a) performed mutational analysis of EmpA and found that mutation in the conserved residues, His346, His350, Glu347 and Glu370, possibly involved in the zinc-binding sites and the active center of EmpA resulted in an almost complete loss of proteolytic activity as well as cytotoxicity against a flounder gill cell line. On the other hand the mutation in the predicted substrate-binding sites (Tyr361, His429 and Asp417) caused smaller reductions in both proteolytic activity and cytotoxicity indicating that mutations in the substrate-binding sites only affect the substrate specificity rather than the proteolytic activity. Zhang et al. (2006) demonstrated that EmpA is exported into the periplasm by using the Sec system (type II secretion system) when it is expressed in E. coli.
Milton et al. (1992) tested the effect of EmpA in V. anguillarum virulence using immersion and intraperitoneal injection. The immersion LD50s were 2 × 102 bacteria/ml for the wild type and 1 × 104 bacteria/ml for the empA mutant, while the intraperitoneal LD50s were one bacterium for the wild type and 20 bacteria for the empA mutant suggesting that empA is somewhat involved in the virulence of V. anguillarum. Denkin and Nelson (2004) performed virulence tests for V. anguillarum NB10 and M93Sm and their empA mutants using intraperitoneal and anal intubation in Atlantic salmon. They only found a reduction of the virulence when the anal intubation route was used for the empA mutant of M93Sm, while the reduction of virulence was observed in both intraperitoneal and anal intubation in case of the empA mutant of NB10. These data suggest that EmpA is a virulence factor during the infection of the gastrointestinal tract of Atlantic salmon by V. anguillarum. Yang et al. (2007b) purified from E. coli a recombinant V. anguillarum EmpA (rEmpA) with a His-tag. They showed that rEmpA displays cytotoxicity to flounder gill cells. Intraperitoneal injection of the purified rEmpA into turbot caused hemorrhage in the peritoneal cavity, necrotic signals at the site of injection in most injected fish, and death.
Another type of metalloprotease PrtV that is a homologue of V. cholerae PrtV has been identified and characterized (Mo et al., 2010). These authors demonstrated that a prtV mutant shows lower enzymatic activities for gelatinase on gelatin agar, protease against azocasein, and glycosidases such as alkaline phosphatase, lucine arylamidase, trypsin and N-acetyl-β-glucosaminidase, while higher activities for esterase and esterase/lipase as compared to the parent strain. The prtV mutant also displayed a lower growth rate in turbot intestinal mucus, and showed lower virulence (at least one log difference) to turbot when the intraperitoneal route was used for inoculation
Motility
The correlation between flagellum and virulence in V. anguillarum has been proposed (Chart, 1983; Norqvist and Wolf-Watz, 1993). Four flagellin genes encoding FlaABCD and a potential flagellin gene encoding FlaE were identified in V. anguillarum NB10 (McGee et al., 1996; Milton et al., 1996). Those genes are located within two clusters, flaEDB and flaAC, and the organization of the two locus are very similar to the V. parahaemolyticus flagellin cluster. Mutations in each gene except for flaE showed a loss of the particular flagellin particle, but obvious structural loss in mutants was not observed. The full deletion of each gene showed that flaA-full gene deletion resulted in almost 50% decrease of motility, while each flaBCDE mutants showed only less than 15% decrease in motility. None of the full gene deletions affected virulence using an intraperitoneal route. However, when immersion challenge was used the flaA-full gene deletion mutant showed a decrease of approximately 103 in virulence, while each flaBCDE mutant showed only a slight decrease in virulence (approximately 10 to 102). Those results indicate that V. anguillarum requires flagellae in the process of invasion but once they passed the integument flagella are no longer required.
O’Toole et al. (1996) identified non-motile mutants by using transposon mutagenesis. Genes interrupted by the transposon are homologues of rpoN encoding the sigma-54 subunit, flgI encoding the flagellar P-ring protein, flgB encoding the flagellar rod protein, flgL encoding the flagellar hook-associated protein HAP3, and flhA encoding one of the components of the flagellum export protein.
The mutations in flgB, flgL or flhA caused a reduction of virulence of 102 to 103 when using the immersion route, while there were no differences using the intraperitoneal route. The flgI mutant showed dramatically lower virulence in both immersion and intraperitoneal routes of infection.
