Abstract
Ubiquinone (Coenzyme Q) plays an important role in the mitochondrial respiratory chain and also acts as an antioxidant in its reduced form, protecting cellular membranes from peroxidation. De novo disulfide bond generation in the E. coli periplasm involves a transient complex consisting of DsbA, DsbB and ubiquinone (UQ). It is hypothesized that a charge-transfer complex intermediate is formed between the UQ ring and the DsbB-C44 thiolate during the reoxidation of DsbA, which gives a distinctive ∼500 nm absorbance band. No enzymological precedent exists for an UQ-protein thiolate charge transfer complex, and definitive evidence of this unique reaction pathway for DsbB has not been fully demonstrated. In order to study the UQ-8-DsbB complex in the presence of native lipids, we have prepared isotopically labeled samples of precipitated DsbB (WT and C41S) with endogenous UQ-8 and lipids, and we have applied advanced multidimensional solid-state NMR methods. Double-quantum filter and dipolar dephasing experiments facilitated assignments of UQ isoprenoid chain resonances not previously observed and headgroup sites important for the characterization of the UQ redox states: methyls (∼ 20 ppm), methoxys (∼ 60 ppm), olefin carbons (120∼140 ppm) and carbonyls (150∼160 ppm). Upon increasing the DsbB(C41S) pH from 5.5 to 8.0, we observed a 10.8 ppm upfield shift for the UQ C1 and C4 carbonyls indicating an increase of electron density on the carbonyls. This observation is consistent with the deprotonation of the DsbB-C44 thiolate at pH 8.0 and provides direct evidence of the charge transfer complex formation. A similar trend was noted for the UQ chemical shifts of the DsbA(C33S)-DsbB(WT) heterodimer, confirming that the charge-transfer complex is unperturbed by the DsbB(C41S) mutant used to mimic the intermediate state of the disulfide bond generating reaction pathway.
Introduction
Ubiquinone (UQ) is a membrane-bound redox-active cofactor that functions in electron transfer pathways in mitochondria and some bacteria.1 UQ is found in all membranes of the mammalian cell, and in its reduced form acts as an antioxidant.2 UQ consists of a polar headgroup and a long hydrophobic tail composed of a species-dependent number (n) of five-carbon isoprenoid units; UQs are described using the nomenclature “UQ-n”, such as UQ-10 in human cells, UQ-9 in rat, UQ-8 in Escherichia coli (Figure 1a) and UQ-6 in Saccharomyces cerevisiae. For DsbB, the isoprenoid chain length affects the rate of ubiquinone reactivity as measured by stopped flow absorbance decay of the ∼500 nm signal, and UQ-1 has a 6-fold slower decay than endogenous UQ-8.44 In general, the binding of quinone to protein can have a dramatic effect upon its properties, with changes in midpoint potential up to 600 mV.3 The modulation of quinone reactivity by the protein matrix is not fully understood, but relevant factors include electrostatics, hydrogen-bonding and aromatic stacking, as well as the ability of the protein to provide or limit proton access.
Figure 1.

(a) UQ-8 structure with IUPAC nomenclature. Expansions of the (b) aliphatic and (c) UQ-8 resonances region of the 13C-13C 2D correlation spectra of U-DsbB(C41S) at 12.5 kHz MAS with 25 ms DARR mixing (B0 = 17.6 T, Tsample = −12 °C, 1.5 s pulse delay, maximum t1 = 15.36 ms, maximum t2 = 20.48 ms). Clearly resolved and amino-acid specific correlations are indicated in rectangle boxes for Ala, Ile, Ser and Thr and ambiguous assignments of the isoprenoid side chain are indicated in dashed circles. C13′∼33′ represent the six methyl groups of 13′, 17′, 21′, 25′, 29′, 33′. C11∼31, C12∼32 and C13∼33 represent the corresponding methylene and olefin groups.
High-resolution structures of quinone binding sites carry enormous potential to clarify the structural and physical bases of quinone reactivity, but can be difficult to achieve; in addition to the problems associated with producing highly-diffracting crystals of membrane proteins, there can be loss of quinone, or disorder at the binding site. Solid-state NMR (SSNMR) has been shown to be an effective and versatile tool for studying function-related cofactors in large protein systems, such as UQ-10,4,5 chlorophyll,6 bacteriochlorophyll,7,8 and retinal.9-14 Resonance assignments of isotopically labeled cofactors are often facilitated by cofactor reincorporation into the binding sites of proteins. For instance, Bushweller and coworkers reincorporated 13C-labeled UQ-2 into detergent solubilized DsbB for their NMR structural investigations.41 In the present work, the endogenous UQ-8 is labeled along with DsbB during growth of E. coli in the isotopic labeling medium. Hence, we employ the sophisticated capabilities of SSNMR techniques to assign the resonances of UQ-8 in isotopically labeled DsbB. As a crucial step toward probing the structure of the DsbB quinone binding site, we show herein the assignment of a 13C-labeled UQ-8 bound to the E. coli membrane protein DsbB and chemical shift changes of UQ related to its charge-transfer complex with DsbB.
