Abstract
We have previously shown that the cell death-promoting protein Bcl-2-interacting mediator of cell death (Bim) is ubiquitinated and degraded following a neuroprotection-conferring episode of brief ischemia (preconditioning). Here, we identify the E3 ligase that ubiquitinates Bim in this model, using a proteomics approach. Using phosphorylated GST-Bim as bait, we precipitated and identified by mass spectrometry tripartite motif protein 2 (TRIM2), a RING (really interesting new gene) domain-containing protein. The reaction between TRIM2 and Bim was confirmed using co-immunoprecipitation followed by immunoblotting. We show that TRIM2 binds to Bim when it is phosphorylated by p42/p44 MAPK but does not interact with a nonphosphorylatable Bim mutant (3ABim). 12-O-tetradecanoylphorbol-13-acetate activation of p42/p44 MAPK drives Bim ubiquitination in mouse embryonic fibroblast cells and is associated with an increased interaction between TRIM2 and Bim. One hour following preconditioning ischemia, the binding of Bim to TRIM2 increased, consistent with the time window of enhanced Bim degradation. Blocking p42/p44 MAPK activation following preconditioning ischemia with U0126 or using the nonphosphorylatable 3ABim reduced the binding between Bim and TRIM2. Immunodepletion of TRIM2 from cell lysates prepared from preconditioned cells reduced Bim ubiquitination. Finally, suppression of TRIM2 expression, using lentivirus transduction of shRNAmir, stabilized Bim protein levels and blocked neuroprotection observed in rapid ischemic tolerance. Taken together, these data support a role for TRIM2 in mediating the p42/p44 MAPK-dependent ubiquitination of Bim in rapid ischemic tolerance.
Keywords: Cell Death, E3 Ubiquitin Ligase, ERK, Ischemia, MAP kinases (MAPKs), Neurobiology, Neuron, Ubiquitin
Introduction
Cerebral ischemia, the deprivation of oxygen and glucose to the brain, can result in neuronal death. However, prior exposure to a brief nonharmful dose of ischemia (preconditioning) activates an endogenous neuroprotective program, rendering the brain protected against subsequent ischemic injury (ischemic tolerance). Rapid (short term) ischemic tolerance in brain and cultured neurons occurs 1 h following a preconditioning stimulus, resulting in profound neuroprotection (1, 2). Although the exact molecular mechanisms of rapid tolerance are not fully resolved, they appear to be mediated by rapid biochemical events, including activation of adenosine receptors, ATP-activated potassium channels, multiple protein kinases, and the ubiquitin-proteasome system (1–7).
The ubiquitin-proteasome system rapidly degrades short lived proteins in the cell. Ubiquitin is added to a target protein following a sequential series of reactions, whereby ubiquitin is bound first to an E1 ligase in a reaction requiring ATP. Ubiquitin is transferred to an E2 protein and then transferred via the E3 ligase to the target protein lysine residue (8). The HECT3 (homologous to the E6-AP carboxyl terminus) class of E3 ligase binds to ubiquitin, prior to conjugating the ubiquitin to the target, whereas the RING (really interesting new gene)-containing E3 ligases appear to mediate the transfer of ubiquitin from the E2 protein directly to the target (9). As such, the specificity of the reaction is determined by the target protein interaction with its E3 ligase.
In our studies of rapid ischemic tolerance, we described two mechanisms whereby the ubiquitination and subsequent degradation of proteins contribute to rapid ischemic tolerance neuroprotection. The degradation of actin-binding structural proteins results in uncoupling of NMDA receptors following brief ischemia, thereby blocking excitotoxicity (6). In addition, we reported the apoptosis-inducing protein Bim (Bcl-2-interacting mediator of cell death) is degraded rapidly following both brief ischemia and adenosine receptor activation in a p42/p44-MAPK dependent manner (1, 7). Following preconditioning ischemia we observed an increase in Bim ubiquitination and a decrease in Bim protein levels, which was blocked by the proteasome inhibitor MG132 (1). Knockdown of Bim in neurons confers protection against ischemia-induced cell injury and other excitotoxic insults (1, 10). The phosphorylation of the Ser65 (Ser69 in rat) residue by p42/p44 MAPK (also known as ERK1/2) is a key step in the degradation of Bim (11). Bim contains a p42/p44 MAPK binding site on its carboxyl terminus, which facilitates its phosphorylation (11). The ubiquitination and degradation of Bim following preconditioning are blocked by inhibiting p42/p44 MAPK activation with U0126, an inhibitor of the upstream protein kinase MEK (1, 11). Blocking p42/p44 MAPK activation with the MEK inhibitors U0126 and PD98059 blocks rapid ischemic tolerance (1). Nevertheless, the E3 ligase mediating the degradation of Bim in neurons following brief ischemia is unknown.
