Abstract

Natural product gene clusters are often tightly regulated, resulting in gene cluster silencing in laboratory fermentation studies. The systematic overexpression of transcription factors (TFs) associated with biosynthetic gene clusters found in the genome of Burkholderia thailandensis E264 identified a set of TFs that, when overexpressed, alter the secondary metabolome of this bacterium. The isolation and characterization of burkholdacs A and B, two new acyldepsitripeptide histone deacetylase inhibitors produced by B. thailandensis overexpressing the TF bhcM is reported.
Burkholderia mallei and Burkholderia pseudomallei are the etiological agents of glanders and melioidosis, respectively, and have been classified as priority pathogens by the NIH/CDC because of their perceived threat as potential bioterrorism agents.1 These pathogens have garnered additional interest because their genomes contain a large number of cryptic small molecule biosynthetic gene clusters that are likely to encode, among other things, unappreciated small molecule pathogenicity determinants and, more generally, small molecules that might specifically interact with the host proteome.2 Burkholderia thailandensis E264 is a close relative of both pathogens, but is not considered a human pathogen.3 B. thailandensis is therefore a convenient model in which to study cryptic secondary metabolism in Burkholderia spp. Natural product gene clusters are often tightly regulated by both positive- and negative-acting transcription factors (TFs) frequently resulting in gene cluster silencing or low-level expression in laboratory fermentation studies. The manipulation of individual secondary metabolite gene cluster-associated TFs has been shown to alter the set of metabolites a microbe produces (Figure 1A).4,5 Here we describe the characterization of burkholdacs A (1) and B (2), two new histone deacetylase (HDAC) inhibitors identified through the systematic overexpression of TFs associated with natural product gene clusters encoded within B. thailandensis.
Figure 1.
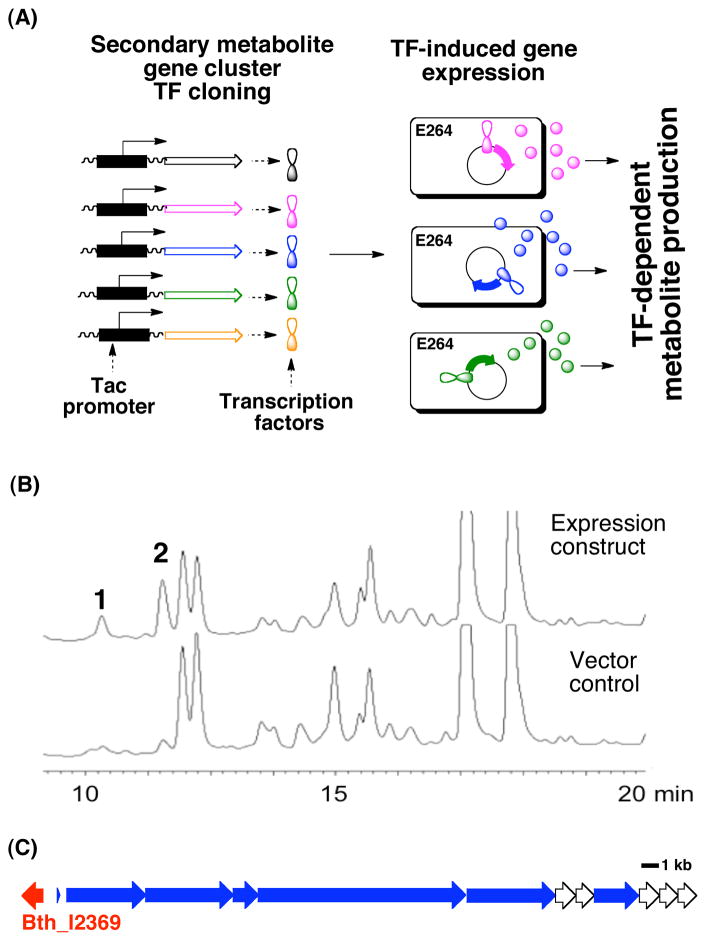
Activating cryptic metabolite production via transcription factor expression: (A) Transcription factors putatively associated with B. thailandensis E264 biosynthetic gene clusters were PCR amplified, individually cloned downstream of a Ptac promoter and the resulting constructs were then transformed back into B. thailandensis E264. Culture broth extracts from E264 strains over-expressing each transcription factor were assessed for the presence of metabolites not seen in extracts from similarly treated vector control cultures. (B) HPLC analysis of the ethyl acetate extract from cultures of E264 overexpressing TF Bth_I2369. Peaks marked 1 and 2 on this trace correspond to burkholdacs A and B, respectively (Figure 2). (C) The predicted NRPS/PKS gene cluster directly adjacent to TF Bth_I2369 is shown (blue, predicted NRPS/PKS genes; red, cloned transcription factor).
