Abstract
The ataxia-telangiectasia mutated and RAD3-related (ATR) kinase initiates DNA damage signaling pathways in human cells after DNA damage such as that induced upon exposure to ultraviolet light by phosphorylating many effector proteins including the checkpoint kinase Chk1. The conventional view of ATR activation involves a universal signal consisting of genomic regions of replication protein A-covered single-stranded DNA. However, there are some indications that the ATR-mediated checkpoint can be activated by other mechanisms. Here, using the well defined Escherichia coli lac repressor/operator system, we have found that directly tethering the ATR activator topoisomerase IIβ-binding protein 1 (TopBP1) to DNA is sufficient to induce ATR phosphorylation of Chk1 in an in vitro system as well as in vivo in mammalian cells. In addition, we find synergistic activation of ATR phosphorylation of Chk1 when the mediator protein Claspin is also tethered to the DNA with TopBP1. Together, these findings indicate that crowding of checkpoint mediator proteins on DNA is sufficient to activate the ATR kinase.
Keywords: Cell Cycle, Checkpoint Control, DNA-binding Protein, DNA Damage, DNA-Protein Interaction, DNA Repair, Enzyme Mechanisms, Nucleic Acid Enzymology, Protein-DNA Interaction, Protein Kinases
Introduction
DNA damage checkpoints delay cell cycle progression in response to DNA damage to maintain genomic integrity. In mammalian cells, checkpoint response signaling pathways are initiated primarily by two large phosphoinositide 3-kinase-related serine-threonine kinases, ataxia-telangiectasia mutated (ATM)2 and ATM and RAD3-related (ATR). ATM is activated mainly, but not exclusively, by double-stranded breaks, and ATR is activated by ultraviolet (UV) and UV-mimetic chemical agents as well as other conditions that result in replication fork stalling. Upon activation, ATR activates the key signal-transducing kinase Chk1 by phosphorylating it on Ser317 and Ser345 (1, 2), and the mediator protein Claspin is required for the efficient phosphorylation of Chk1 (3–5). ATR activation also requires topoisomerase IIβ-binding protein 1 (TopBP1), which has been shown to activate ATR directly in defined systems in vitro (6–11) as well as in vivo (6, 12).
The current model for ATR activation is as follows. Single-stranded DNA (ssDNA) generated at sites of DNA damage during repair, transcription, or replication is bound by replication protein A (RPA), which then recruits ATR through a physical interaction between RPA and the ATR-binding partner, ATR-interacting protein (ATRIP). Independently, Rad17-RFC loads the 9-1-1 (Rad9-Rad1-Hus1) checkpoint complex at primer/template junctions, where it recruits TopBP1 in the proximity of ATR. We have previously described an in vitro system that recapitulates ATR phosphorylation of Chk1 dependent on RPA-coated ssDNA and TopBP1 (8). However, RPA is not required for maximal ATR kinase activity in this system when the ssDNA is replaced with DNA containing bulky DNA base adducts (9, 10). In fact, we found that TopBP1 binds directly to damaged DNA and that the DNA binding activity of TopBP1 is required to confer damaged DNA-dependent activation of ATR. Thus, we hypothesized that TopBP1 may directly recognize damaged DNA in the cell to activate ATR. It is difficult to assess the direct contribution of DNA damage in the activation of ATR because of the large number of cellular processes that generate ssDNA when DNA damage is encountered (i.e. replication, transcription, and repair). Therefore, to address the question of whether direct binding of TopBP1 to DNA can activate ATR, we have tethered TopBP1 to DNA by fusing TopBP1 to the lac repressor (LacR), which has high binding affinity for DNA containing the lac operator (LacO) sequence. We find that in the presence of LacO DNA, LacR-TopBP1 activates ATR phosphorylation of Chk1 both in vitro and in vivo. In addition, when LacR-Claspin is also tethered to the DNA it functions synergistically with LacR-TopBP1. These findings indicate that concentrating TopBP1 and Claspin on DNA is sufficient for Chk1 phosphorylation by ATR.
EXPERIMENTAL PROCEDURES
Antibodies, Reagents, and DNA
Chk1 Ser(P)345 antibodies were purchased from Cell Signaling Technology (2348), TopBP1 antibodies were from Millipore (AB3245), RPA2 antibodies were from Calbiochem (NA18), and Claspin (sc-48771) and Chk1 (sc-8408) antibodies were purchased from Santa Cruz Biotechnology. Isopropyl β-d-thiogalactopyranoside (IPTG) was purchased from Promega (V3955) and was dissolved in water. LacO plasmid is a ∼15-kb plasmid that contains 256 Lac operator repeats cloned into pBluescript (Addgene plasmid 17655, named Lac-I-SceI-Tet) (13). The control plasmid was selected because of its similar size of 14.15 kb and was generated by cloning the ATR gene into the pcDNA5-FRT/TO-LacI plasmid.