The identification of the enzyme that ubiquitinates Bim, thereby promoting its degradation (the Bim E3 ligase), would make an attractive target for antistroke therapy; however, the identification of the E3 ligase for Bim has proved to be a controversial matter. To date the tyrosine kinase c-Cbl (12) and CIS, a member of the SKIP-Cullen E3 ligase family (13), have been suggested to function as a Bim E3 ligase; however, other studies suggest that c-Cbl may not mediate p42/p44 MAPK-dependent Bim ubiquitination (14). Taken together, this would suggest that, at least, an additional protein may function as a Bim E3 ligase regulating p42/p44 MAPK-dependent Bim degradation in neurons following brief ischemia.
Here, we provide evidence that TRIM2 may function as the Bim E3 ligase in rapid ischemic tolerance. TRIM2 was identified as a Bim-interacting protein in a proteomic study. We show in multiple systems how TRIM2 interacts with Bim in a p42/p44 MAPK-dependent manner, under conditions of enhanced Bim ubiquitination. Further studies confirm TRIM2-mediated Bim degradation as the mechanism of rapid ischemic tolerance-induced neuroprotection.
EXPERIMENTAL PROCEDURES
Drugs and Materials
Carbobenzoxy-l-leucyl-l-leucyl-l-leucinal (MG132) was from Calbiochem. 12-O-Tetradecanoylphorbol-13-acetate (TPA) and Emetine dihydrochloride hydrate waere from Sigma. Anti-Bim (rabbit monoclonal and polyclonal), phospho-Bim (Ser69), total p42/p44 MAPK, and phospho-p42/p44 MAPK (Thr202/Tyr204) were from Cell Signaling Technologies. Goat anti-TRIM2 was from Novus Biological. Anti-ubiquitin, GST, α-tubulin, protein A-agarose, and protein G-agarose beads were from Santa Cruz Biochemicals.
Primary Neuronal Cell Culture and Oxygen Glucose Deprivation-modeled Ischemia
Primary neuronal cultures were prepared from postnatal day 1 rat pups of mixed sex, using a modification of the method described previously (27). All experiments were performed in accordance with the American animal protection legislation and approved by the Legacy Health System or Morehouse School of Medicine Institutional Animal Care and Use Committee. Cortices were dissected from the pups and chopped and incubated for 10 min in a papain solution (40 units/ml). Cells were dissociated in Dulbecco's modified Eagle's medium containing 50 μm d-2-amino-5-phosphonovalerate and 10% FBS. Cells were grown in neurobasal A medium containing 2% B27 supplement and 1% GlutaMAX (Invitrogen). Medium was replaced every 2–3 days. Cells were maintained in a humidified incubator (Forma Scientific) gassed with 5% CO2/95% O2 for 10–14 days in vitro. Cells were typically 80–90% neuronal (NeuN-positive), and the morphology was assessed on a batch of sister cultures using DiO staining as described in Ref. 6. Cultures showing signs of excitotoxicity were discarded (excessive membrane blebbing/varicosities).
Ischemia was modeled by oxygen and glucose deprivation (OGD) as previously described (6). Cells were removed from a normoxic incubator and washed twice with PBS (1.37 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.7 mm KH2PO4, pH 7.4) containing 1.0 mm MgCl2 and 0.5 mm CaCl2. Cells were placed into an anoxic chamber (Forma Scientific/ShelLabs Bactron) and maintained at 35 °C for the duration of the ischemic insult. Following ischemia, cells were replenished with normal neurobasal A medium and returned to the normoxic incubator.