The flgI or flgB mutants showed expression of at least one of the flagellin subunits FlaA and FlaB, whereas the flgL mutant shows wild-type production of flagellin subunits whereas the flhA mutant did not produce any flagellin. Electron microscopic observations revealed that the flgI, flgB or flhA mutants did not show any flagellum, while the flgL mutant showed a short cylindrical projection from one of the poles of the bacterium. Furthermore, they also identified a chemotaxis gene cheR located in upstream of flgB. Motility assays in broth showed that the cheR mutant swarmed rapidly but it did not exhibit the wild type tumbling motion associated with re-orientation of the direction of swimming. When soft agar was used for the assay, the cheR mutant showed similar phenotype with the aflagellate mutants although having wild-type expression of the flagellin subunits, and the presence of wild-type flagellae was confirmed by electron microscopy. Those results indicated that the cheR mutant possesses a flagellate non-chemotactic phenotype. Furthermore, they found that the cheR mutant exhibited a reduction of virulence of approximately 102 in immersion but not in intraperitoneal injection.
Ormonde et al. (2000) tested whether the mutation in cheR affects the adherence to CHSE cells, and found that the cheR mutant shows stronger adherence to the CHSE cells than the wild type strain. Thus the smooth swimming phenotype of the cheR mutant may increase the chance for V. anguillarum to find the attachment site on the cells.
O’Toole et al. (1999) isolated skin and intestinal mucus from rainbow trout, and showed that the wild type V. anguillarum displayed stronger chemotactic response to intestinal mucus than to skin mucus. The chemotactic response to both type of mucus was abolished when the cheR gene was mutated. By analyzing components of the intestinal mucus they demonstrated that free amino acids (glutamic acid, glutamine, glycine, histidine, isoleucine, leucine, serine and threonine), carbohydrates (fucose, glucose, mannose and xylose) and lipid compounds (bile acids such as taurocholic acid and taurochenodeoxycholic acid) induce chemotactic responses. They also found that the concentration of those amino aids and carbohydrates identified as chemoatractants were much lower in skin mucus than in intestinal mucus suggesting an explanation for why intestinal mucus causes stronger chemotactis than the skin mucus. Furthermore, the mixture of all chemoattractants identified from intestinal mucus reconstituted a high level of chemotactic activity similar to that present in the intestinal mucus homogenate, while none of the single compounds showed similar level of chemotactic activity to the homogenate.
The importance of the rpoN sigma factor gene for V. anguillarum motility was also determined by screening of non-motile transposon mutants (O’Toole et al., 1996). The rpoN gene was cloned and the nucleotide sequence obtained (O’Toole et al., 1997). They found that the V. anguillarum rpoN gene consists of 1,458 bp and potentially encodes 486 amino acids. RpoN exhibited strong homology to the alternative sigma-54 factor of many bacteria. The rpoN mutant did not express any flagellin subunits and it also lost the motility phenotype (O’Toole et al., 1996; O’Toole et al., 1997). The mutant showed an attenuation of virulence as compared with the wild type by immersion challenge but not by the intraperitoneal route suggesting that RpoN does not regulate virulence genes that are necessary after passing the integument (O’Toole et al., 1997).
Ormonde et al. (2000) identified the motY gene encoding a stator for the sodium-driven motor of the polar flagella. Mutations in the motY gene did not affect the structure of the flagellum, even that the mutant was non-motile. The motY mutant of V. anguillarum showed a 750-fold decrease in virulence in rainbow trout only by the immersion assay. Those results are very similar to the results observed with flagellum mutants. Therefore motility but not flagellin proteins are necessary for virulence of V. anguillarum. They also observed that the flagellum might not affect the adherence to the CHSE cells. The invasion analysis of the CHSE cells using either wild type or the mutants motY or flaA, demonstrated that motility might increase the frequency of invasion by increasing the chance of collision between the pathogen and the host cells. However, the flhA mutant missing the flagellum showed similar value to the wild type indicating that the flagellum might physically hinder the internalization of the nonmotile bacterium.
Lipopolysaccharides (LPS)
Aoki et al. (1985) demonstrated that V. anguillarum PT479 isolated from ayu (Plecoglossus altivelis) exhibits high virulence to ayu when it was passed seven times through ayu, while it exhibited low virulence when it was cultured 20 times in brain heart infusion medium. The difference of these high- and low-virulent strains was found in the LPS structure, the high-virulence strain possessing an extra high molecular weight band. Furthermore, they observed that the high-virulence strain transports iron more efficiently than the low-virulent strain.