The B. thailandensis E264 genome contains at least 11 gene clusters with predicted nonribosomal peptide synthetase (NRPS) or polyketide synthase (PKS) genes. Thailandamide and bactobolin D, the products encoded by two of these gene clusters, were identified through the examination of metabolites produced by wild type strains of B. thailandensis.2 The metabolites encoded by the remaining NRPS/PKS gene clusters have not yet been identified. A bioinformatics analysis of the B. thailandensis E264 genome identified 30 TFs either within or directly adjacent to predicted NRPS/PKS gene clusters. Each TF was PCR amplified from B. thailandensis genomic DNA and cloned behind the Ptac promoter found in the broad host range expression vector pJWC1tac (Table S1). These TF expression constructs were then conjugated into B. thailandensis E264, and ethyl acetate extracts from IPTG-induced cultures of the resulting exconjugates were examined by TLC and LCMS. We observed two instances where the induction of a secondary metabolite gene cluster-associated TF, Bth_I2369 and Bth_II1681, altered the metabolic profile found in the culture broth organic extracts (Figures 1B and S1). Each of these TFs lies directly adjacent to a predicted small molecule encoding gene cluster and is presumed to stimulate the production of metabolites encoded by an adjacent gene cluster. Bth_II1681 encodes for a LysR-type TF associated with the previously characterized thailandamide biosynthetic cluster.2a Ishida et al. recently showed that disruption of Bth_II1681 induces the production of a novel thailandamide lactone variant.5 For this study, we pursued the metabolites whose biosynthesis is stimulated by Bth_I2369 (bhcM), an AraC-type TF directly adjacent to an uncharacterized hybrid-NRPS/PKS biosynthetic cluster (Figure 1B, C).
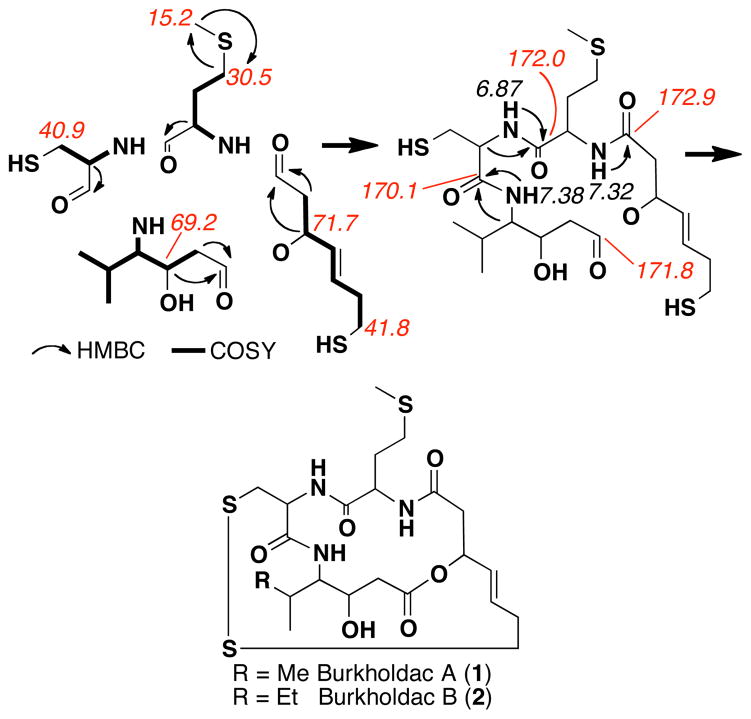
Two clone specific metabolites (1 and 2), produced in a 1:3 ratio, were purified from extracts of B. thailandensis cultures overexpressing bhcM (Figure 1B). The structures of compounds 1 and 2 were then elucidated using HRMS and NMR data. The 1H-1H COSY spectrum of 1 defined four spin systems (Figure 2). Three of these spin systems contain COSY couplings to partially exchangeable amide protons and show HMBC correlations to carbonyl carbons, indicating they are amino acids. Based on 13C and 1H chemical shift data and HMBC correlations, these amino acids were determined to be methionine, cysteine and the γ-amino acid statine. The fourth COSY spin system contains six-carbons, including one trans olefin (J=15 Hz) and an oxygen-substituted methine (δ 71.7 ppm). The last carbonyl carbon (δ 172.9 ppm) found in the 13C spectra is connected to one end of this spin system by HMBC correlations. The other end of the spin system is predicted, based on 13C and 1H chemical shift data and the predicted molecular formula, to be sulfur substituted, thus yielding a ζ-thio-β-hydroxyacid substructure. The three amino acids and the ζ-thio-β-hydroxyacid can be connected into an acylated tripeptide