Proteomic Identification of TRIM2
GST-Bim (Millipore) was phosphorylated by incubating with active p42/p44 MAPK (ERK1/2) (Millipore) for 30 min at 37 °C. Active MAPK was then removed by incubating with an excess of immobilized phospho-p42/p44 MAPK antibody (Cell Signaling). No active p42/p44 MAPK was detected following immunoprecipitation (data not shown). Phosphorylated GST-Bim was then bound to glutathione-Sepharose beads (Amersham Biosciences) for 30 min at room temperature and then washed twice with PBS/0.1% Nonidet P-40. Cells lysates were prepared from neuronal cells (1.0 mg), cleared, and then incubated for 2 h at 4 °C with the phosphorylated GST-Bim/Sepharose beads. MG132 was present during the incubation to prevent degradation of ubiquitinated Bim. The beads were separated by centrifugation and washed three times with PBS/0.1% Nonidet P-40. The beads were boiled in loading buffer, and released proteins were subjected to PAGE. Gel sections (approximate 10–30-kDa gel fragments) were analyzed at the proteomics facility at the Fred Hutchinson Cancer Research Center (Seattle, WA). Unstained gel slices were washed twice with water, and “in-gel” protein digestions were carried out as described (28) but omitting the reduction and alkylation steps. Samples were desalted using micro-C18 ZipTips (Millipore) and dried and resuspended in 5 μl of 0.1% trifluoroacetic acid and analyzed by liquid chromatography electrospray ionization tandem mass spectrometry with a LCQ “classic” mass spectrometer (ThermoElectron) using a previously described instrument configuration (29). Data were collected in a data-dependent mode in which a MS scan was followed by tandem MS scans of the three most abundant ions from the preceding MS scan. Mass spectrometry data were searched against rat or human IPI protein databases using the protein database search algorithm X!Tandem. False discovery rates were assigned using Peptide Prophet, and results were filtered and sorted using the Computational Proteomic Analysis System. A cutoff criterion of Peptide Prophet minimum probability scores of >0.9 (false discovery rate of <5%) was used to filter the peptide identification results. The IPI database of identified peptides was searched to identify proteins with either RING or HECT domain motifs. Only proteins containing a RING or HECT domain were considered for further analysis.
Mouse Embryonic Fibroblast Cell Culture and Stimulation
Mouse embryonic fibroblast cultures were grown and maintained in Advanced DMEM with 10% FBS and 1% GlutaMAX. The cultures were split twice weekly at a 1:4 ratio, and 6-cm dishes were plated for experimentation. To split cultures, 0.25% trypsin/EDTA was added to the flasks for 3–6 min or until cells visibly dislodged from the bottom of the flask. The cells were then transferred to another tube with Advanced DMEM containing FBS (10%) and GlutaMAX. Then the tube containing the cells and medium was spun at 2000 rpm for 5 min. Once the spin was complete the supernatant was removed, and the cell pellet was then resuspended in the feeding medium and plated back into flasks for cultivation or dishes for experimentation.
To stimulate mouse embryonic fibroblast (MEF) cells two protocols were used. The first was similar to that of Ley et al. (11) whereby cells were starved of serum for 24 h and pretreated with 10 μm emetine for 10 min prior to incubating with serum (10% FCS) for 60 min. In the second protocol, cells were starved of serum for 24 h. Then the cultures were stimulated using 10 nm TPA (Sigma) for 1 h in the presence of 10 μm emetine (pretreated for 10 min).
Immunoprecipitation
Cells were lysed in a buffer containing 150 mm NaCl, 50 mm Tris-HCl, 1.0% Nonidet P-40, 1 mm EDTA containing protease inhibitors (1.0 μg/ml aprotinin, 0.5 μg/ml leupeptin, 1.0 μg/ml pepstatin, 10 μg/ml PMSF (Sigma)), and 10 μl/ml phosphatase inhibitor mixture (Sigma). Cells were cleared at 6000 rpm for 5 min at 4 °C and protein levels determined using Bradford reagent. Protein levels were normalized to ∼500 μg/sample. For exogenous Bim immunoprecipitations, samples were incubated with either GST-Bim or GST-Bim 3A (Millipore) for 20 min at 30 °C. Samples were then incubated with protein A-agarose beads preconjugated with anti-TRIM2 antibody (Novus). For endogenous Bim immunoprecipitations, samples were incubated with rabbit monoclonal anti-Bim antibody (Cell Signaling) preconjugated to protein A beads (Santa Cruz Biotechnology) with a bridging antibody (Active Motif, 10 μl/reaction). Samples were incubated with immunoprecipitation antibodies for 1 h at room temperature. Proteins were precipitated by the addition of 50 μl of protein-A agarose beads (Santa Cruz Biotechnology) followed by centrifugation at 1000 rpm for 1 min at 4 °C. Proteins were washed with PBS/0.1% Nonidet P-40 before being boiled and subjected to SDS-PAGE. Immunoprecipitation experiments were performed at least twice.