A cluster consisting of four genes, rmlBADC, predicted to function in the biosynthesis of the O side chain precursor dTDP-rhamnose from its own precursor glucose 1-phosphate has been identified in the V. anguillarum O1 serotype strain 531A (pJHC1, a pJM1-like plasmid) (Welch and Crosa, 2005). A polar mutant in rmlC and a non polar mutant in rmlD resulted in the loss of the O1 side chain as well as loss of siderophore anguibactin mediated iron transport. Further studies using the rmlD mutant unveiled that the loss of ferric-anguibactin transport was due to instability of the ferric anguibactin outer membrane protein FatA in the rmlD mutant. The rmlD gene was also essential for the resistance of V. anguillarum to the bactericidal action of nonimmune serum from rainbow trout. From those results it was concluded that the lipopolysaccharide O1 side chain is indispensable for serum resistance as well as anguibactin-mediated iron transport (Welch and Crosa, 2005).
Norqvist and Wolf-Watz (1993) constructed a Tn5–132 transposon library of V. anguillarum 775.17B and they screened the mutants using an experimental infection of rainbow trout. They found that two strains, VAN20 and VAN70, showed lower virulence as compared with the wild type. VAN70 was identified as the Tn5–132 transposon inserted in the virB gene. They also identified another gene, virA in the same locus. Mutations in virA or virB caused an attenuation of the virulence of V. anguillarum by both intraperitoneal and immersion experimental infections. They also determined that both the virA and virB mutants did not react with a rabbit polyclonal antibody against whole cells of the wild type strain indicating that both the virA and virB products are major surface antigens, and found that the difference between wild type and mutants are in the LPS profiles. The virA and virB genes have been identified in different serotype O1 strains, while no expression was detected by Western blot analysis in O2 serotype strains. The mutation in each of the genes in an O1 clinical strain showed reduction of virulence as observed in 775.17B indicating that virA and virB are serotype O1 specific genes. Jedani et al. (2000) reported that virA and virB are located in the rfb region of V. anguillarum O1, a homologue of the V. cholerae rfb region involved in O-antigen biosynthesis, thus they renamed virA and virB as wbhS and wbhR, respectively. Milton et al. (1995) analyzed VAN20 in which Tn5–132 transposed into two sites, and identified two genes. Each of the two genes thus identified were mutated, and virulence test were performed. One mutant in the virC gene showed reduction of virulence (a 104-fold for immersion and 106-fold for intraperitoneal) as compared with the wild type whereas the other mutant did not change the LD50 value indicating that only the virC gene is a virulence factor in V. anguillarum.
Exopolysaccharides (EPS)
Croxatto et al. (2007) identified two divergently transcribed operons, orf1-wbfD-wbfC-wbfB and wza-wzb-wzc. wzabc genes are conserved in bacteria that produce group-1 capsular polysaccharides. Wza is a surface-located outer membrane lipoprotein, Wzb is a protein tyrosine phosphatase and Wzc is a protein tyrosine kinase. The wbfDCB genes are similar to those of V. cholerae O139, which map in the rfb/capsule DNA locus. They demonstrated that the mutation in the orf1, wbfD and wza genes caused a decrease in the level of EPS but not LPS indicating that the wza-wzb-wzc locus is involved in EPS but not LPS biosyntheses in V. anguillarum. They also showed that the EPS may form a loose capsule or a slime layer that is not anchored to the cell and easily shed into the culture supernatant. The orf1 and the wza mutants showed reduction of the virulence in both immersion and intraperitoneal inoculation routes, while the wbfD mutant exhibited attenuation of virulence only when challenging by immersion. These results indicated that the EPS biosynthesis gene may have different functions during infection. The authors also demonstrated that the wild type strain penetrates the skin mucosal layer within 5 hours after infection, proliferate and attach to the scales at the 12 hours time point forming a biofilm by 24–48 hours. On the contrary the wza mutant never proliferates in the fish mucus and was not detected after 24 hours post infection. Each orf1, wbfD and wza mutants showed less efficient penetration of the skin mucus as compared with the wild type. Biofilm formation on fish scales was compared between mutants and wild type. It is clear that orf1 and wza are required to make biofilms, while the wbfD mutation did not affect biofilm formation. Furthermore, they found that wza and orf1 are required for protease as well as mucinase activity, while the wbfD mutant showed again similar results to the wild type strain. Weber et al. (2010) demonstrated that wzb and wzc are essential for EPS transport as well as virulence to rainbow trout in both the immersion and intraperitoneal routes. The authors also showed that exopolysaccharide mutants such as Δwza, Δwzb and Δwzc were impaired to colonize the skin (or colonized only 5–10% of the fish) and decreased in motility through the mucus as compared with the wild type strain, even that those mutants are still able to colonize the intestine. Moreover, they found that exopolysaccharide mutants are more sensitive not only to lysozyme but also to antimicrobial peptides such as polymixin B and parasin as compared with the wild type.