Plasmid Construction
The lac repressor sequence from pKM208 (14) (Addgene plasmid 13077) was the template for PCR with LacI forward oligonucletide (5′-GGCCAGATCTAAGCTTACCATGCCAGTAACGTTATACGATGTCG-3′)and LacI reverse oligonucleotide (5′-GGCCGTCGACGCGGCCGCGGTACCAGGCCTGCTAGCGGATCCCACCTTCCTCTTCTTCTTGGGTCGGGAAACCTGTCGTGCCAG-3′) which was digested with restriction enzymes HindIII and XhoI and cloned into these sites of pcDNA5-FRT/TO and pcDNA3 (Invitrogen). FLAG-TopBP1, FLAG-Claspin-C (amino acids 679–1332), FLAG-Claspin-FL, FLAG-Claspin 3A mutants (where amino acids Thr916, Ser945, Ser982 were mutated to alanine) (11) and FLAG-ATR were subcloned into the BamHI and NotI sites of these vectors. The fusion proteins produced from these plasmids contain the following: the N-terminal LacR, which is able to dimerize but not form a tetramer; a canonical nuclear localization sequence; a FLAG epitope; and the indicated checkpoint protein.
Purification of Checkpoint Proteins
Native ATR-ATRIP, GST-TopBP1-His, RPA, and His-Chk1 kinase-dead (His-Chk1-kd) were all purified as described previously (8, 9, 15). LacR-TopBP1 and LacR-Claspin-C WT and 3A were expressed in 293 FlpIn T-REX cells as described in the manufacturer's directions (Invitrogen), and purified with anti-FLAG-agarose (Sigma) as described previously (16).
Kinase Assays
The procedure was essentially as described previously (11). Briefly, kinase assay reactions contained 14 mm Hepes, pH 7.9, 30 mm KCl, 1.2 mm MgCl2, 0.4 mm ATP, 0.3 mm DTT, 1% glycerol, and 1 μm microcystin in a 10-μl final volume. Purified ATR-ATRIP (0.25 nm) was incubated in the reaction buffer for 20 min at 30 °C with 12 nm His-Chk1-kd and the indicated amounts of DNA and recombinant TopBP1 and Claspin. The reactions were terminated by the addition of SDS-PAGE loading buffer and separated by 10% SDS-PAGE. Chk1 phosphorylation was detected by immunoblotting using Ser(P)345 antibodies, and the levels of total Chk1 protein were subsequently detected by immunoblotting the same membrane. Levels of phosphorylation were quantified using ImageQuant 5.2 software after scanning the immunoblots. The highest level of Chk1 phosphorylation in each experiment was set equal to 100, and the levels of phosphorylated Chk1 in the other lanes were determined relative to this value. The averages from at least three independent experiments were graphed, and the error bars indicate the S.D.
DNA Binding Assay
LacO oligonucleotide 5′-TGTGGAATTGTGAGCGCTCACAATTCCACA-3′ or control oligonucletide 5′-ACACCTTAACACTCGCGAGTGTTAAGGTGT-3′ was 5′ end-labeled with γ-32P by polynucleotide kinase according to the manufacturer's instructions (New England Biolabs), self-annealed, and used in electrophoretic mobility shift assays (EMSAs). The indicated amounts of LacR-TopBP1 or LacR-Claspin-C were preincubated for 90 min on ice with or without 150 μm IPTG (concentration in final reactions) and then further incubated for 30 min at room temperature with 10 nm labeled oligonucleotides in 0.5× TBE (25 mm Tris, 25 mm boric acid, 0.6 mm EDTA), 3% glycerol, and 1 mm DTT. The reactions were loaded directly onto a 5% w/v polyacrylamide (30:1 acrylamide:bisacrylamide) gel, cast, and run in 0.5× TBE. Electrophoresis was carried out for 60 min at 25 mA. After drying, the gel was analyzed by phosphorimaging.
Cell Lines
NIH2/4 (LacO array-containing NIH3T3 cells) (13) and NIH3T3 cells were cultured in DMEM-H supplemented with 10% fetal calf serum, penicillin/streptomycin, and l-glutamine. The NIH2/4 cells were also cultured with 100 μg/ml hygromycin. For transient expression of LacR fusion proteins, 1 × 105 cells in 24-well plates were transfected with the indicated amount of plasmid using 1.25 μl of Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The cells were harvested 18 h after transfection by rinsing twice with 1× PBS and lysing with 50 μl of SDS sample buffer (62.5 mm Tris-HCl, pH 6.8, 2% w/v SDS, 10% glycerol, 50 mm DTT, 0.01% bromphenol blue). After brief sonication and boiling, 15 μl of the sample was loaded onto 10% SDS-PAGE for immunoblotting as described above.