Immunoblotting
Samples for immunoblotting were lysed in a buffer (0.1 m NaCl, 0.02 m Tris-HCl, pH 7.6, 1 mm EDTA, pH 8.0, 1.0% Nonidet P-40) containing protease inhibitors (1.0 μg/ml aprotinin, 0.5 μg/ml leupeptin, 1.0 μg/ml pepstatin, 10 μg/ml PMSF, and a phosphatase inhibitor mixture (Sigma). Protein levels were determined using Bradford reagent and 50-μg samples of lysate were prepared. Samples were boiled in a sample loading buffer (0.14 m Tris-HCl, pH 6.8, 20% glycerol, 2.2% SDS, and 200 mm DTT), quenched on ice for 1 min, and then loaded onto SDS-polyacrylamide gels (10–15%, depending on the experiment). Proteins were transferred onto PMSF membrane, blocked with Tris-buffered saline containing 5% nonfat milk or 0.1% BSA for phopho-specific antibodies. Primary antibodies were incubated for 2 h at room temperature followed by washing in Tris-buffered saline and 0.1% Tween 20. Secondary antibodies (Cell Signaling) were incubated at a 1:2000 dilution for 1 h at room temperature, followed by further washes. Membranes were incubated with chemiluminescence reagent (Visualizer; Upstate) for 2 min prior to digital determination of luminescence (Kodak 1D image station). Chemiluminescence was quantified using the Kodak 1D software.
Ubiquitination Assay
Ubiquitination assays were performed according to the manufacturer's instructions (BioMol). Briefly, normalized cell lysates were incubated with GST-Bim, E1 ligase, 11 E2 ligases, ATP, DTT, MG132, and ubiquitin for 20 min at 37 °C. Bim was immunoprecipitated with preconjugated anti-GST antibody (Santa Cruz Biotechnology), and proteins were immunoblotted and probed for ubiquitin.
c-myc-TRIM2 Synthesis
The construct encoding the human cDNA TRIM2 (pGBK-T7-c-myc-TRIM2) was synthesized by amplification of the human cDNA region encoding TRIM2 from pCMV-Sport6-TRIM2 (MHS1010-7429489; Clone ID 4156234; Open Biosystems, Huntsville, AL) using the inverse PCR method (30). The forward primer (gcc AAc ATA Tgg ccA gTg Aag gcA ccA Ac) introduced an NdeI restriction site at methionine 1. The reverse primer (gTc gAc cTg TAA gTA Tcg ATA gAc TTT gAA A) introduced a SalI restriction site at the stop codon (relative position amino acid 745). The amplified region was cloned 5′ NdeI-3′ SalI under control of the T7 promoter of pGBK-T7 (Clontech) and completely sequenced prior to use as a template. c-myc-TRIM2 was synthesized by in vitro transcription of the cDNA clone (pGBKT7-c-myc-TRIM2) using T7 RNA polymerase (Invitrogen). This was followed by in vitro translation in a nuclease-treated rabbit reticulocyte lysate system (Promega, Madison, WI) of the capped transcripts following previously described methods (31).
TRIM2 shRNA Synthesis
We expressed four TRIM2 (NM_015271) specific Expression ArrestTM microRNA-adapted shRNA (shRNAmir) vectors (V2LHS_80589, V2LHS_254587, V2LHS_80545, and V2LHS_80547; Open Biosystems) and one nonsilencing GIPZ lentiviral shRNAmir control (RHS4346) using the commercially available Trans-LentiviralTM shRNA Packaging System (TLP4614; Open Biosystems). Manufacturers' protocols were followed throughout, but briefly the day prior to transfection, HEK293T cells were plated at a density of 5.5 × 106. The following day a 1:5 DNA:Arrest-in transfection reagent ratio was used with serum-free DMEM (Invitrogen) for transfection. After a 6-h incubation in a CO2 incubator at 37 °C, 1 ml of standard medium was added to the cultures. Following a further 48-h incubation, the medium was withdrawn and stored at 4 °C, and fresh medium was applied to the cells for 24 h (72 h after transfection). The growth medium (from both 48- and 72-h time points) was then centrifuged at 3000 rpm for 20 min (4 °C). The supernatant was then ultracentrifuged at 73,800 × g for 90 min, and the pellet was resuspended in 250 μl of serum-free DMEM. This was centrifuged at 13,000 rpm for 5 min, prior to aliquoting. The prepared viral particles were then titered following the manufacturers' protocols throughout, prior to use at a multiplicity of infection of 10.