Conclusions
We have described that virulence factors in V. anguillarum can be classified into those that are required to gain access to the fish host and those that are necessary for bacterial proliferation. In the latter classification one of the most important factors concerns the ability of bacteria to use the otherwise unavailable iron that is bound to the specific iron binding proteins such as transferrin and lactoferrin and that is mostly encoded in the 65 kb plasmid pJM1. It is obvious that many factors such as proteases, hemolysins, LPS, and EPS are also important in allowing access to the fish. Demonstrating the power of the modern approaches 40 putative virulence factors have been identified by random genome sequencing of the V. anguillarum strain H775–3 (Rodkhum et al., 2006). The identified putative virulence factors include a repeat toxin gene cluster rtxABCD, four hemolysin genes, an adheshin gene, 13 flagellum genes, four type IV pilus genes, three protease genes, seven LPS biosynthesis genes, a siderophore biosynthesis gene and rpoN in which some of them were originally identified by more genetic classical approaches.
Acknowledgements
The research in our laboratory discussed here was supported by grants AI19018 and GM64600 from the National Institutes of Health to J. H. C.
References
- Actis LA, Potter SA, Crosa JH. Iron-regulated outer membrane protein OM2 of Vibrio anguillarum is encoded by virulence plasmid pJM1. J. Bacteriol. 1985;161:736–742. doi: 10.1128/jb.161.2.736-742.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Actis LA, Tolmasky ME, Farrell DH, Crosa JH. Genetic and molecular characterization of essential components of the Vibrio anguillarum plasmid-mediated iron-transport system. J. Biol. Chem. 1988;263:2853–2860. [PubMed] [Google Scholar]
- Actis LA, Tolmasky ME, Crosa LM, Crosa JH. Characterization and regulation of the expression of FatB, an iron transport protein encoded by the pJM1 virulence plasmid. Mol. Microbiol. 1995;17:197–204. doi: 10.1111/j.1365-2958.1995.mmi_17010197.x. [DOI] [PubMed] [Google Scholar]
- Actis LA, Tolmasky ME, Crosa JH. Vibriosis. In: Woo PTK, Bruno DW, editors. Fish Diseases and Disorders, vol. 3: Viral, Bacterial, and Fungal Infections, 2nd edition. 2nd edition. vol. 3. Oxfordshire, UK: CABI International; 2011. pp. 570–605. Viral, Bacterial, and Fungal Infections. [Google Scholar]
- Alice AF, Lopez CS, Crosa JH. Plasmid- and chromosome-encoded redundant and specific functions are involved in biosynthesis of the siderophore anguibactin in Vibrio anguillarum 775: a case of chance and necessity? J. Bacteriol. 2005;187:2209–2214. doi: 10.1128/JB.187.6.2209-2214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki T, Nomura J, Crosa JH. Virulence of Vibrio anguillarum with particular emphasis on the outer membrane components. Bull. Jpn. Soc. Sci. Fish. 1985;51:1249–1254. [Google Scholar]
- Balado M, Osorio CR, Lemos ML. A gene cluster involved in the biosynthesis of vanchrobactin, a chromosome-encoded siderophore produced by Vibrio anguillarum. Microbiology. 2006;152:3517–3528. doi: 10.1099/mic.0.29298-0. [DOI] [PubMed] [Google Scholar]
- Balado M, Osorio CR, Lemos ML. Biosynthetic and regulatory elements involved in the production of the siderophore vanchrobactin in Vibrio anguillarum. Microbiology. 2008;154:1400–1413. doi: 10.1099/mic.0.2008/016618-0. [DOI] [PubMed] [Google Scholar]
- Balado M, Osorio CR, Lemos ML. FvtA is the receptor for the siderophore vanchrobactin in Vibrio anguillarum: utility as a route of entry for vanchrobactin analogues. Appl. Environ. Microbiol. 2009;75:2775–2783. doi: 10.1128/AEM.02897-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman AM. Die rote Beulenkrankheit des Aals. Bericht aus der Königlichen Bayerischen Versuchsstation. 1909;2:10–54. [Google Scholar]
- Canestrini G. La malatti dominate delle anguille. Atti Institute Veneto Service. 1893;7:809–814. [Google Scholar]
- Chart H. Multiflagellate variants of Vibrio anguillarum. J. Gen. Microbiol. 1983;129:2193–2197. doi: 10.1099/00221287-129-7-2193. [DOI] [PubMed] [Google Scholar]