DsbB, a 20 kDa protein with four transmembrane helices, catalyzes the transfer of two electrons from a pair of free cysteine residues (C41, C44) to UQ, resulting in a C41-C44 disulfide bond.15,16 The disulfide bond is subsequently transferred to a second pair of cysteine residues (C104, C130), and then to the soluble periplasmic protein DsbA. DsbA in turn oxidizes cysteine pairs in target proteins, promoting folding. The UQ binding site in DsbB is unique among known quinone binding sites: under certain conditions a ∼500 nm absorbance appears. This absorbance band has been attributed to the formation of a thiolate-quinone charge-transfer complex, and it disappears upon reduction of the quinone or decrease in pH.17,18 In this study, we used the DsbB mutant C41S, which allowed the stable formation of the ∼500 nm absorbance band, mimicking the key catalytic intermediate during the disulfide bond generation process.18 Using [U-13C, 15N]DsbB(C41S) and reverse-labeled DsbB(C41S) with natural abundance Phe, Leu, Tyr, Pro, Trp (FLYPW-DsbB(C41S)), we were able to identify and assign the resonances of the endogenous protein-bound UQ-8. We also observed a 10.8 ppm upfield shift for UQ carbonyl upon pH change from 5.5 to 8.0, indicating an increase of the electron density on the UQ carbonyl as evidence of the formation of the charge-transfer complex. This chemical shift change is consistent with the trend of 13C chemical shift changes induced by the distinct charge distributions observed for varied aromatic systems.19 The downfield shift of DsbB C44 Cβ also indicates charge transfer from C44 to UQ. We also acquired two-dimensional 13C-13C correlation spectrum of the complex of natural abundance DsbA(C33S) and [U-13C, 15N] DsbB (DsbA(C33S)/DsbB*) which also stabilizes the ∼500 nm absorbance band and resembles the intermediate state in DsbA-DsbB system. We found that the chemical shifts of UQ-8 in,DsbA(C33S)/DsbB* are almost identical to those in DsbB(C41S), indicating that both samples have the same thiolate-quinone charge-transfer complex.
Materials and Methods
Sample Preparation
DsbB was expressed from E. coli C43 (DE3) containing a plasmid encoding either DsbB “wild type” with a 6-His tag and mutation of two nonessential cysteines, C8A and C49V, or DsbB(C41S), containing an additional mutation of the conserved C41.17 Preparation of DsbB(C41S) for SSNMR has been described previously.20 The solid-state NMR samples contain isotopically labeled DsbB, endogenous UQ-8 and lipids in addition to residual dodecylmaltoside detergent. The protein-lipid ratio is around 1:24. 27
To prepare natural abundance DsbA for DsbA(C33S)/DsbB* complex formation, a plasmid encoding DsbA(C33S) was obtained from K. Inaba and K. Ito, Kyoto.21 The plasmid was transformed into in E. coli BL21(DE3)/pREP4, and the cells grown in LB medium containing 2 mM MgSO4 and 60 μg/ml ampicillin, at 37 °C. Protein expression was induced with 0.2 mM IPTG, and growth continued for 8 hr at 25 °C. DsbA was released from the cells using osmotic shock, and purified by QFF anion exchange chromatography (GE Healthcare, Piscataway, NJ) in 10 mM MOPS pH 7.22 The protein eluted at 30-50 mM NaCl. The yield of DsbA was over 100 mg protein/L of culture.
To prepare the DsbA(C33S)/DsbB* complex, a small amount of [U-13C,15N]DsbB was titrated with DsbA(C33S) to observe complex formation at ∼500 nm.17 The sample for SSNMR was then prepared with a substoichiometric amount of [U-13C,15N]DsbB so that all the labeled material would be covalently bound. The sample was concentrated, dialyzed for several days against 25 mM Tris, pH 8.0, and centrifuged for 1 h at 100,000 g to remove a small amount of contaminating protein. The supernatant was centrifuged for 20 h at 100,000 g to pellet the dark red DsbA(C33S)/DsbB* complex, which was packed into a 3.2 mm thin wall NMR rotor.
To prepare [U-13C,15N]DsbB(C41S) with natural abundance Phe, Leu, Tyr, Pro, and Trp through reverse-labeling,23,24 freshly transformed colonies of E. coli C43(DE3) containing DsbB C41S expression plasmid17 were transferred to LB containing 2 mM MgSO4 and 60 μg/ml ampicillin and grown overnight. The overnight culture was used to inoculate 4 L of the same media. This culture was incubated at 37 °C with shaking until a culture density of A600 = 0.8 was reached. Following the procedure of Marley and coworkers,25 the cells were harvested, washed, and resuspended in 1 L of labeling medium. This medium was a modified version of Studier medium “P”26 containing 50 mM Na2HPO4, 50 mM KH2PO4, 5 mM Na2SO4, 2 mM MgSO4, 10 μg/ml thiamine, 10 μg/ml biotin, 0.3% 15N-NH4Cl, 0.4% U-13C-glucose, 60 μg/ml ampicillin, 0.2 × Studier trace metals (10 μM FeCl3, 4 μM CaCl2, 2 μM MnCl2, 2 μM ZnSO4, and 0.4 μM each of CoCl2, CuCl2, NiCl2, Na2MoO4, Na2SeO3, and H3BO3.), and 1 mM each of natural abundance Phe, Leu, Tyr, Pro, and Trp. Following resuspension, the cells were incubated at 25 °C with shaking for 1 hour, and protein expression was induced with 0.8 mM IPTG. After overnight growth (16 h) at 25 °C, the cells were harvested, washed, and stored as a pellet at -80 °C until use. Since this reverse-labeling procedure did not avoid substantial labeling of Pro in DsbB, in subsequent growths we reverse-labeled only Phe, Leu, Tyr, and Trp.