RESULTS
Proteomic Identification of a Phospho-Bim-interacting Protein with the Molecular Signature of an E3 Ligase
A nonbiased proteomic approach was used to identify Bim-binding proteins with the molecular signature of an E3 ligase (i.e. containing a RING or HECT domain). Because Bim ubiquitination is dependent on its phosphorylation by p42/p44 MAPK (11), we first phosphorylated GST-Bim using exogenous activated p42/p44 MAPK. Phosphorylation of the Bim was confirmed by immunoblotting with a phospho-Bim-specific antibody (anti-phospho-Bim Ser55; Upstate; data not shown). Lysates prepared from rat cortical neurons were incubated with phospho-Bim-GST bound to glutathione-conjugated agarose beads. Precipitated proteins were resolved by 10% SDS-PAGE and gel sections subjected to proteomic analysis. Mass spectrometry data were analyzed using Peptide Prophet, and a criterion of a p value > 0.9 was selected.
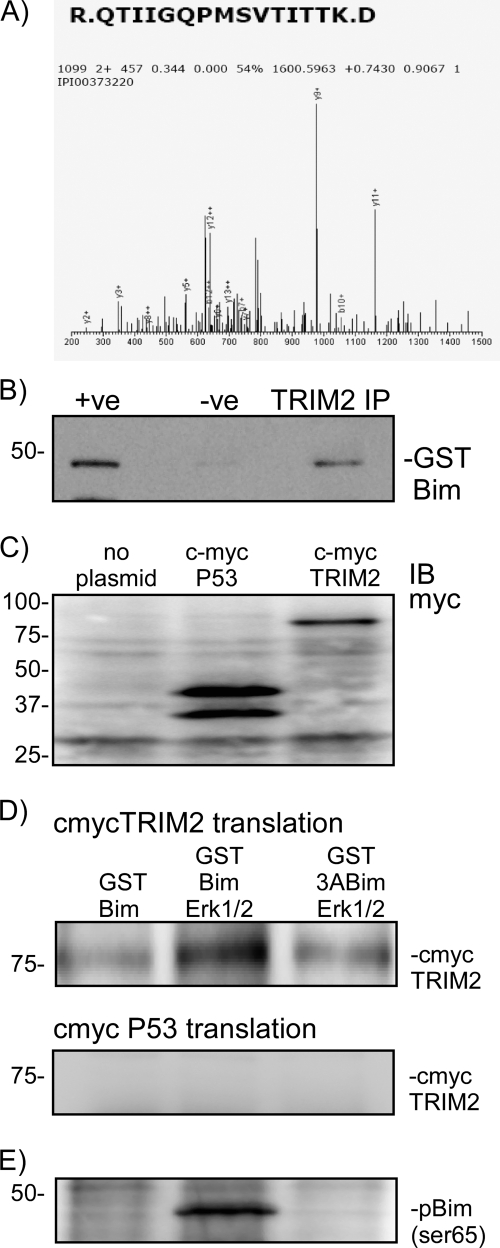
Our mass spectrometry data identified two previously identified Bim-binding proteins, dynein (15) and a 14-3-3 isoform (14-3-3 ζ) (16) (data not shown). Interestingly, only one peptide was identified which matched a protein in the IPI database containing either a RING or HECT domain (Fig. 1A). (Note that the IPI database number for this peptide shown in Fig. 1 is no longer active.) Blast searching the peptide sequence in Fig. 1 revealed a 100% match to the rat isoform of TRIM2. TRIM2 is a member of the TRIM family of RING domain containing E3 ligases (17).
FIGURE 1.