- Crosa JH. A plasmid associated with virulence in the marine fish pathogen Vibrio anguillarum specifies an iron-sequestering system. Nature. 1980;284:566–568. doi: 10.1038/284566a0. [DOI] [PubMed] [Google Scholar]
- Crosa JH, Walsh CT. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev. 2002;66:223–249. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croxatto A, Lauritz J, Chen C, Milton DL. Vibrio anguillarum colonization of rainbow trout integument requires a DNA locus involved in exopolysaccharide transport and biosynthesis. Environ. Microbiol. 2007;9:370–382. doi: 10.1111/j.1462-2920.2006.01147.x. [DOI] [PubMed] [Google Scholar]
- Denkin SM, Nelson DR. Regulation of Vibrio anguillarum empA metalloprotease expression and its role in virulence. Appl. Environ. Microbiol. 2004;70:4193–4204. doi: 10.1128/AEM.70.7.4193-4204.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lorenzo M, Stork M, Tolmasky ME, Actis LA, Farrell D, Welch TJ, Crosa LM, Wertheimer AM, Chen Q, Salinas P, Waldbeser L, Crosa JH. Complete sequence of virulence plasmid pJM1 from the marine fish pathogen Vibrio anguillarum strain 775. J. Bacteriol. 2003;185:5822–5830. doi: 10.1128/JB.185.19.5822-5830.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lorenzo M, Poppelaars S, Stork M, Nagasawa M, Tolmasky ME, Crosa JH. A nonribosomal peptide synthetase with a novel domain organization is essential for siderophore biosynthesis in Vibrio anguillarum. J. Bacteriol. 2004;186:7327–7336. doi: 10.1128/JB.186.21.7327-7336.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lorenzo M, Stork M, Naka H, Tolmasky ME, Crosa JH. Tandem heterocyclization domains in a non-ribosomal peptide synthetase essential for siderophore biosynthesis in Vibrio anguillarum. Biometals. 2008;21:635–648. doi: 10.1007/s10534-008-9149-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell DH, Crosa JH. Purification and characterization of a secreted protease from the pathogenic marine bacterium Vibrio anguillarum. Biochemistry. 1991;30:3432–3436. doi: 10.1021/bi00228a012. [DOI] [PubMed] [Google Scholar]
- Grisez L, Ollevier F. Comparative Serology of the Marine Fish Pathogen Vibrio anguillarum. Appl. Environ. Microbiol. 1995;61:4367–4373. doi: 10.1128/aem.61.12.4367-4373.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrell L, Novotny AJ, Schiewe MH, Hodgins H. Isolation and description of two vibrios pathogenic to Pacific salmon in Puget Sound, Washington. Fisheries Bulletin. 1976;74:447–449. [Google Scholar]
- Hirono I, Masuda T, Aoki T. Cloning and detection of the hemolysin gene of Vibrio anguillarum. Microb. Pathog. 1996;21:173–182. doi: 10.1006/mpat.1996.0052. [DOI] [PubMed] [Google Scholar]
- Jedani KE, Stroeher UH, Manning PA. Distribution of IS1358 and linkage to rfb-related genes in Vibrio anguillarum. Microbiology. 2000;146:323–331. doi: 10.1099/00221287-146-2-323. [DOI] [PubMed] [Google Scholar]
- Koster WL, Actis LA, Waldbeser LS, Tolmasky ME, Crosa JH. Molecular characterization of the iron transport system mediated by the pJM1 plasmid in Vibrio anguillarum 775. J. Biol. Chem. 1991;266:23829-23829–23833. [PubMed] [Google Scholar]
- Kuehl CJ, Crosa JH. Molecular and genetic characterization of the TonB2-cluster TtpC protein in pathogenic vibrios. Biometals. 2009;22:109–115. doi: 10.1007/s10534-008-9194-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehl CJ, Crosa JH. The TonB energy transduction systems in Vibrio species. Future Microbiol. 2010;5:1403–1412. doi: 10.2217/fmb.10.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen JL, Pedersen K, Dalsgaard I. Vibrio anguillarum serovars associated with vibrioses in fish. J. Fish. Dis. 1994;17:259–267. [Google Scholar]
- Lee JH, Kim MW, Kim BS, Kim SM, Lee BC, Kim TS, Choi SH. Identification and characterization of the Vibrio vulnificus rtxA essential for cytotoxicity in vitro and virulence in mice. J. Microbiol. 2007;45:146–152. [PubMed] [Google Scholar]