Membrane isolation, solubilization of DsbB in 1% dodecylmaltoside (DDM), and purification of the His-tagged DsbB on Talon cobalt resin (Clontech, Mountain View, CA) were described previously.27 Fractions containing DsbB were pooled, concentrated to 15 ml with a 50 kDa MWCO Centriprep™ concentrator, and then dialyzed overnight at 4 °C against 25 mM Tris-HCl, pH 8.0. This dialysis resulted in precipitation of contaminating proteins, which were pelleted at 8000 g for 20 min. The DsbB was further concentrated and 100 mM NaCl and 5 mM CaCl2 were added. Factor Xa (Novagen, Madison, WI) was added to cleave the 6-His tag, and the solution was stirred at 22 °C for 16 h. Contaminating protein was again removed by centrifugation (80000 g, 20 min) and the solution was concentrated. For preparation of a DsbB (C41S) sample at pH 5.5, an aliquot of the preparation was slowly titrated with 1 M MES, pH 5.0, with stirring, and then dialyzed against 25 mM MES pH 5.5. The pH 8 sample was dialyzed against 25 mM Tris, pH 8.0. The samples were then assayed to determine the concentration of DDM using the colorimetric assay described in Lau and Bowie (1997).28 The DDM was removed over several days, by first adding an aliquot of methyl-β-cyclodextrin (MBCD) sufficient to remove 50% of the DDM, and stirring at 4 C for 12-24 h. MBCD binds an equimolar amount of DDM.29,30 Increasingly smaller aliquots of MBCD were added, until the solution became slightly turbid, at which point it was centrifuged at 100,000 g for 4 h. Hard and soft portions of the DsbB pellet were combined and again resuspended in 25 mM buffer with addition of MBCD as above until the 4 h centrifugation produced only a hard pellet. At this point the pellet was washed with 25 mM buffer, and centrifuged for 20 h. The DsbB pellets were packed in 3.2 mm thin wall SSNMR rotors. The mass of labeled protein packed into these rotors was 8 mg and 5 mg for pH 5.5 and 8.0, respectively, calculated from the 13C signals detected in DP experiments, which were calibrated by comparing to the total integrated signals of a NMR standard of known quantity (adamantane). The UV-Vis spectra of DsbB(C41S) were acquired on an Evolution 600 Spectrophotometer (Thermo Scientific, Waltham, MA).
NMR Spectroscopy
SSNMR experiments were performed on a 750 MHz Varian Inova spectrometer, a 600 MHz Varian InfinityPlus spectrometer and a 500 MHz Varian VNMRS spectrometer (1H frequency). All spectrometers were equipped with 3.2 mm HCN MAS probes. Typical π/2 pulse widths were 2.0-2.5 μs for 1H, 2.6-3.4 μs for 13C and 3.8 μs for 15N. The π pulse width for 15N REDOR dephasing was 14.4 μs. All experiments utilized tangent ramped cross polarization (CP)31 with TPPM32 or SPINAL33 decoupling of the protons applied during acquisition and evolution periods on average at ∼75 kHz. Detailed parameters are given in the figure captions. Sample temperatures were determined by ethylene glycol calibration.34 Chemical shifts were referenced to DSS, using adamantane as a secondary standard.35 Spectra were processed with NMRPipe36 and peak picking and assignments were performed in Sparky (T.D. Goddard and D.G. Kneller, University of California, San Francisco).
Results and Discussion
The solution NMR chemical shifts of ubiquinone serve as a guide for solid-state NMR assignments, and a comprehensive spectral analysis of both ubiquinone and ubiquinol in organic solvents is available.37,38 Through the use of a mild, one-step purification for [U-13C, 15N]DsbB(C41S), which we will refer to as U-DsbB(C41S), most of its bound, fully 13C-labeled UQ-8 is retained. Figure 1a shows the molecular structure of UQ-8. Based on the solution chemical shifts, we expected the unique ubiquinone crosspeaks between methyl (10∼25 ppm) and olefin carbon (120∼140 ppm) in 2D 13C-13C correlation spectra, which differ from the aromatic residues that lack side-chain methyls in DsbB. Indeed, we observed such crosspeaks in the U-DsbB(C41S) 13C-13C spectrum near the phase transition temperature of the sample (∼ −12 °C), which gave the best resolution and sensitivity for UQ-8 signals. Figure 1b shows the aliphatic region of U-DsbB(C41S) 13C-13C spectrum with 25 ms DARR39 mixing at pH 8.0, where ∼500 nm absorbance appears. I45, which is adjacent to C44 implicated in the formation of a charge-transfer complex with UQ-8,17,18 also shows unique Cδ1 to Cα, β, γ1 and γ2 crosspeaks. Figure 1c shows the expanded region with unique UQ-8 crosspeaks. In the group of peaks on the upper left, corresponding to the one-bond correlations of methyl and olefin carbon, we assign the most upfield shift in the F1 dimension to C5′, attached to the quinone ring. The rest of peaks in that group are assigned to the methyls and olefin carbons on the isoprenoid side chain. Using a similar analysis strategy based on the chemical shift patterns in solution NMR, other crosspeaks were assigned accordingly.
Assignments resulting from 2D DARR spectra are non-specific and can be inaccurate due to the overlap of signals from aromatic residues. Therefore, to confirm our assignments, we carried out double-quantum (DQ) 13C-13C 2D experiments with the POST-C7 recoupling scheme40 to get specific one-bond and two-bond correlations. Figure 2a and 2b show DQ spectra of U-DsbB(C41S) at −12 °C with 0.8 and 1.2 ms POST-C7 mixing times. In DQ experiments with POST-C7 mixing, one-bond and two-bond coherences have opposite intensities, and thus are straightforward to identify. At the short mixing time (Figure 2a), only one-bond correlations appear in the spectrum. Here, the one unique peak (28.2 ppm, 135.2 ppm) can only be assigned to (C38, C37) because other methylenes (C11∼35) with chemical shifts of around 29 ppm are two bonds away from the olefin carbons (C13∼33) with chemical shifts of 135∼140 ppm. At the longer mixing time (Figure 2b), two-bond correlations with negative intensity emerge at the positions as expected to reflect the coherences illustrated in red in Figure 2c. Overall, the DQ spectra here provide better resolution and selectivity than DARR, and the interference from lipid signals and the greater-than-two-bond correlations from aromatic residues have been removed. Additionally, these spectra contain peaks from the quinone isoprenoid chains, which are most likely to bind DsbB to give reasonable CP intensities. This is especially interesting since it is often the case in either X-ray crystallographic or solution NMR structural studies of membrane proteins that a quinone binding site is empty or only partially occupied, and is then reconstituted with a more hydrophilic analog (e.g. UQ-1, UQ-2, or an inhibitor) of the endogenous quinone, for example in the solution NMR structure of DsbB.41 In some crystal structures, as with the DsbA/DsbB complex, the full-length isoprenoid chain is present, but is too disordered to appear in the electron density map.42,43 Kinetic studies of DsbB have shown that reduction of added UQ-1 is six-fold slower than the endogenous UQ-8, suggesting that the longer isoprenoid chain has a role in properly positioning or anchoring UQ-8 within the DsbB active site.44 It will be of interest in the future to explore whether resolved quinone–to-protein correlations can be observed using SSNMR for the full length of the UQ-8 isoprenoid chain in DsbB.