Proteomic identification of TRIM2 as a Bim-binding protein and confirmation in a cell-free transcription/translation assay. A, phospho-Bim-binding proteins were precipitated from rat cortical neurons and subjected to one-dimensional PAGE followed by mass spectroscopy. The peptide R.QTIIGQPMSVTITK.D was identified by mass spectrometry as a phospho-Bim-binding protein. This peptide corresponds to the rat homologue of TRIM2. Because this was the only E3 ligase protein identified, it was considered further. B, binding of TRIM2 and Bim was confirmed by immunoprecipitation. Rat brain was incubated with GST-Bim and then subjected to TRIM2 immunoprecipitation. GST-Bim was identified by immunoblotting precipitated (IP) proteins with a GST-specific antibody. As a control the TRIM2 antibody was omitted from the reaction (−ve). C, using a cell-free expression system (Promega), TRIM2 and a control protein (p53 binding domain) DNA clones were synthesized into proteins. Expression was confirmed by immunoblotting (IB) lysate samples with a c-myc-specific antibody. D, GST-Bim was incubated in the absence or presence of MAPK with either myc-TRIM2 or myc-P53. As a control, a nonphosphorylatable mutant Bim (3ABim) was incubated with myc-TRIM2. Specific binding was determined by performing a GST pulldown and immunoblotting precipitated proteins for myc. Binding of myc-tagged TRIM2 increases when activated MAPK is added to the reaction. Incubation with a nonphosphorylatable BIM (3ABim) reduces this interaction. E, phosphorylation of Bim by MAPK was confirmed by blotting lysates with a Ser(P)69-specific antibody.
To confirm our observation, we performed an immunoprecipitation experiment in reverse of our proteomic experiment. We prepared a protein A-agarose-immobilized TRIM2 antibody and incubated it with 500 μg of rat brain lysate and GST-Bim. The blot was then probed with an anti-GST antibody (Santa Cruz Biotechnology). As a negative control, cells were incubated with protein A-agarose beads. GST-Bim was immunoprecipitated from rat brain lysates using a TRIM2 antibody, hence confirming our observation that TRIM2 is a Bim-interacting protein (Fig. 1B).
Bim-TRIM2 Interactions in a Cell-free Expression System Require p42/p44 MAPK
We used a cell-free transcription/translation assay to confirm the binding of TRIM2 to Bim. c-myc-tagged TRIM2 was synthesized by in vitro transcription/translation of the cDNA clone (pGBKT7-TRIM2) (Fig. 1C). As a positive control we also synthesized a c-myc-tagged p53 binding domain (35-kDa translation product). Under basal conditions, c-myc TRIM2 weakly immunoprecipitated with GST-Bim (Fig. 1D); however, the binding was stronger when active p42/p44 MAPK was added to the reaction (Fig. 1D). Bim phosphorylation by p42/p44 MAPK was confirmed by immunoblotting with antibodies raised against Ser(P)65 Bim (Fig. 1E). To show that the binding of TRIM2 with Bim was specific for phosphorylated Bim, we also incubated TRIM2 with the nonphosphorylatable mutant 3ABim and p42/p44 MAPK. The 3ABim mutant contains point mutations S55A, S65A, and S73A. Binding of myc-TRIM2 with 3ABim in the presence of active MAPK was weak (Fig. 1D). As a negative control, we performed an immunoprecipitation using myc-p53. As can be seen in Fig. 1D, no binding of c-myc p53 was observed (Fig. 1D). These data show that the binding of Bim to TRIM2 requires phosphorylation of Bim by p42/p44 MAPK on Ser55, Ser65, or Ser73.
TRIM2 Interacts with Bim following p42/p44 MAPK Activation in the MEF Cell Line
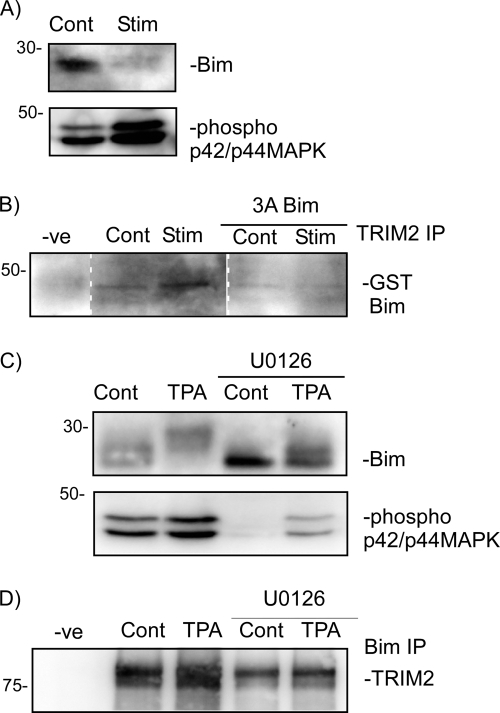
We used a MEF cell line to investigate further whether TRIM2 may function as a Bim E3 ligase. MEF cells have previously been used to study Bim ubiquitination (11, 18). MEF cells express both Bim and our candidate Bim E3 ligase TRIM2. Under basal conditions, MAPK activity in MEF cells is low, as determined by phospho-p42/p44 MAPK levels (Fig. 2A). Serum starvation of MEFs followed by serum stimulation has been shown to activate p42/p44 MAPK kinase and promote Bim ubiquitination (11, 14). Phospho-p42/p44 MAPK levels were increased following serum stimulation of cells, and Bim protein levels were decreased accordingly (Fig. 2A).