- Lemos ML, Salinas P, Toranzo AE, Barja JL, Crosa JH. Chromosome-mediated iron uptake system in pathogenic strains of Vibrio anguillarum. J. Bacteriol. 1988;170:1920–1925. doi: 10.1128/jb.170.4.1920-1925.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Rock JL, Nelson DR. Identification and characterization of a repeat-in-toxin gene cluster in Vibrio anguillarum. Infect. Immun. 2008;76:2620–2632. doi: 10.1128/IAI.01308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Alice AF, Naka H, Crosa JH. The HlyU protein is a positive regulator of rtxA 1, a gene responsible for cytotoxicity and virulence in the human pathogen Vibrio vulnificus. Infect. Immun. 2007;75:3282–3289. doi: 10.1128/IAI.00045-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Ma Y, Wu H, Shao M, Liu H, Zhang Y. Cloning, identification and expression of an entE homologue angE from Vibrio anguillarum serotype O1. Arch. Microbiol. 2004;181:287–293. doi: 10.1007/s00203-004-0652-x. [DOI] [PubMed] [Google Scholar]
- Liu Q, Ma Y, Zou L, Zang Y. Gene cloning, expression and functional characterization of a phosphopantetheinyl transferase from Vibrio anguillarum serotype O1. Arch. Microbiol. 2005;183:34–44. doi: 10.1007/s00203-004-0745-6. [DOI] [PubMed] [Google Scholar]
- Lopez CS, Alice AF, Chakraborty R, Crosa JH. Identification of amino acid residues required for ferric-anguibactin transport in the outer-membrane receptor FatA of Vibrio anguillarum. Microbiology. 2007;153:570–584. doi: 10.1099/mic.0.2006/001735-0. [DOI] [PubMed] [Google Scholar]
- Lopez CS, Crosa JH. Characterization of ferric-anguibactin transport in Vibrio anguillarum. Biometals. 2007;20:393–403. doi: 10.1007/s10534-007-9084-9. [DOI] [PubMed] [Google Scholar]
- Mazoy R, Osorio CR, Toranzo AE, Lemos ML. Isolation of mutants of Vibrio anguillarum defective in haeme utilisation and cloning of huvA, a gene coding for an outer membrane protein involved in the use of haeme as iron source. Arch. Microbiol. 2003;179:329–338. doi: 10.1007/s00203-003-0529-4. [DOI] [PubMed] [Google Scholar]
- McGee K, Horstedt P, Milton DL. Identification and characterization of additional flagellin genes from Vibrio anguillarum. J. Bacteriol. 1996;178:5188–5198. doi: 10.1128/jb.178.17.5188-5198.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milton DL, Norqvist A, Wolf-Watz H. Cloning of a metalloprotease gene involved in the virulence mechanism of Vibrio anguillarum. J Bacteriol. 1992;174:7235–7244. doi: 10.1128/jb.174.22.7235-7244.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milton DL, Norqvist A, Wolf-Watz H. Sequence of a novel virulence-mediating gene, virC, from Vibrio anguillarum. Gene. 1995;164:95–100. doi: 10.1016/0378-1119(95)00538-h. [DOI] [PubMed] [Google Scholar]
- Milton DL, O’Toole R, Horstedt P, Wolf-Watz H. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 1996;178:1310–1319. doi: 10.1128/jb.178.5.1310-1319.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo Z, Guo D, Mao Y, Ye X, Zou Y, Xiao P, Hao B. Identification and characterization of the Vibrio anguillarum prtV gene encoding a new metalloprotease. Chin. J. Oceanol. Limnol. 2010;28:55–61. [Google Scholar]
- Mourino S, Osorio CR, Lemos ML. Characterization of heme uptake cluster genes in the fish pathogen Vibrio anguillarum. J. Bacteriol. 2004;186:6159–6167. doi: 10.1128/JB.186.18.6159-6167.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourino S, Rodriguez-Ares I, Osorio CR, Lemos ML. Genetic variability of the heme uptake system among different strains of the fish pathogen Vibrio anguillarum: identification of a new heme receptor. Appl. Environ. Microbiol. 2005;71:8434–8441. doi: 10.1128/AEM.71.12.8434-8441.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naka H, Lopez CS, Crosa JH. Reactivation of the vanchrobactin siderophore system of Vibrio anguillarum by removal of a chromosomal insertion sequence originated in plasmid pJM1 encoding the anguibactin siderophore system. Environ. Microbiol. 2008;10:265–277. doi: 10.1111/j.1462-2920.2007.01450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naka H, López CS, Crosa JH. Role of the pJM1 plasmid-encoded transport proteins FatB, C and D in ferric anguibactin uptake in the fish pathogen Vibrio anguillarum. Environ. Microbiol. Rep. 2010;2:104–111. doi: 10.1111/j.1758-2229.2009.00110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norqvist A, Norrman B, Wolf-Watz H. Identification and characterization of a zinc metalloprotease associated with invasion by the fish pathogen Vibrio anguillarum. Infect. Immun. 