Figure 2.
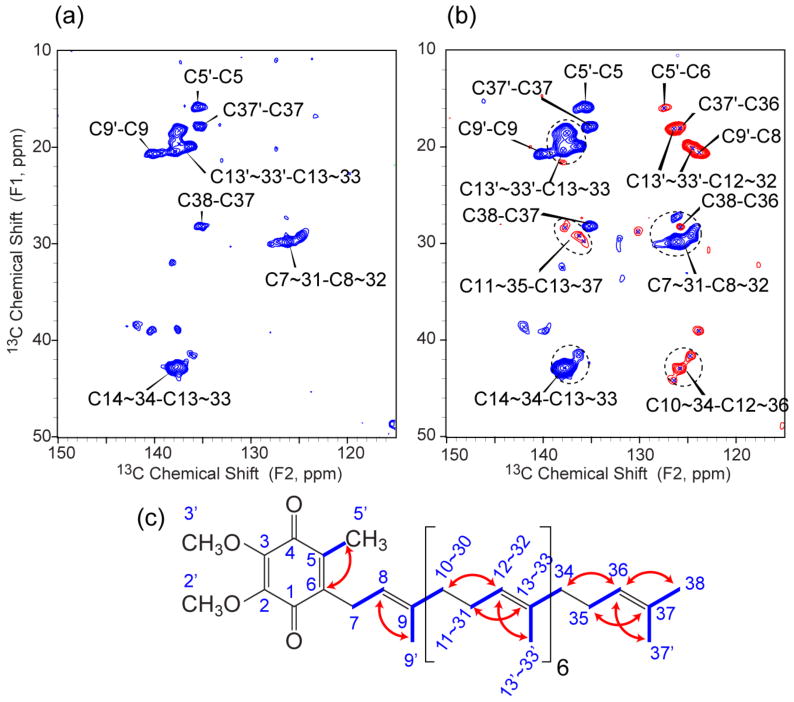
13C-13C DQ spectra of U-DsbB(C41S) at 10.0 kHz MAS with (a) 0.8 ms and (b) 1.2 ms POST-C7 mixing (B0 = 14.1 T, Tsample = −12 °C, 2 s pulse delay, maximum t1 = 10.24 ms, maximum t2 = 20.48 ms). Blue contours represent one-bond correlations and red contours represent two-bond correlations with the opposite sign. (c) UQ-8 structure with highlights of one-bond (blue lines) and two-bond (red arrows) correlations observed in DQ experiments.
Two dimensional data of U-DsbB(C41S) yielded the assignments of the full isoprenoid side chain and part of the quinone headgroup. However, the other parts of the quinone headgroup could not be assigned due to the overlap with aromatic residues and the inefficient DQ excitation for those sites. Hence we prepared a reverse-labeled DsbB(C41S) sample with natural abundance Phe, Leu, Tyr, Pro, Trp (FLYPW-DsbB), which simplifies 2D spectra in the aromatic region. Half of this purified DsbB sample was packed into the rotor from a solution in 25 mM Tris at pH 8.0; the other half from a solution in 25 mM MES at pH 5.5. Previous studies have shown that the presence of both oxidized quinone and C44 are necessary for the appearance of the ∼500 nm band, and this band disappears with a pKa of 6.6, suggesting a C44-thiolate-quinone complex as the source of the ∼500 nm absorbance.17,18 We confirmed that our purified DsbB(C41S) showed a loss of the ∼500 nm band upon titration to pH 5.5 (Figure 3). Thus, we expected a chemical shift perturbation due to the pH change from the quinone headgroup carbons. In fact, a 2D DARR experiment was able to provide resolved crosspeaks for UQ-8 headgroup carbons (Supporting Information, Figure S1), but there were still peaks from two His residues in the UQ region overlapping the peaks of interest. Hence, 15N REDOR dephasing was implemented to remove His signals in subsequent 2D correlation experiments.
Figure 3.

UV-Vis spectra of DsbB(C41S) from pH 5.53 to 7.85. DsbB in 25 mM Tris, pH 7.8 was titrated with 1 M MES (pH 5.5) to adjust the pH. The spectral change was complete within 30 s. Each spectrum was corrected for the change in volume upon addition of MES. Inset shows the absorbance at maximum 488 nm as a function of pH. pKa is determined to be 6.62 ± 0.02 by numerical fitting of the titration curve.