RESULTS
Chk1 Phosphorylation by ATR Is Stimulated When TopBP1 Is Tethered to DNA in a Defined System
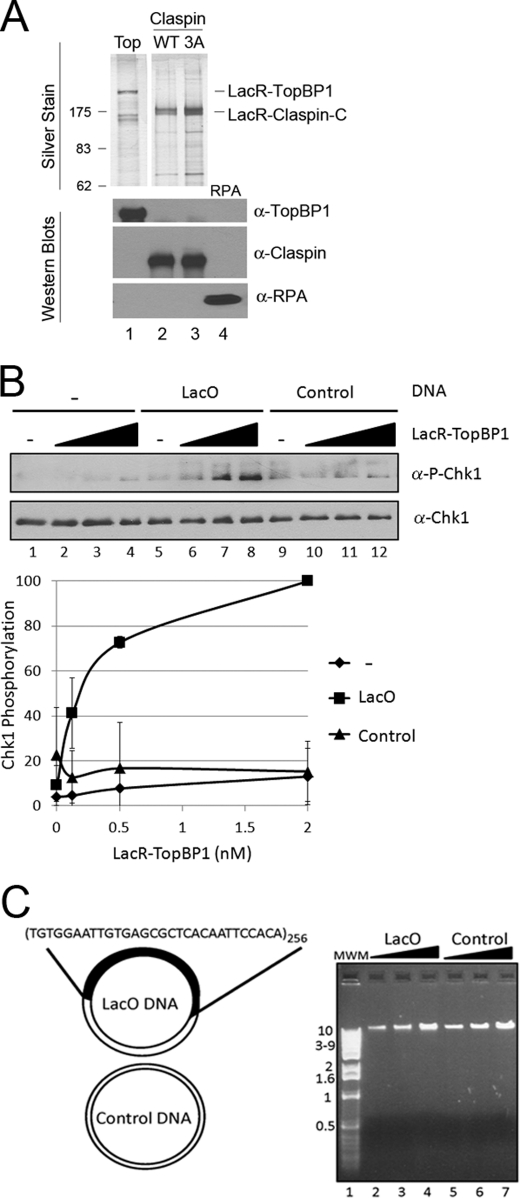
Because we have previously shown that DNA stimulates ATR kinase activity in a manner dependent on TopBP1 binding to DNA (9), we reasoned that when fused to LacR, TopBP1 should stimulate ATR kinase activity specifically when in the presence of LacO DNA even in the absence of DNA damage. To test this directly, we purified LacR-TopBP1 (Fig. 1A) and added it to kinase reactions where we can analyze the site-specific phosphorylation of Chk1 by ATR. Fig. 1B shows the results from adding increasing concentrations of LacR-TopBP1 to ATR kinase reactions without DNA (lanes 1–4), with LacO DNA (lanes 5–8), or with control DNA (lanes 9–12). LacR-TopBP1 stimulates ATR phosphorylation of Chk1 in reactions containing the LacO DNA up to 10-fold more than in reactions lacking DNA (compare lanes 8 and 4). The activity is specific for DNA containing the LacO sequence because an equal concentration of DNA lacking the LacO repeats does not stimulate ATR kinase activity (lanes 9–12). We observe the highest LacO sequence dependence at low concentrations of DNA, such as used in this experiment, where there is a negligible effect of adding DNA lacking LacO (see Fig. 3B for higher DNA concentrations). Because we have previously observed an effect of DNA length on the activation of ATR (10), it is important to note that the LacO-containing and control DNAs used in this study are of similar size (Fig. 1C). These data, along with our previous finding that recruitment of ATR to DNA by TopBP1 bound to bulky base adducts was sufficient for its activation, lead us to conclude that recruitment of ATR to DNA by whatever means is sufficient to activate it.
FIGURE 1.
Tethering TopBP1 to DNA activates ATR phosphorylation of Chk1 in a defined system. A, 5 ng of purified LacR-TopBP1 (lane 1), LacR-Claspin-C WT (lane 2), LacR-Claspin-C 3A (Thr916, Ser945, Ser982 to alanine) (lane 3), and RPA (lane 4) were fractionated by SDS-PAGE and then either stained with silver (top panels) or Western blotted with the indicated antibodies (bottom panels). B, LacR-TopBP1 (0, 0.125, 0.5, 2 nm) was added to kinase reactions containing 0.25 nm ATR and 12 nm His-Chk1. Reactions 1–4 contain no DNA, reactions 5–8 contain 3 pm LacO DNA plasmid, and reactions 9–12 contain 3 pm control DNA plasmid. ATR kinase activity was determined by immunoblotting for phospho-Chk1 and Chk1 as indicated. The graph shows quantitative analysis of the data. C, LacO and control DNA by ethidium bromide staining after electrophoresis on a 0.8% agarose gel. 100, 200, 400 ng of LacO DNA (lanes 2–4) or control DNA (lanes 5–7), previously linearized by digestion with the single cutting restriction endonucleases SacI and BamHI, respectively, were loaded onto the gel. The plasmids were linearized to simplify visualization, but linearization does not affect ATR activation in our system.
FIGURE 3.
DNA binding by TopBP1 and Claspin is required for the synergistic stimulation of ATR phosphorylation of Chk1 in a defined system. A, LacR-TopBP1 and LacR-Claspin-C DNA binding in the presence or absence of IPTG is analyzed by EMSAs. The binding reactions contained 10 nm 32P-labeled 30-bp LacO DNA (lanes 1–3 and 6–11) or control DNA (lanes 4 and 5) and 50 nm LacR-Claspin-C (lanes 2, 3, and 5) or 5 nm, 10 nm, 20 nm LacR-TopBP1 (lanes 6–8 and 9–11). In lanes 3 and 9–11, the LacR fusion protein was preincubated on ice with IPTG at a final concentration of 150 μm. The unbound DNA is indicated by a gray arrow, and the protein-bound DNA is indicated by a black arrow. The graph shows quantitative analysis of the data. B, ATR kinase reactions contained 0.125 nm LacR-TopBP1 and 6 nm LacR-Claspin-C and either no DNA (lanes 1 and 8), or 3, 12, or 48 pm LacO DNA (lanes 2–4 and 9–11), or 3, 12, or 48 pm control DNA (lanes 5–7 and 12–14) without (lanes 1–7) or with 150 μm IPTG (lanes 8–14).