FIGURE 2.
Bim degradation in MEF cells is associated with an increase interaction between TRIM2 and Bim. MEF cells were serum-starved for 24 h and then incubated for 1 h with serum and emetine. A, lysates were immunoblotted for phospho-p42/p44 MAPK and Bim protein levels. B, immunoprecipitation of TRIM2 with either GST-Bim or GST3ABim following serum stimulation (Stim) of MEF cells. Pulldown is determined by immunoblotting precipitated proteins with a GST-specific antibody. Representative blots of two independent observations are shown. C, MEF cells were incubated with TPA for 1 h, and cell lysates were prepared. Some cells were co-incubated with 10 μm U0126. Bim protein levels and phospho-MAPK levels were determined by immunoblotting. D, lysates from TPA-treated MEF cells were subjected to endogenous Bim immunoprecipitation. Precipitated proteins were immunoblotted for TRIM2 with a TRIM2-specific antibody (Novus). TRIM2 pulldown with Bim increased following incubation of MEF cells with TPA and was reduced by U0126.
We next investigated whether TRIM2 interacts with Bim under conditions where Bim ubiquitination occurs. GST-Bim binding to TRIM2 increased in serum-stimulated cells compared with nonstimulated cells (Fig. 2B). When the nonphosphorylatable Bim mutant 3ABim was used in the immunoprecipitation reactions, we did not observe pulldown of 3ABim with TRIM2 in either control or stimulated cells (Fig. 2B). This suggests that Bim degradation in MEF cells requires a p42/p44 MAPK-dependent interaction between Bim and TRIM2.
We further investigated the role of p42/p44 MAPK in regulating the interaction of Bim and TRIM2 using an additional stimulation of p42/p44 MAPK, TPA. Incubation of cells with TPA (10 nm, 1 h) resulted in an increase in MAPK phosphorylation and reduced Bim protein levels (Fig. 2C). Immunoprecipitation of endogenous Bim with endogenous TRIM2 increased following TPA treatment of cells. The enhanced interaction between TRIM2 and Bim, as well as MAPK phosphorylation, was reduced in cells pretreated with U0126 (10 μm) prior to TPA stimulation (Fig. 2D). Bim protein levels were increased in U0126-treated cells.
Immunoprecipitation of Bim with TRIM2 following Preconditioning Ischemia
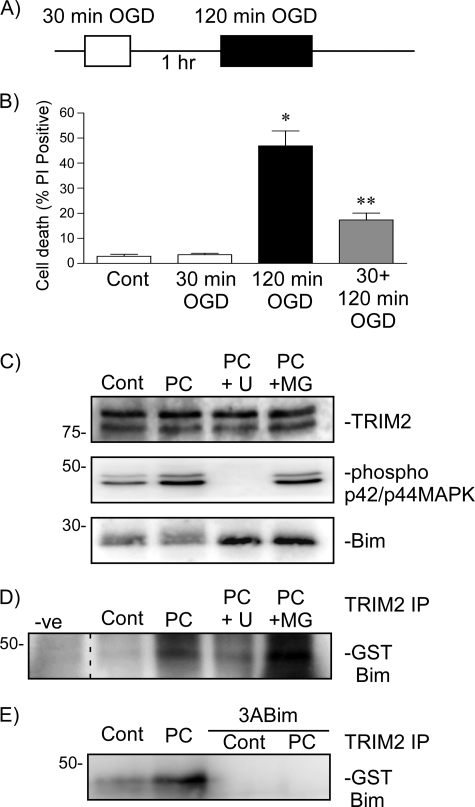
Rapid ischemic tolerance was modeled in vitro, using 14-day rat cortical neurons subjected to OGD ischemia, as described previously (1) (Fig. 3A). Exposure to 30-min ischemia was not harmful (Fig. 3B), whereas 120-min OGD induces ∼50% cell death (p < 0.001, 120 min OGD versus control). Preconditioning cells with 30-min OGD, 1 h prior to harmful ischemia, reduced cell death by 60% compared with cells subject to 120-min OGD alone (p < 0.001 120 min OGD versus 30 + 120 min OGD) (Fig. 3B). We previously have shown that three proteasome inhibitors MG132, carbobenzoxy-l-leucyl-l-leucyl-l-norvalinal (MG115), and clasto-lactacystin-lactone all block rapid ischemic tolerance (6). One hour following preconditioning ischemia Bim protein levels were decreased compared with control values (75.2 ± 6% of control values (p < 0.02, n = 8, paired Student's t test of logn-transformed data) (Fig. 3C) (1).