Both FLYPW-DsbB(C41S) samples at pH 5.5 and 8.0 showed a similar crosspeak pattern as for the U-DsbB(C41S) for the UQ isoprenoid chain (Figure S1b, c). Figure 4 shows a comparison of the expanded regions of 13C-13C 2D spectra of FLYPW-DsbB(C41S) at pH 5.5 and 8.0. Additional crosspeaks show up at a F1 frequency of 64 ppm corresponding to the correlations from methoxy to carbonyl and olefin carbon on the quinone ring. The sample at pH 5.5 shows one set of peaks, which may result from bound UQ-8 in the presence of a neutral, protonated C44 (Figure 4a). The sample at pH 8.0 shows a very different set of peaks (UQ C2′,3′−C2,3 and C2′,3′−C1,4), which corresponds to UQ-8 in a thiolate-quinone charge-transfer complex (Figure 4b).17 Table 1 summarizes 13C chemical shifts of UQ headgroup in solution and bound to DsbB (The full chemical shift list is in Supporting Information Table S1). Interestingly, the carbonyl chemical shifts of UQ-8 in pH 5.5 are 20 ppm upfield from free UQ in solution38 or bound UQ stabilized by hydrogen bonding and π-stacking interactions in other membrane protein systems,4 suggesting that the binding of UQ-8 in DsbB changes the electron distribution of UQ headgroup significantly. Furthermore, there is an additional 10.8 ppm upfield shift of carbonyl resonances from pH 5.5 to 8.0, indicating that the electron population on UQ headgroup carbonyl groups is increased by the negative charge from deprotonated C44 thiolate. This is derived from the fact that 13C chemical shifts move upfield significantly with negative charge on aromatic systems.19 Figure 5 shows the schematic diagram of interactions between DsbB C44 and UQ at pH 5.5 and 8.0. At pH 5.5, we hypothesize that even though there is no full negative charge on C44 thiol, the sulfur could be still close enough to have a partial charge separation with UQ carbonyls to influence the electron density (Figure 5a). Similar interactions have been reported between nitrogen or sulfur and carbonyl in cyclooctanone compounds in a transannular fashion.45 In comparison, deprotonated C44 thiolate with full negative charge at pH 8.0 transfer electron to UQ carbonyls to stabilize the complex (Figure 5b). As shown in Supporting Information, Figure S2 of U-DsbB(C41S) 15N-13C-13C 3D spectra, the chemical shift assignments of C44 in U-DsbB(C41S) at pH 8.0 also support the hypothesis, where DsbB C44 Cα (60.6 ppm) and Cβ (33.7 ppm) chemical shifts are between the typical chemical shifts of oxidized and reduced Cys residues in proteins (oxidized: Cα 58.0 ppm, Cβ 39.4 ppm; reduced: Cα 61.3 ppm, Cβ 27.8 ppm).46 Such chemical shifts of C44 Cα and Cβ are also similar to those in Zn-ligated cysteines of many metal-bound proteins in Protein Data Bank.47 This indicates that negative charge moves from C44 thiolate towards UQ through the charge-transfer complexation, which in turn causes the de-shielding effect on C44 Cβ accounting for the downfield shift, similar to the situation where a Zn ion draws electron density from a nucleophilic thiolate.
Figure 4.
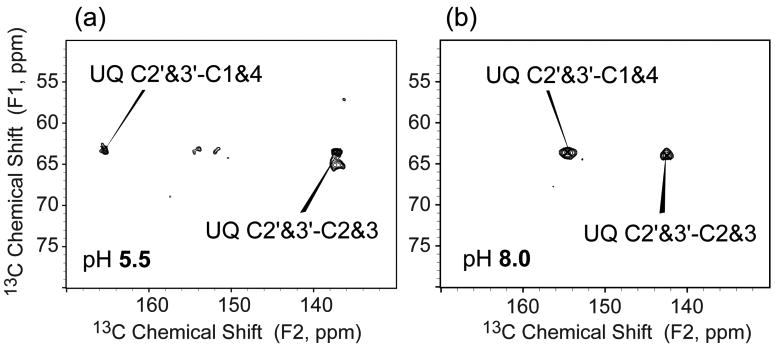
Expansions of the 13C-13C 2D correlation spectra of FLYPW-DsbB(C41S) samples at pH (a) 5.5 and (b) 8.0 with 100 ms DARR mixing and 1.8 ms 15N REDOR dephasing under 13.333 kHz MAS (B0 = 14.1 T, Tsample = −16.6 °C, 1.5 s pulse delay, maximum t1 = 10.24 ms, maximum t2 = 20.48 ms). Crosspeaks between methoxys (C2′ and C3′) and carbonyls (C1 and C4) or double-bonded carbons (C2 and C3) are marked.
Table 1.
13C chemical shifts of UQ headgroup in solution and bound to DsbB. (DSS scale, the solution shifts in TMS scale are converted by +2.0 ppm35). Carbonyl chemical shifts in DsbB with significant difference are bolded.
| Chemical Shift (ppm) | UQ-1038 | Ubiquinol-1037 | UQ-8 in DsbB(C41S) pH 5.5 | UQ-8 in DsbB(C41S) pH 8.0 |
|---|---|---|---|---|
| C=O (1, 4) | 185.9, 186.7 | 141.9, 142.1 | 165.5 | 154.7 |
| C=C (2, 3) | 146.4, 146.2 | 138.9, 138.8 | 137.5 | 142.4 |
| C=C (5, 6) | 140.8, 143.7 | 120.0, 124.0 | 126.8, 137.5 | 127.4, 135.6 |
| CH3O (2′, 3′) | 63.0 | 62.8, 62.7 | 63.4, 65.2 | 63.6, 64.0 |
| CH3 (5′) | 13.9 | 14.0 | 15.0 | 15.9 |
Figure 5.
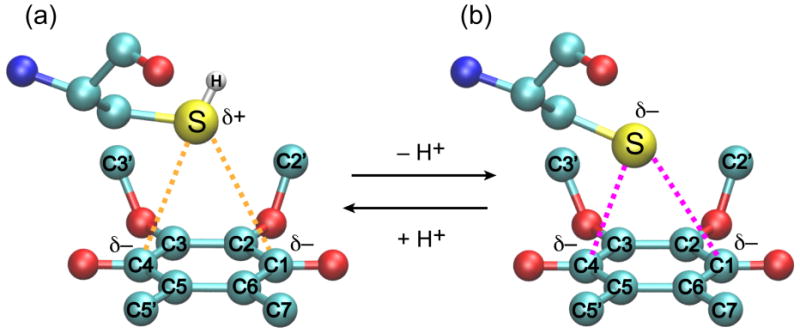
Schematic representations of DsbB C44 and UQ complex at (a) pH 5.5 and (b) pH 8.0. Atoms are color-coded: carbons (cyan), oxygens (red), sulfurs (yellow), hydrogen (white) and nitrogens (blue). Dashed lines indicate the interactions between thiolate/thiol and UQ carbonyl groups.