DNA-tethered TopBP1 and Claspin Synergistically Stimulate ATR Phosphorylation of Chk1 in a Defined System
Consistent with data in vivo, we have previously reported that the phosphorylation of Chk1 by ATR is highly dependent on Claspin in vitro (11). Furthermore, a C-terminal fragment of Claspin is sufficient for its activity in vitro. We, therefore, purified the C-terminal fragment of Claspin fused to the LacR (Fig. 1A) to determine whether tethering Claspin to DNA would affect ATR phosphorylation of Chk1. Fig. 2A shows the results from adding LacR-Claspin to reactions with or without LacR-TopBP1 in the presence or absence of LacO-containing DNA. Under these more limiting reaction conditions we only observe significant ATR kinase activity in reactions containing LacR-TopBP1, LacR-Claspin, and LacO-containing DNA (lane 8). The limiting concentration of LacR-TopBP1 used in these reactions is insufficient to activate ATR even in the presence of LacO DNA (lane 5). Reactions lacking TopBP1 do not have significant Chk1 phosphorylation even in the presence of LacR-Claspin and LacO DNA (lane 11), suggesting that the dimerization of LacR is not simply bringing multiple DNAs together to activate ATR. These results indicate that LacR-Claspin works synergistically with LacR-TopBP1 in the ATR-Chk1 kinase reaction by recruiting the ATR substrate Chk1 in the presence of LacO-containing DNA and further reinforce the concept that ATR recruitment to DNA by any means is sufficient to activate its kinase function.
FIGURE 2.
TopBP1 and Claspin function synergistically to stimulate ATR phosphorylation of Chk1 when tethered to DNA in a defined system. A, ATR kinase reactions contained no mediator proteins (lanes 1–3), 0.125 nm LacR-TopBP1 (lanes 4–6), 6 nm LacR-Claspin-C (lanes 10–12), or both (lanes 7–9). Reactions 1, 4, 7, and 10 contain no DNA (−), reactions 2, 5, 8, and 11 contain 3 pm LacO DNA (L), and reactions 3, 6, 9, and 12 contain 3 pm control DNA (C). B, GST-TopBP1 is not synergistic with LacR-Claspin, indicating that the mediators must co-localize to stimulate ATR effectively. ATR kinase reactions contained 6 nm LacR-Claspin-C and either 3 pm LacO DNA (lanes 1–7) or 3 pm control DNA (lanes 8–14), and either no TopBP1 (lanes 1 and 8), 0.0417, 0.125, or 0.375 nm LacR-TopBP1 (lanes 2–4 and 9–11), or 0.417, 1.25, or 3.75 nm GST-TopBP1 (lanes 5–7 and 12–14). C, mutations in the Chk1 binding domain of LacR-Claspin-C (Thr916, Ser945, Ser982 changed to alanine) abolish its ability to synergize with LacR-TopBP1 to activate ATR phosphorylation of Chk1. ATR kinase reactions contained 0.125 nm LacR-TopBP1, 3 pm LacO DNA (lanes 1–7), or 3 pm control DNA (lanes 8–14), and either no LacR-Claspin (lanes 1 and 8), 0.7, 2, or 6 nm LacR-Claspin-C WT (lanes 2–4 and 9–11), or mutant (3A, lanes 5–7 and 12–14).
To measure the specific contribution of tethering Claspin to DNA to activation of ATR, we added LacR-Claspin to reactions containing TopBP1 fused to either GST or LacR (Fig. 2B). We observe 10-fold more Chk1 phosphorylation in reactions containing LacR-Claspin together with LacR-TopBP1 in the presence of LacO-containing DNA than in the presence of control DNA (compare lanes 4 and 11). However, in reactions containing LacR-Claspin together with GST-TopBP1 there is no significant difference in the kinase activity with or without DNA containing the LacO sequence (compare lanes 5–7 and 12–14). Note that a higher concentration of GST-TopBP1 was used in these reactions so that the signal would be above background. These results indicate that GST-TopB1 does not function synergistically with LacR-Claspin and LacO DNA, suggesting that both checkpoint mediators must be co-localized on the DNA to stimulate ATR phosphorylation of Chk1 effectively.
Mutations in the Chk1 binding domain of Claspin abolish the ability of Claspin to mediate ATR phosphorylation of Chk1 in vivo (17) and in vitro (11, 18). We purified mutated LacR-Claspin (3A) (Fig. 1A) to test whether mutations in the Chk1 binding domain of LacR-Claspin (Thr916, Ser945, Ser982 to alanine) would disrupt its function in our defined system. Fig. 2C shows the results from adding increasing amounts of wild-type (WT) or mutant (3A) LacR-Claspin to reactions containing LacR-TopBP1 and either LacO or control DNA. We find that there is up to 10-fold more Chk1 phosphorylation in reactions containing wild-type LacR-Claspin and LacR-TopBP1 with LacO DNA compared with control DNA (lanes 2–4 versus lanes 9–11); however, there is no significant kinase activity in reactions containing mutant LacR-Claspin in either condition (lanes 5–7 and 12–14). These results further illustrate the essential function of Claspin in mediating Chk1 phosphorylation by ATR and the similarity between our in vitro system and the in vivo reaction.