using HMBC correlations from the partially exchangeable amide protons and the α-carbon protons to each adjacent carbonyl carbon (Figure 2). To satisfy the HRMS-predicted molecular formula (C22H35N3O6S3), two additional unsaturations must be added to the linear substructure. One unsaturation can be satisfied with the formation of an ester bond between the fourth carbonyl (δ 171.8 ppm) and the oxygen-substituted methine (δ 71.7 ppm) of the ζ-thio-β-hydroxyacid. The incorporation of a disulfide bond then satisfies the final unsaturation to give burkholdac A (1). The presence of a disulfide bond was confirmed by the gain of two protons upon dithiothreitol reduction. The structure of burkholdac B (2 ) was elucidated using identical arguments with one exception. The molecular formula predicted by HRMS indicates 2 contains an additional CH2, which is seen in the COSY as an additional methylene in the γ-amino acid of the tripeptide. Compounds 1 and 2 are new members of a small class of acyldepsipeptides that includes spiruchostatins A (3) and B (4 ), FK228 ( 5 ) and FR901375 (6) (Figure 3).6 FK228 and FR901375 are tetrapeptides while the spiruchostatins are γ-amino acid containing tripeptides. Burkholdacs A and B differ from spiruchostatins A and B by the substitution of a methionine for an alanine.7
Figure 2.
NMR assignments of the burholdac core structure.
Figure 3.
Natural depsipeptides related to burkholdacs A and B.
The structures of burkholdacs A and B are supported by biosynthetic arguments based on the gene cluster that is directly adjacent to bhcM, the TF that was overexpressed in this study (Figure 4). This gene cluster, which we have named the BHC (B urkholderia HDAC inhibitor) cluster, resembles the FK228 (5) (or DEP) gene cluster from Chromobacterium violaceum.6b Both clusters contain genes that are predicted to encode the biosynthesis of the chain-initiating ζ–thio-β-hydroxyacid (bhcABCF), a disulfide bridge-forming oxidoreductase (bhcH) and putative resistance/proofreading enzymes (bhcG, I, J) (Table S2).6b,d Unique to the BHC cluster however are the modular NPRS and PKS genes, bhcDEK, which are predicted to encode the biosynthesis of the tripeptide. BhcD contains three NRPS modules that are expected to afford the tandem addition of Met, Cys and Val/Ile to theζ-thio-β-hydroxyacid starter. Transformation of the terminal Val/Ile to the γ-amino acid derivative could then be carried out by the ketosynthase (KS) and ketoreductase (KR) domains on BhcE. BhcE does not contain a predicted acyltransferase (AT) domain and therefore likely uses a yet-unknown AT domain in trans. The biosynthesis of the ζ.–thio-β-hydroxyacid starter unit of both the DEP and the BHC clusters is also predicted to use trans AT domains.6b The final NRPS gene in this cluster, BhcK, contains an additional thiolation domain and has the only thioesterase domain present in the gene cluster. BhcK is therefore predicted to carry out the cyclization and release of the acyldepsipeptide to yield the reduced form of 1 (Figure 4).