The chemical shift assignments of UQ-8 in DsbB(C41S) provide a basis for investigating UQ-8 in the covalently-bound DsbA(C33S)/DsbB complex. This complex mimics a transient reaction intermediate in the catalytic cycle of the DsbA-DsbB system.42 Like DsbB(C41S), and the transient intermediate itself, DsbA(C33S)/DsbB shows the pH-dependent ∼500 nm absorbance band. 2D 13C-13C DARR experiments were carried out on the complex of natural abundance DsbA(C33S) and [U-13C, 15N]DsbB (DsbA(C33S)/DsbB*). Most of the crosspeaks in the 2D 13C-13C correlation spectrum for DsbA(C33S)/DsbB* are also observed for U-DsbB(C41S), while T103 is only observed in the complex sample (Figure S3a). Since T103 is adjacent to C104, which is covalently bonded to DsbA C30, the appearance of T103 can be attributed to the binding of DsbA reducing the dynamics and/or disorder of the DsbB loop regions, which in turn enhances CP signals. Figure 6 shows the direct comparison of expansions of UQ-8 resonances in U-DsbB(C41S) and DsbA(C33S)/DsbB*. The crosspeak pattern is the same as in the spectrum of U-DsbB. The similarity in SSNMR spectra correlates with ∼500 nm absorbance found in both samples, and strongly suggests that the interaction between C44 and UQ-8 is the same in DsbB(C41S) and DsbA(C33S)/DsbB. Actually, the fact that both samples exhibit only one set of chemical shifts corresponding to UQ headgroup at pH 8.0 rules out the possibility that the ∼500 nm absorbance is due to quinhydrone-like charge-transfer complex between UQ and hydroquinone,48 or otherwise we would expect to observe another set of peaks from hydroquinone.
Figure 6.
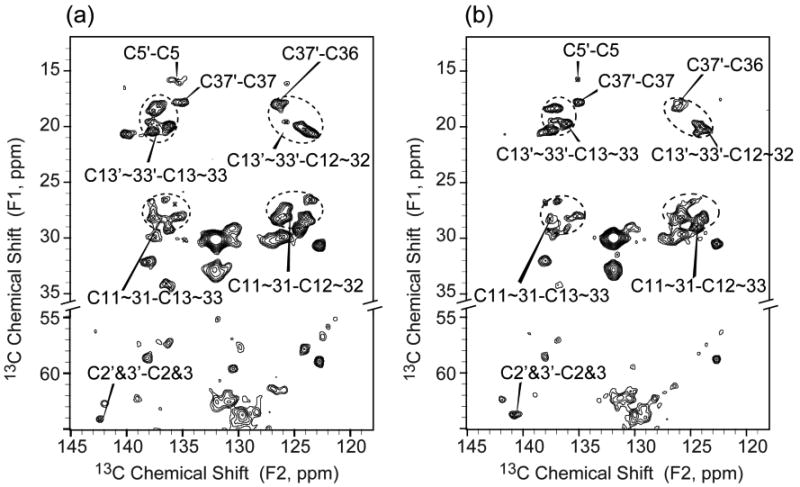
Comparison of expansions of UQ-8 resonances regions from a 13C-13C 2D correlation spectra of (a) U-DsbB(C41S) and (b) DsbA(C33S)/DsbB* complex at 12.5 kHz MAS with 25 ms DARR mixing (B0 = 17.6 T, Tsample = −12 °C, 1.5 s pulse delay, maximum t1 = 15.36 ms, maximum t2 = 20.48 ms). Both samples exhibit the similar crosspeaks of UQ isoprenoid chain and headgroup.
In the process of obtaining high-resolution SSNMR spectra of DsbB and DsbA/DsbB complex, we found that the temperature plays an important role in the spectral quality due to the dynamic nature of these membrane protein samples (Figure 7). We have previously determined that our samples contained ∼24 phospholipids per DsbB.27 Based on the DDM assays, there are minimal amounts of residual detergent in SSNMR samples. This combination of residual lipids and detergent is sufficient to cause a phase transition of membrane samples at ∼ −10 °C. In addition, the ability of this residual lipid and detergent to mimic the native membrane environment is most likely responsible for the microscopic order, manifested as narrow 13C linewidths (∼0.5 ppm), in our precipitated DsbB samples. At −12 °C (Figure 7a and e), close to the phase transition temperature, the crosspeaks from UQ-8 and the residues in the loop region or close to UQ-8 binding site are well resolved. As the temperature is decreased, these peaks broaden. Some peaks, such as I45 and UQ-8 C5′-C5, disappear entirely, indicating that these sites become disordered at low temperature (Figure 7).
Figure 7.