1990;58:3731–3736. doi: 10.1128/iai.58.11.3731-3736.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norqvist A, Wolf-Watz H. Characterization of a novel chromosomal virulence locus involved in expression of a major surface flagellar sheath antigen of the fish pathogen Vibrio anguillarum. Infect. Immun. 1993;61:2434–2444. doi: 10.1128/iai.61.6.2434-2444.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole R, Milton DL, Wolf-Watz H. Chemotactic motility is required for invasion of the host by the fish pathogen Vibrio anguillarum. Mol. Microbiol. 1996;19:625–637. doi: 10.1046/j.1365-2958.1996.412927.x. [DOI] [PubMed] [Google Scholar]
- O’Toole R, Milton DL, Horstedt P, Wolf-Watz H. RpoN of the fish pathogen Vibrio (Listonella) anguillarum is essential for flagellum production and virulence by the water-borne but not intraperitoneal route of inoculation. Microbiology. 1997;143:3849–3859. doi: 10.1099/00221287-143-12-3849. [DOI] [PubMed] [Google Scholar]
- O’Toole R, Lundberg S, Fredriksson SA, Jansson A, Nilsson B, Wolf-Watz H. The chemotactic response of Vibrio anguillarum to fish intestinal mucus is mediated by a combination of multiple mucus components. J. Bacteriol. 1999;181:4308–4317. doi: 10.1128/jb.181.14.4308-4317.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier V, Queen J, Satchell KJ. Successful small intestine colonization of adult mice by Vibrio cholerae requires ketamine anesthesia and accessory toxins. PLoS One. 2009;4:e7352. doi: 10.1371/journal.pone.0007352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormonde P, Horstedt P, O’Toole R, Milton DL. Role of motility in adherence to and invasion of a fish cell line by Vibrio anguillarum. J. Bacteriol. 2000;182:2326–2328. doi: 10.1128/jb.182.8.2326-2328.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen K, Grisez L, van Houdt R, Tiainen T, Ollevier F, Larsen JL. Extended serotyping scheme for Vibrio anguillarum with the definition and characterization of seven provisional O-serogroups. Curr. Microbiol. 1999;38:183–189. doi: 10.1007/pl00006784. [DOI] [PubMed] [Google Scholar]
- Rock JL, Nelson DR. Identification and characterization of a hemolysin gene cluster in Vibrio anguillarum. Infect. Immun. 2006;74:2777–2786. doi: 10.1128/IAI.74.5.2777-2786.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodkhum C, Hirono I, Crosa JH, Aoki T. Four novel hemolysin genes of Vibrio anguillarum and their virulence to rainbow trout. Microb. Pathog. 2005;39:109–119. doi: 10.1016/j.micpath.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Rodkhum C, Hirono I, Stork M, Di Lorenzo M, Crosa JH, Aoki T. Putative virulence-related genes in Vibrio anguillarum identified by random genome sequencing. J. Fish Dis. 2006;29:157–166. doi: 10.1111/j.1365-2761.2006.00692.x. [DOI] [PubMed] [Google Scholar]
- Schiewe MH, Crosa JH, Ordal EJ. Deoxyribo-nucleic acid relationships among marine vibrios pathogenic to fish. Can. J. Microbiol. 1977;23:954–958. doi: 10.1139/m77-142. [DOI] [PubMed] [Google Scholar]
- Schiewe MH, Trust TJ, Crosa JH. Vibrio ordalii sp. nov.: A causative agent of vibriosis in fish. Can. J. Microbiol. 1981;6:343–348. [Google Scholar]
- Soengas RG, Larrosa M, Balado M, Rodriguez J, Lemos ML, Jimenez C. Synthesis and biological activity of analogues of vanchrobactin, a siderophore from Vibrio anguillarum serotype O2. Org. Biomol. Chem. 2008;6:1278–1287. doi: 10.1039/b719713f. [DOI] [PubMed] [Google Scholar]