DNA Binding by LacR-TopBP1 and LacR-Claspin Is Required for the Synergistic Stimulation of ATR Phosphorylation of Chk1 in a Defined System
To confirm further that the synergistic stimulation of ATR kinase activity is due to the binding of the LacR fusion proteins to the LacO-containing DNA, we performed the kinase reactions in the presence or absence of IPTG. The binding of IPTG to LacR reduces its affinity for LacO 1000-fold (19). We first measured DNA binding by EMSA of LacR-Claspin and LacR-TopBP1 to DNA oligonucleotides containing the LacO sequence or to control oligonucleotides in the presence or absence of IPTG. The results in Fig. 3A indicate that LacR-Claspin has high affinity for LacO-containing DNA (lane 2) relative to the control DNA (lane 5) and that the DNA binding is lost in the presence of IPTG (lane 3). Similarly, LacR-TopBP1 binds to LacO DNA (lanes 6–8), and the binding is eliminated in the presence of IPTG (lanes 9–11). Therefore, we performed kinase assays in the presence or absence of the same concentration of IPTG and found that the synergistic ATR activation by LacR-TopBP1 and LacR-Claspin in the presence of LacO-containing DNA was eliminated in the presence of IPTG (Fig. 3B, compare lanes 2 and 3 with lanes 9 and 10). We added increasing amounts of DNA to these reactions to assess whether there were nonspecific effects of adding IPTG to the kinase reactions. In fact, we do not observe any nonspecific inhibition of the reactions with the IPTG concentration used in our experiments (compare lanes 4 and 11, and 7 and 14). Together, these results indicate that the tethering of TopBP1 and Claspin to DNA is required for the synergistic stimulation of ATR phosphorylation of Chk1.
Tethering TopBP1 to DNA Induces Chk1 Phosphorylation in Mammalian Cells
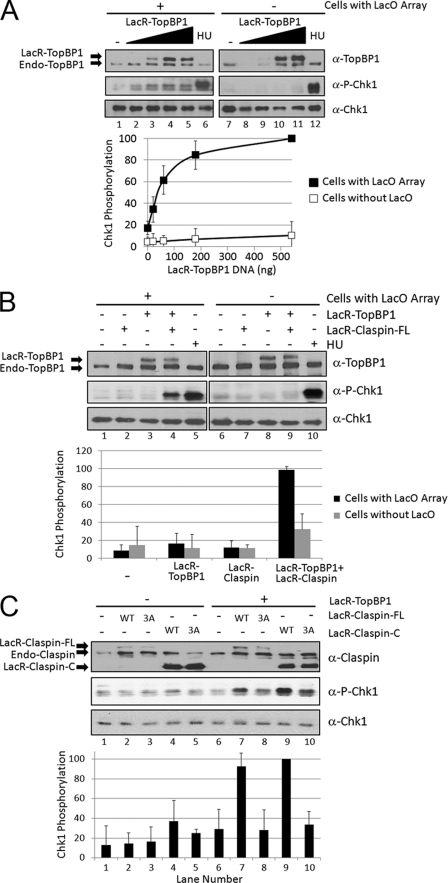
To address the physiological relevance of the ATR activation we observe when TopBP1 is tethered to DNA in our in vitro system, we examined the ability of DNA-tethered TopBP1 to induce Chk1 phosphorylation in mammalian cells. We transiently expressed LacR-TopBP1 in a NIH3T3 cell line that contains the LacO array stably integrated into the genome, named NIH2/4 (13). Fig. 4A shows results from transfecting these cells with increasing amounts of an expression vector encoding LacR-TopBP1. At the highest concentration of transfected LacR-TopBP1 (where the levels of LacR-TopBP1 are similar to endogenous levels of TopBP1), there is 5-fold more Chk1 phosphorylation than the control condition in which cells were transfected with empty vector (compare lanes 5 and 1). Therefore, DNA-tethered TopBP1 activates ATR phosphorylation of Chk1 in vivo, similar to what we have observed in vitro.
FIGURE 4.