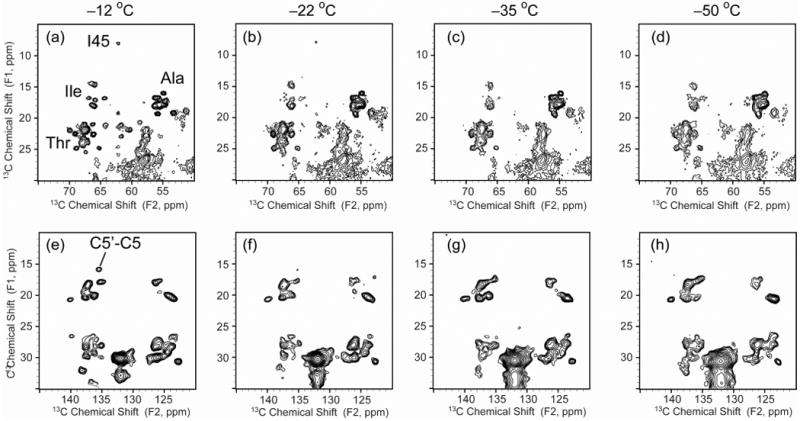
13C-13C 2D correlation spectra of U-DsbB(C41S) under 12.5 kHz MAS with 25 ms DARR mixing at various temperatures (B0 = 17.6 T, Tsample = −12 ∼ −50 °C, 1.5 s pulse delay, maximum t1 = 15.36 ms, maximum t2 = 20.48 ms). (a-d) Expanded regions with Ala, Ile and Thr crosspeaks at various temperatures. (e-h) Expanded regions with UQ-8 crosspeaks at various temperatures. Signals of residues that are close to the membrane surface get broader linewidths when the temperature decreases (I45, Ala, Ile and Thr in the loop regions, UQ C5′).
Conclusions
In summary, we have successfully characterized the endogenous UQ-8 in the disulfide bond generating membrane protein DsbB by obtaining high-resolution SSNMR spectra of several differently labeled membrane protein samples. The data have shown the chemical shift perturbation of UQ headgroup upon the unique binding to DsbB(C41S), which suggests that DsbB changes the electron distribution of UQ headgroup remarkably. In particular, the observation of a 10.8 ppm upfield shift for UQ carbonyl upon pH change from 5.5 to 8.0 is consistent with the increased electron population on UQ as evidence of the charge-transfer complex. We identified the same UQ resonances in the 41 kDa membrane protein complex, DsbA(C33S)/DsbB, which also forms the charge-transfer complex with UQ. These results open up the opportunity of atomic-level resolution of the mechanism of de novo disulfide bond generation in E. coli, and also demonstrate the ability of SSNMR to effectively probe functional UQ redox chemistry in a noncrystalline integral membrane protein.
Supplementary Material
Acknowledgments
This research was supported by the National Institutes of Health (NIGMS and Roadmap Initiative R01GM075937) and the Molecular Biophysics Training Grant (PHS 5 T32 GM008276) and Ullyot Fellowship to LJS. The authors thank School of Chemical Sciences NMR Facility at the University of Illinois at Urbana-Champaign for assistance with data acquisition and Dr. Jakob Lopez and Gemma Comellas for helpful discussions.
Footnotes
Supporting Information Available: 13C-13C 2D correlation spectra of FLYPW-DsbB(C41S) with and without 15N REDOR dephasing pulses; 13C-13C 2D correlation spectrum of DsbA(C33S)/DsbB*; a full list of chemical shifts of UQ. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Cramer WA, Knaff DB. Energy Transduction in Biological Membranes. Springer; New York: 1990. [Google Scholar]
- 2.Turunen M, Olsson J, Dallner G. Biochim Biophys Acta, Biomembr. 2004;1660:171. doi: 10.1016/j.bbamem.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 3.Srinivasan N, Golbeck JH. Biochim Biophys Acta, Bioenerg. 2009;1787:1057. doi: 10.1016/j.bbabio.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 4.van Liemt WB, Boender GJ, Gast P, Hoff AJ, Lugtenburg J, de Groot HJ. Biochemistry. 1995;34:10229. doi: 10.1021/bi00032a017. [DOI] [PubMed] [Google Scholar]
- 5.Boers RB, Gast P, Hoff AJ, de Groot HJM, Lugtenburg J. Eur J Org Chem. 2002;2002:189. [Google Scholar]
- 6.Matysik J, Alia, Gast P, van Gorkom HJ, Hoff AJ, de Groot HJ. Proc Natl Acad Sci U S A. 2000;97:9865. doi: 10.1073/pnas.170138797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akutsu H, Egawa A, Fujiwara T. Photosynth Res. 2010;104:221. doi: 10.1007/s11120-009-9523-2. [DOI] [PubMed] [Google Scholar]
- 8.Ganapathy S, Oostergetel GT, Wawrzyniak PK, Reus M, Gomez Maqueo Chew A, Buda F, Boekema EJ, Bryant DA, Holzwarth AR, de Groot HJ. Proc Natl Acad Sci U S A. 2009;106:8525. doi: 10.1073/pnas.0903534106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bajaj VS, Mak-Jurkauskas ML, Belenky M, Herzfeld J, Griffin RG. Proc Natl Acad Sci U S A. 2009;106:9244. doi: 10.1073/pnas.0900908106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Struts AV, Salgado GF, Tanaka K, Krane S, Nakanishi K, Brown MF. J Mol Biol. 2007;372:50. doi: 10.1016/j.jmb.2007.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahuja S, Crocker E, Eilers M, Hornak V, Hirshfeld A, Ziliox M, Syrett N, Reeves PJ, Khorana HG, Sheves M, Smith SO. J Biol Chem. 2009;284:10190. doi: 10.1074/jbc.M805725200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawamura I, Degawa Y, Yamaguchi S, Nishimura K, Tuzi S, Saito H, Naito A. Photochem Photobiol. 2007;83:346. doi: 10.1562/2006-06-20-RC-941. [DOI] [PubMed] [Google Scholar]
- 13.Kamihira M, Vosegaard T, Mason AJ, Straus SK, Nielsen NC, Watts A. J Struct Biol. 2005;149:7. doi: 10.1016/j.jsb.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 14.Ahuja S, Eilers M, Hirshfeld A, Yan EC, Ziliox M, Sakmar TP, Sheves M, Smith SO. J Am Chem Soc. 2009;131:15160. doi: 10.1021/ja9034768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kadokura H, Katzen F, Beckwith J. Annu Rev Biochem. 2003;72:111. doi: 10.1146/annurev.biochem.72.121801.161459. [DOI] [PubMed] [Google Scholar]