- Sorensen UB, Larsen JL. Serotyping of Vibrio anguillarum. Appl. Environ. Microbiol. 1986;51:593–597. doi: 10.1128/aem.51.3.593-597.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stork M, Di Lorenzo M, Mourino S, Osorio CR, Lemos ML, Crosa JH. Two tonB systems function in iron transport in Vibrio anguillarum, but only one is essential for virulence. Infect. Immun. 2004;72:7326–7329. doi: 10.1128/IAI.72.12.7326-7329.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stork M, Otto BR, Crosa JH. A novel protein, TtpC, is a required component of the TonB2 complex for specific iron transport in the pathogens Vibrio anguillarum and Vibrio cholerae. J. Bacteriol. 2007;189:1803–1815. doi: 10.1128/JB.00451-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiainen T, Pedersen K, Larsen JL. Vibrio anguillarum serogroup O3 and V. anguillarum-like serogroup O3 cross-reactive species—comparison and characterization. J. Appl. Microbiol. 1997;82:211–218. [PubMed] [Google Scholar]
- Tolmasky ME, Actis LA, Crosa JH. Genetic analysis of the iron uptake region of the Vibrio anguillarum plasmid pJM1: molecular cloning of genetic determinants encoding a novel trans activator of siderophore biosynthesis. J. Bacteriol. 1988;170:1913–1919. doi: 10.1128/jb.170.4.1913-1919.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toranzo AE, Barja JL. A review of the taxonomy and seroepizootiology of Vibrio anguillarum, with special reference to aquaculture in the northwest Spain. Dis. Aquat. Org. 1990;9:73–82. [Google Scholar]
- Toranzo AE, Magarinos B, Romalde JL. A review of the main bacterial fish diseases in mariculture systems. Aquaculture. 2005;246:37–61. [Google Scholar]
- Trust TJ, Courtice ID, Khouri AG, Crosa JH, Schiewe MH. Serum resistance and hemagglutination ability of marine vibrios pathogenic for fish. Infect. Immun. 1981;34:702–707. doi: 10.1128/iai.34.3.702-707.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varina M, Denkin SM, Staroscik AM, Nelson DR. Identification and characterization of Epp, the secreted processing protease for the Vibrio anguillarum EmpA metalloprotease. J. Bacteriol. 2008;190:6589–6597. doi: 10.1128/JB.00535-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber B, Chen C, Milton DL. Colonization of fish skin is vital for Vibrio anguillarum to cause disease. Environ. Microbiol. Rep. 2010;2:133–139. doi: 10.1111/j.1758-2229.2009.00120.x. [DOI] [PubMed] [Google Scholar]
- Welch TJ, Chai S, Crosa JH. The overlapping angB and angG genes are encoded within the trans-acting factor region of the virulence plasmid in Vibrio anguillarum: essential role in siderophore biosynthesis. J. Bacteriol. 2000;182:6762–6773. doi: 10.1128/jb.182.23.6762-6773.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch TJ, Crosa JH. Novel role of the lipopolysaccharide O1 side chain in ferric siderophore transport and virulence of Vibrio anguillarum. Infect. Immun. 2005;73:5864–5872. doi: 10.1128/IAI.73.9.5864-5872.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheimer AM, Verweij W, Chen Q, Crosa LM, Nagasawa M, Tolmasky ME, Actis LA, Crosa JH. Characterization of the angR gene of Vibrio anguillarum: essential role in virulence. Infect. Immun. 1999;67:6496–6509. doi: 10.1128/iai.67.12.6496-6509.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winton J, Rohovec J, Fryer J. Bacterial and viral diseases of cultured salmonids in the Pacific Northwest. In: Crosa JH, editor. Bacterial and Viral Diseases of Fish. Seattle: Washington Sea Grant; 1983. pp. 1–20. [Google Scholar]
- Xu Z, Wang Y, Han Y, Chen J, Zhang XH. Mutation of a novel virulence-related gene mltD in Vibrio anguillarum enhances lethality in zebra fish. Res. Microbiol. 2010 doi: 10.1016/j.resmic.2010.08.003. [DOI] [PubMed] [Google Scholar]
- Yang H, Chen J, Yang G, Zhang XH, Li Y. Mutational analysis of the zinc metalloprotease EmpA of Vibrio anguillarum. FEMS Microbiol. Lett. 2007a;267:56–63. doi: 10.1111/j.1574-6968.2006.00533.x. [DOI] [PubMed] [Google Scholar]
- Yang H, Chen J, Yang G, Zhang XH, Li Y, Wang M. Characterization and pathogenicity of the zinc metalloprotease empA of Vibrio anguillarum expressed in Escherichia coli. Curr. Microbiol. 2007b;54:244–248. doi: 10.1007/s00284-006-0495-6. [DOI] [PubMed] [Google Scholar]
- Zhang F, Chen J, Chi Z, Wu LF. Expression and processing of Vibrio anguillarum zinc-metalloprotease in Escherichia coli. Arch. Microbiol. 2006;186:11–20. doi: 10.1007/s00203-006-0118-4. [DOI] [PubMed] [Google Scholar]