Figure 4.
(a) DEP and BHC gene clusters. (b) The late stages of burkholdac biosynthesis (bhcDEK) are unique to the BHC cluster. ACP/PCP, acyl or peptidyl carrier protein; C, condensation; A, adenylation; E, epimerization; KS, ketosynthase; KR, ketoreductase; TE, thioesterase. Superscript “nf” notes nonfunctional domain. Gene and domain functional predictions appear in Table S2.
Of the three NRPS modules found in the BHC gene cluster, only the Cys incorporating module contains an epimerizaton domain. Based on biosynthetic arguments, burkholdacs A and B are therefore predicted to contain L-Met, D-Cys and L-Val/Ile moieties. The stereochemicial details shown in Figure 4 reflict these predictions. NMR studies designed to confirm our stereochemical predictions were inconclusive. This may be a general phenomena with molecules of this type as X-ray crystallography was used to assign the relative stereochemistry within FK228, and the majority of the stereochemicial assignments within spiruchostatin A and B were ultimately determined by total synthesis.6,8
Metabolites in this family (3 – 6) are potent HDAC inhibitors, and FK228 has been approved for treatment of cutaneous T cell lymphomas.6,8 They occur as pro-drugs that are active in vivo upon reduction of the disulfide bond. The depsipeptide “cap” is thought to allow this class of inhibitors to discriminate between individual HDAC family members.8 The major metabolite (2) was selected for HDAC analysis. Dithiothreitol-reduced 2 shows potent activity against HDACs 1–3 (IC50 ~30 nM) and is essentially inactive against HDACs 4–10 at nM concentrations (Figure S2). Recent gene profiling and in vivo studies indicate HDAC inhibition may aid in microbial virulence.9 Although B. thailandensis is not a human pathogen, it is infectious to other organisms.3 The role spiruchostatins play in pathogenesis remains to be determined.
Systematic expression of secondary metabolite gene cluster associated TFs is a simple and generalizable strategy by which a subset of previously cryptic biosynthetic gene clusters can be activated within sequenced bacterial genomes. Sequenced eukaryotic pathogens in particular are likely to be rich sources of cryptic gene clusters that encode metabolites capable of interacting specifically within the human proteome.
Supplementary Material
Acknowledgments
This work was supported by the Northeast Biodefense Center (U54-AI057158) and NIH GM077516. SFB is a Howard Hughes Medical Institute early career scientist.
Footnotes
Supporting Information Available. Molecular biology, molecule production and isolation protocols as well as NMR spectra are available free of charge at http://pubs.acs.org.