- 16.Ito K, Inaba K. Curr Opin Struct Biol. 2008;18:450. doi: 10.1016/j.sbi.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 17.Inaba K, Takahashi YH, Fujieda N, Kano K, Miyoshi H, Ito K. J Biol Chem. 2004;279:6761. doi: 10.1074/jbc.M310765200. [DOI] [PubMed] [Google Scholar]
- 18.Inaba K, Takahashi YH, Ito K, Hayashi S. Proc Natl Acad Sci U S A. 2006;103:287. doi: 10.1073/pnas.0507570103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fliszar S, Cardinal G, Beraldin MT. J Am Chem Soc. 1982;104:5287. [Google Scholar]
- 20.Li Y, Berthold DA, Frericks HL, Gennis RB, Rienstra CM. ChemBioChem. 2007;8:434. doi: 10.1002/cbic.200600484. [DOI] [PubMed] [Google Scholar]
- 21.Inaba K, Ito K. EMBO J. 2002;21:2646. doi: 10.1093/emboj/21.11.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bardwell JCA, Mcgovern K, Beckwith J. Cell. 1991;67:581. doi: 10.1016/0092-8674(91)90532-4. [DOI] [PubMed] [Google Scholar]
- 23.Vuister GW, Kim SJ, Wu C, Bax A. J Am Chem Soc. 1994;116:9206. [Google Scholar]
- 24.Shi LC, Ahmed MAM, Zhang WR, Whited G, Brown LS, Ladizhansky V. J Mol Biol. 2009;386:1078. doi: 10.1016/j.jmb.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 25.Marley J, Lu M, Bracken C. J Biomol NMR. 2001;20:71. doi: 10.1023/a:1011254402785. [DOI] [PubMed] [Google Scholar]
- 26.Studier FW. Protein Expr Purif. 2005;41:207. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, Berthold DA, Gennis RB, Rienstra CM. Protein Sci. 2008;17:199. doi: 10.1110/ps.073225008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lau FW, Bowie JU. Biochemistry. 1997;36:5884. doi: 10.1021/bi963095j. [DOI] [PubMed] [Google Scholar]
- 29.Degrip WJ, Vanoostrum J, Bovee-Geurts PHM. Biochem J. 1998;330:667. doi: 10.1042/bj3300667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li L, Nachtergaele S, Seddon AM, Tereshko V, Ponomarenko N, Ismagilov RF. J Am Chem Soc. 2008;130:14324. doi: 10.1021/ja805361j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hediger S, Meier BH, Kurur ND, Bodenhausen G, Ernst RR. Chem Phys Lett. 1994;223:283. [Google Scholar]
- 32.Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG. J Chem Phys. 1995;103:6951. [Google Scholar]
- 33.Fung BM, Khitrin AK, Ermolaev K. J Magn Reson. 2000;142:97. doi: 10.1006/jmre.1999.1896. [DOI] [PubMed] [Google Scholar]
- 34.Raiford DS, Fisk CL, Becker ED. Anal Chem. 1979;51:2050. [Google Scholar]
- 35.Morcombe CR, Zilm KW. J Magn Reson. 2003;162:479. doi: 10.1016/s1090-7807(03)00082-x. [DOI] [PubMed] [Google Scholar]
- 36.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. J Biomol NMR. 1995;6:277. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 37.Afri M, Ehrenberg B, Talmon Y, Schmidt J, Cohen Y, Frimer AA. Chem Phys Lipids. 2004;131:107. doi: 10.1016/j.chemphyslip.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 38.Vanliemt WBS, Steggerda WF, Esmeijer R, Lugtenburg J. Recl Trav Chim Pays-Bas. 1994;113:153. [Google Scholar]
- 39.Takegoshi K, Nakamura S, Terao T. Chem Phys Lett. 2001;344:631. [Google Scholar]
- 40.Hohwy M, Jakobsen HJ, Edén M, Levitt MH, Nielsen NC. J Chem Phys. 1998;108:2686. [Google Scholar]
- 41.Zhou YP, Cierpicki T, Jimenez RHF, Lukasik SM, Ellena JF, Cafiso DS, Kadokura H, Beckwith J, Bushweller JH. Mol Cell. 2008;31:896. doi: 10.1016/j.molcel.2008.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Inaba K, Murakami S, Suzuki M, Nakagawa A, Yamashita E, Okada K, Ito K. Cell. 2006;127:789. doi: 10.1016/j.cell.2006.10.034. [DOI] [PubMed] [Google Scholar]
- 43.Inaba K, Murakami S, Nakagawa A, Iida H, Kinjo M, Ito K, Suzuki M. EMBO J. 2009;28:779. doi: 10.1038/emboj.2009.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tapley TL, Eichner T, Gleiter S, Ballou DP, Bardwell JCA. J Biol Chem. 2007;282:10263. doi: 10.1074/jbc.M611541200. [DOI] [PubMed] [Google Scholar]
- 45.Nakashima TT, Maciel GE. Org Magn Res. 1972;4:321. [Google Scholar]
- 46.Zhang H, Neal S, Wishart DS. J Biomol NMR. 2003;25:173. doi: 10.1023/a:1022836027055. [DOI] [PubMed] [Google Scholar]
- 47.Kornhaber GJ, Snyder D, Moseley HNB, Montelione GT. J Biomol NMR. 2006;34:259. doi: 10.1007/s10858-006-0027-5. [DOI] [PubMed] [Google Scholar]
- 48.Regeimbal J, Gleiter S, Trumpower BL, Yu CA, Diwakar M, Ballou DP, Bardwell JCA. Proc Natl Acad Sci U S A. 2003;100:13779. doi: 10.1073/pnas.1935988100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