Tethering both TopBP1 and Claspin to DNA causes synergistic induction of Chk1 phosphorylation in mammalian cells. A, LacO array-containing NIH2/4 cells (lanes 1–6) or control NIH3T3 cells (lanes 7–12) were transfected with 0, 20, 60, 180, or 540 ng of pcDNA3-LacR-TopBP1 plasmid and empty pcDNA3 plasmid such that all of the transfections had equal amounts of DNA. The cells were lysed 18 h later, and the levels of TopBP1, phospho-Chk1, and Chk1 were analyzed by immunoblotting as indicated. Quantitation of the amount of Chk1 phosphorylation after transfection is graphed below. As a positive control, cells were treated for 15 min with 2 mm hydroxyurea (HU) to induce Chk1 phosphorylation (lanes 6 and 12). B, LacO array-containing NIH2/4 cells (lanes 1–5) or NIH3T3 control cells (lanes 6–10) were transfected with 20 ng of pcDNA3-LacR-TopBP1 (lanes 3, 4, 8, and 9), 50 ng of pcDNA3-LacR-Claspin-FL (lanes 2, 4, 7, and 9), or treated with hydroxyurea (lanes 5 and 10) as in A. C, LacO array-containing NIH2/4 cells were transfected with 20 ng of pcDNA3-LacR-TopBP1 (lanes 6–10), 50 ng of pcDNA3-LacR-Claspin-FL WT (lanes 2 and 7), 50 ng of pcDNA3-LacR-Claspin-FL 3A mutant (lanes 3 and 8), 50 ng pcDNA3-LacR-Claspin-C WT (lanes 4 and 9), 50 ng pcDNA3-LacR-Claspin-C 3A mutant (lanes 5 and 10) and empty pcDNA3 as in A.
Tethering Both TopBP1 and Claspin to DNA Results in Synergistic Induction of Chk1 Phosphorylation in Mammalian Cells
Because we observed synergistic effects of tethering both TopBP1 and Claspin to DNA in our in vitro system, we wanted to determine whether this synergy could be recapitulated in vivo. Fig. 4B shows the results from transfecting LacR-Claspin with or without LacR-TopBP1 in cells containing the LacO array or in cells without the array. Because transfecting LacO array-containing cells with sufficiently high concentrations of LacR-TopBP1-expressing plasmid activates ATR (Fig. 4A), we transfected a limited amount of LacR-TopBP1 so that there was no significant Chk1 phosphorylation in cells expressing LacR-TopBP1 alone (lane 3). However, co-expressing LacR-Claspin together with LacR-TopBP1 resulted in 5-fold more Chk1 phosphorylation than either alone (compare lane 4 with lanes 2 and 3). Importantly, this effect is only observed in cells containing the LacO array (lane 4) because we do not detect significant Chk1 phosphorylation when LacR-TopBP1 and LacR-Claspin are co-expressed in NIH3T3 control cells that do not have the LacO array (lane 9). Therefore, as was observed in vitro, crowding ATR on DNA with two different tethers that are also known to be an activator (TopBP1) and a mediator (Claspin) is sufficient to activate ATR phosphorylation of Chk1 in vivo.
When Tethered Together on DNA with TopBP1, the C Terminus of Claspin Is Sufficient for Synergistic Stimulation of Chk1 Phosphorylation, and Mutations Abrogate the Function
We have observed that a C-terminal fragment of Claspin (amino acids 679–1332) is sufficient for its function in our in vitro system and that mutations of three amino acids in the Chk1 binding domain of this fragment (Thr916, Ser945, and Ser982) abrogate its ability to mediate ATR phosphorylation of Chk1. We wished to test whether this fragment was also sufficient for the synergistic effect we observe in vivo and whether the mutated Claspin was functional. Fig. 4C shows the results from transfecting NIH2/4 LacO array-containing cells with full-length LacR-Claspin (-FL) or with the C-terminal fragment (-C), which is WT or mutated (3A), with or without LacR-TopBP1. We observe similar levels of Chk1 phosphorylation when either -FL or -C LacR-Claspin is co-transfected with LacR-TopBP1 (lanes 7 and 9). Importantly, the Chk1 phosphorylation is 3-fold more than with LacR-TopBP1 alone (lane 6) or without LacR-TopBP1 (lanes 2 and 4). However, the 3A mutation in the LacR-Claspin-FL (lane 8) or LacR-Claspin-C fragment (lane 10) diminishes the ability of Claspin to function synergistically with TopBP1 to induce Chk1 phosphorylation. Therefore, as is the case in vitro, the C terminus of Claspin is sufficient in vivo for synergistic stimulation of Chk1 phosphorylation when tethered to DNA with TopBP1, and mutations in the Chk1 binding domain of Claspin abrogate this function.
DISCUSSION
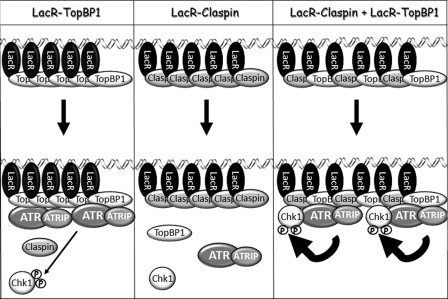
In Fig. 5 we present a summary model for the activation of Chk1 phosphorylation by ATR upon tethering an activator and mediator protein to DNA. Here, we report that tethering the checkpoint activator TopBP1 to DNA is sufficient to activate ATR phosphorylation of Chk1 both in vitro and in vivo. We have shown previously that the higher binding affinity of TopBP1 for damaged DNA results in specific activation of ATR in the presence of damaged DNA (9). In the current report, we have bypassed the requirement for damaged DNA both in vitro and in vivo by fusing TopBP1 to LacR and using DNA containing LacR binding sites. The results from our in vivo studies are somewhat unexpected. It has been reported previously that overexpressing a fragment of TopBP1 containing the ATR-activating domain causes ATR activation in mammalian cells in the absence of DNA damage (6, 12). However, simply tethering the ATR activating domain of TopBP1 to chromatin by fusing it to histone H2B was not sufficient to induce Chk1 phosphorylation in DT40 chicken cells in the absence of DNA damage (20). In our study, LacR-TopBP1 is not overexpressed relative to endogenous TopBP1 (Fig. 4, A and B); however, it is highly concentrated on the chromatin because of the repeated LacO sequence. Thus, there may be a TopBP1 concentration threshold required for ATR activation. In fact, we do observe LacR-TopBP1 concentration-dependent activation of Chk1 both in vitro (Fig. 1B) and in vivo (Fig. 4A).