References
- 1.Adler NR, Govan B, Cullinane M, Harper M, Adler B, Boyce JD. FEMS Microbiol Rev. 2009;33:1079. doi: 10.1111/j.1574-6976.2009.00189.x. [DOI] [PubMed] [Google Scholar]
- 2.(a) Nguyen T, Ishida K, Jenke-Kodama H, Dittmann E, Gurgui C, Hochmuth T, Taudien S, Platzer M, Hertweck C, Piel J. Nat Biotechnol. 2008;26:225. doi: 10.1038/nbt1379. [DOI] [PubMed] [Google Scholar]; (b) Duerkop BA, Varga J, Chandler JR, Peterson SB, Herman JP, Churchill ME, Parsek MR, Nierman WC, Greenberg EP. J Bacteriol. 2009;191:3909. doi: 10.1128/JB.00200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Seyedsayamdost MR, Chandler JR, Blodgett JA, Lima PS, Duerkop BA, Oinuma K, Greenberg EP, Clardy J. Org Lett. 2010;12:716. doi: 10.1021/ol902751x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Kim HS, Schell MA, Yu Y, Ulrich RL, Sarria SH, Nierman WC, DeShazer D. BMC Genomics. 2005;6:174. doi: 10.1186/1471-2164-6-174. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Yu Y, Kim HS, Chua HH, Lin CH, Sim SH, Lin D, Derr A, Engels R, DeShazer D, Birren B, Nierman WC, Tan P. BMC Microbiol. 2006;6:46. doi: 10.1186/1471-2180-6-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Holden MT, McGowan SJ, Bycroft BW, Stewart GS, Williams P, Salmond GP. Microbiology. 1998;144:1495. doi: 10.1099/00221287-144-6-1495. [DOI] [PubMed] [Google Scholar]; (b) Bergmann S, Schumann J, Scherlach K, Lange C, Brakhage AA, Hertweck C. Nat Chem Biol. 2007;3:213. doi: 10.1038/nchembio869. [DOI] [PubMed] [Google Scholar]; (c) Challis GL. J Med Chem. 2008;51:2618. doi: 10.1021/jm700948z. [DOI] [PubMed] [Google Scholar]; (d) Scherlach K, Hertweck C. Org Biomol Chem. 2009;7:1753. doi: 10.1039/b821578b. [DOI] [PubMed] [Google Scholar]
- 5.Ishida K, Lincke T, Behnken S, Hertweck C. J Am Chem Soc. 2010;132:13966. doi: 10.1021/ja105003g. [DOI] [PubMed] [Google Scholar]
- 6.(a) Chen Y, Gambs C, Abe Y, Wentworth P, Jr, Janda KD. J Org Chem. 2003;68:8902. doi: 10.1021/jo034765b. [DOI] [PubMed] [Google Scholar]; (b) Cheng YQ, Yang M, Matter AM. Appl Environ Microbiol. 2007;73:3460. doi: 10.1128/AEM.01751-06. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Narita K, Kikuchi T, Watanabe K, Takizawa T, Oguchi T, Kudo K, Matsuhara K, Abe H, Yamori T, Yoshida M, Katoh T. Chemistry. 2009;15:11174. doi: 10.1002/chem.200901552. [DOI] [PubMed] [Google Scholar]; (d) Wang C, Wesener SR, Zhang H, Cheng YQ. Chem Biol. 2009;16:585. doi: 10.1016/j.chembiol.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 7.This appears to be the first sequenced methionine-selective adenylation domain and is only the second bacterial NRPS product with a Met residue: Caboche S, Leclere V, Pupin M, Kucherov G, Jacques P. J Bacteriol. 2010;192:5143. doi: 10.1128/JB.00315-10.
- 8.(a) Shigematsu N, Ueda H, Takase S, Tanaka H, Yamamoto K, Tada T. J Antibiot (Tokyo) 1994;47:311. doi: 10.7164/antibiotics.47.311. [DOI] [PubMed] [Google Scholar]; (b) Masuoka Y, Nagai A, Shin-ya K, Furihata K, Nagai K, Suzuki K, Hayakawa Y, Seto H. Tetrahedron Lett. 2001;42:41. [Google Scholar]; (c) Yurek-George A, Habens F, Brimmell M, Packham G, Ganesan A. J Am Chem Soc. 2004;126:1030. doi: 10.1021/ja039258q. [DOI] [PubMed] [Google Scholar]; (d) Yurek-George A, Cecil ARL, Mo AHK, Wen S, Rogers H, Habens F, Maeda S, Yoshida M, Packham G, Ganesan A. J Med Chem. 2007;50:5720. doi: 10.1021/jm0703800. [DOI] [PubMed] [Google Scholar]
- 9.Roger T, Lugrin J, Le Roy D, Goy G, Mombelli M, Koessler T, Ding XC, Chanson AL, Reymond MK, Miconnet I, Schrenzel J, Francois P, Calandra T. Blood. 2011;117:1205. doi: 10.1182/blood-2010-05-284711. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.