FIGURE 5.
Model for activation of phosphorylation of Chk1 by ATR upon tethering the TopBP1 activator and Claspin mediator proteins to DNA. Tethering the ATR activator TopBP1 to DNA induces cooperative binding of ATR and is sufficient to activate ATR phosphorylation of Chk1 (left), whereas tethering Claspin alone is not sufficient (middle). Claspin increases the affinity of ATR for Chk1 and thus functions synergistically when tethered to DNA together with TopBP1 (right). Note that this is an artificial mechanism of crowding-induced activation of ATR-ATRIP favoring the collision between TopBP1/ATR-ATRIP/Claspin/Chk1. Physiologically relevant modes of regulating the binding equilibria between ATR-ATRIP and mediator/activator proteins and substrates include direct binding of these proteins to RPA-coated ssDNA, DNA structures or base adducts, repair proteins, and other factors.
We have found that even though tethering the mediator Claspin to DNA is insufficient to activate ATR phosphorylation of Chk1 either in vitro or in vivo, tethering Claspin to the DNA together with TopBP1 functions synergistically to activate ATR phosphorylation of Chk1 both in vitro and in vivo. Claspin increases the affinity of ATR for Chk1 (11). Mutations in the Chk1 binding domain of Claspin abrogate the phosphorylation of Chk1 by ATR (17, 18, 21, 22). Here, we report that the same mutations also abrogate the ability of Claspin to function when tethered to DNA both in vitro and in vivo. Interestingly, we find that the C-terminal half of Claspin is sufficient for the synergistic activation of ATR phosphorylation of Chk1 both in vitro and in vivo. It is known that the N-terminal half of Claspin has DNA binding activity (23) and is involved in protein-protein interactions required for chromatin association (24); yet, these requirements are bypassed when Claspin is tethered to the DNA.
The lac repressor/operator system has previously been used to study ATM activation in mammalian cells (25) and activation of the ATR homolog, Mec1, in yeast cells (26). Tethering individual mediators in the ATM pathway such as members of the MRN complex (Mre11-Rad50-Nbs1) or Mdc1 was sufficient to activate ATM in the absence of DNA damage (25). In yeast, however, checkpoint activation was observed when a member of the 9-1-1 complex was tethered together with the Mec1-binding partner Ddc2, but not when either was tethered alone (26). Here, we show that in mammalian cells, simply tethering the activator of ATR is sufficient to induce the checkpoint response and that the effect is amplified when the mediator Claspin is also tethered. Taken together, these studies illustrate the generality that activation of the cellular DNA damage response pathways does not require DNA damage but can be triggered by stable association of checkpoint proteins with chromatin. For the ATR pathway, there are many potential mechanisms for achieving stable chromatin association of the checkpoint proteins, such as by RPA-coated ssDNA, which, through interactions with ATRIP (27) and the checkpoint mediator protein Tipin (28), bring ATR and Claspin into close proximity to one another to facilitate Chk1 phosphorylation. Similarly, interactions between ATR and DNA repair factors (29–32) as well as direct binding of checkpoint proteins to DNA structures such as branched DNA (23) or DNA-containing bulky adducts (9, 33), may also facilitate activation of the ATR pathway.
In conclusion, we have found that tethering TopBP1 to DNA is sufficient to activate ATR phosphorylation of Chk1 both in vitro and in vivo and that when Claspin is also tethered to the DNA there is a synergistic effect on ATR activation. Our findings taken together with the in vivo studies with recruiting ATM by tethering in human cells and recruiting Mec1 by tethering in Saccharomyces cerevisiae, bring into focus that recruitment of phosphoinositide 3-kinase-related serine-threonine kinases members to chromatin by whatever means is sufficient to institute checkpoint signaling. We believe this realization will help define more generally the mechanism by which checkpoint signaling is activated.
Acknowledgments
We thank M. Kemp and C. Capp for critical reading and useful comments, J.-H. Choi for help with purifying native ATR-ATRIP and recombinant GST-TopBP1, and T. Misteli for the NIH2/4 cell line.
This work was supported, in whole or in part, by National Institutes of Health Grant GM32833 (to A. S.).
- ATM
- ataxia-telangiectasia mutated
- ATR
- ATM and RAD3-related
- ATRIP
- ATR-interacting protein
- -C
- C-terminal
- Chk1
- checkpoint kinase 1
- -FL
- full-length
- IPTG
- isopropyl β-d-thiogalactopyranoside
- kd
- kinase-dead
- LacO
- lac operator
- LacR
- lac repressor
- RPA
- replication protein A
- ssDNA
- single-stranded DNA
- TopBP1
- topoisomerase IIβ-binding protein 1.
REFERENCES
- 1. Liu Q., Guntuku S., Cui X. S., Matsuoka S., Cortez D., Tamai K., Luo G., Carattini-Rivera S., DeMayo F., Bradley A., Donehower L. A., Elledge S. J. (2000) Genes Dev. 14, 1448–1459 [PMC free article] [PubMed] [Google Scholar]
- 2. Zhao H., Piwnica-Worms H. (2001) Mol. Cell. Biol. 21, 4129–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chini C. C., Chen J. (2003) J. Biol. Chem. 278, 30057–30062 [DOI] [PubMed] [Google Scholar]
- 4. Lin S. Y., Li K., Stewart G. S., Elledge S. J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 6484–6489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu S., Bekker-Jensen S., Mailand N., Lukas C., Bartek J., Lukas J. (2006) Mol. Cell. Biol. 26, 6056–6064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kumagai A., Lee J., Yoo H. Y., Dunphy W. G. (2006) Cell 124, 943–955 [DOI] [PubMed] [Google Scholar]
- 7. Ball H. L., Ehrhardt M. R., Mordes D. A., Glick G. G., Chazin W. J., Cortez D. (2007) Mol. Cell. Biol. 27, 3367–3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Choi J. H., Lindsey-Boltz L. A., Kemp M., Mason A. C., Wold M. S., Sancar A. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 13660–13665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Choi J. H., Lindsey-Boltz L. A., Sancar A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 13301–13306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Choi J. H., Lindsey-Boltz L. A., Sancar A. (2009) Nucleic Acids Res. 37, 1501–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lindsey-Boltz L. A., Serçin O., Choi J. H., Sancar A. (2009) J. Biol. Chem. 284, 33107–33114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Toledo L. I., Murga M., Gutierrez-Martinez P., Soria R., Fernandez-Capetillo O. (2008) Genes Dev. 22, 297–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Soutoglou E., Dorn J. F., Sengupta K., Jasin M., Nussenzweig A., Ried T., Danuser G., Misteli T. (2007) Nat. Cell Biol. 9, 675–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Murphy K. C., Campellone K. G. (2003) BMC Mol. Biol. 4, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Choi J. H., Sancar A., Lindsey-Boltz L. A. (2009) Methods 48, 3–7 [DOI] [PubMed] [Google Scholar]
- 16. Kang T. H., Lindsey-Boltz L. A., Reardon J. T., Sancar A. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 4890–4895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chini C. C., Chen J. (2006) J. Biol. Chem. 281, 33276–33282 [DOI] [PubMed] [Google Scholar]
- 18. Kumagai A., Dunphy W. G. (2003) Nat. Cell Biol. 5, 161–165 [DOI] [PubMed] [Google Scholar]
- 19. Riggs A. D., Newby R. F., Bourgeois S. (1970) J. Mol. Biol. 51, 303–314 [DOI] [PubMed] [Google Scholar]
- 20. Delacroix S., Wagner J. M., Kobayashi M., Yamamoto K., Karnitz L. M. (2007) Genes Dev. 21, 1472–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kumagai A., Dunphy W. G. (2000) Mol. Cell 6, 839–849 [DOI] [PubMed] [Google Scholar]
- 22. Clarke C. A., Clarke P. R. (2005) Biochem. J. 388, 705–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sar F., Lindsey-Boltz L. A., Subramanian D., Croteau D. L., Hutsell S. Q., Griffith J. D., Sancar A. (2004) J. Biol. Chem. 279, 39289–39295 [DOI] [PubMed] [Google Scholar]
- 24. Lee J., Gold D. A., Shevchenko A., Schevchenko A., Dunphy W. G. (2005) Mol. Biol. Cell 16, 5269–5282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Soutoglou E., Misteli T. (2008) Science 320, 1507–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bonilla C. Y., Melo J. A., Toczyski D. P. (2008) Mol. Cell 30, 267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zou L., Elledge S. J. (2003) Science 300, 1542–1548 [DOI] [PubMed] [Google Scholar]
- 28. Kemp M. G., Akan Z., Yilmaz S., Grillo M., Smith-Roe S. L., Kang T. H., Cordeiro-Stone M., Kaufmann W. K., Abraham R. T., Sancar A., Unsal-Kaçmaz K. (2010) J. Biol. Chem. 285, 16562–16571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shell S. M., Li Z., Shkriabai N., Kvaratskhelia M., Brosey C., Serrano M. A., Chazin W. J., Musich P. R., Zou Y. (2009) J. Biol. Chem. 284, 24213–24222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang Y., Qin J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 15387–15392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yoshioka K., Yoshioka Y., Hsieh P. (2006) Mol. Cell 22, 501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu Y., Fang Y., Shao H., Lindsey-Boltz L., Sancar A., Modrich P. (2010) J. Biol. Chem. 285, 5974–5982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Unsal-Kaçmaz K., Makhov A. M., Griffith J. D., Sancar A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 6673–6678 [DOI] [PMC free article] [PubMed] [Google Scholar]