FIGURE 3.
TRIM2 binding to Bim following preconditioning ischemia is dependent on Bim phosphorylation by p42/p44 MAPK. A, schematic shows the rapid ischemic tolerance paradigm. Neuronal cultures were subjected to preconditioning ischemia (30-min OGD PC) 1 h prior to harmful ischemia (120 min OGD). B, viability was assessed in cultures subjected to 30-min, 120-min, or 30-min OGD followed by 1-h recovery and a subsequent 120-min OGD. Cell death was assessed by propidium iodide assay 24 h following the last ischemic event. Propidium iodide-positive cells are expressed as the percent of total (DAPI-stained) cells in the culture. Data shown are mean ± S.E. (error bars), n = 12. *, p < 0.001 versus control; **, p < 0.001 versus 120-min OGD (one-way ANOVA with the Bonferroni post hoc test). C, cells were subjected to 30-min OGD preconditioning ischemia and then recovered in the presence of 10 μm MG132 (MG) or 10 μm U0126 (U) for 1 h. TRIM2, Bim, and phospho-MAPK levels were assessed using immunoblotting. D, cells were subjected to 30-min OGD preconditioning ischemia and then recovered in the presence of 10 μm MG132 or 10 μm U0126 for 1 h. Lysates were incubated with GST-Bim and then subjected to TRIM2 immunoprecipitation (TRIM2 IP). Precipitated proteins were immunoblotted with a GST-specific antibody. Binding of Bim with TRIM2 increases following 30-min OGD and is reduced by U0126. E, cells were subjected to 30-min preconditioning ischemia. Lysates were prepared and incubated with GST-Bim or GST-3ABim. Following TRIM2 immunoprecipitation (TRIM2 IP), precipitated proteins were immunoblotted with a GST-specific antibody. The pulldown of GST-Bim from cell lysates by TRIM2 (TRIM2 IP) is increased following preconditioning ischemia. Nonphosphorylatable 3ABim did not pull down.
We used immunoprecipitation to determine whether TRIM2 binding to Bim correlates with the degradation of Bim following preconditioning ischemia. Both U0126 and MG132 stabilize Bim protein levels following preconditioning ischemia (Fig. 3C) and (1). U0126 and MG132 had no affect on TRIM2 protein levels (Fig. 3C). Phosphorylation of MAPK was increased following ischemia, and this increase was blocked by U0126 (Fig. 3C). In contrast, blocking the proteasome with MG132, which also blocks preconditioning-induced Bim degradation, did not block the phosphorylation of p42/p44 MAPK (Fig. 3C).
We performed immunoprecipitations on preconditioned cell lysates incubated with GST-Bim. Binding of GST-Bim to TRIM2 increased following preconditioning (Fig. 3D). The binding of TRIM2 to GST-Bim following preconditioning ischemia was reduced in the presence of U0126, but not MG132 (Fig. 3D). In addition, immunoprecipitation of endogenous Bim with TRIM2 increased following preconditioning ischemia and was blocked by U0126 (data not shown).
As a final test of the MAPK sensitivity of the Bim-TRIM2 interaction following preconditioning, we incubated control and preconditioned cell lysates with either GST-Bim or the nonphosphorylatable 3ABim mutant. The immunoprecipitation of GST-Bim by TRIM2 was low in control cells but increased 1 h following 30-min preconditioning ischemia (Fig. 3E). The immunoprecipitation of Bim with TRIM2 was blocked when cells were incubated with the nonphosphorylatable Bim mutant 3ABim (Fig. 3E). This suggests that following preconditioning ischemia, Bim phosphorylation regulates the binding of Bim to TRIM2 in a manner consistent with the ubiquitination and degradation of Bim.
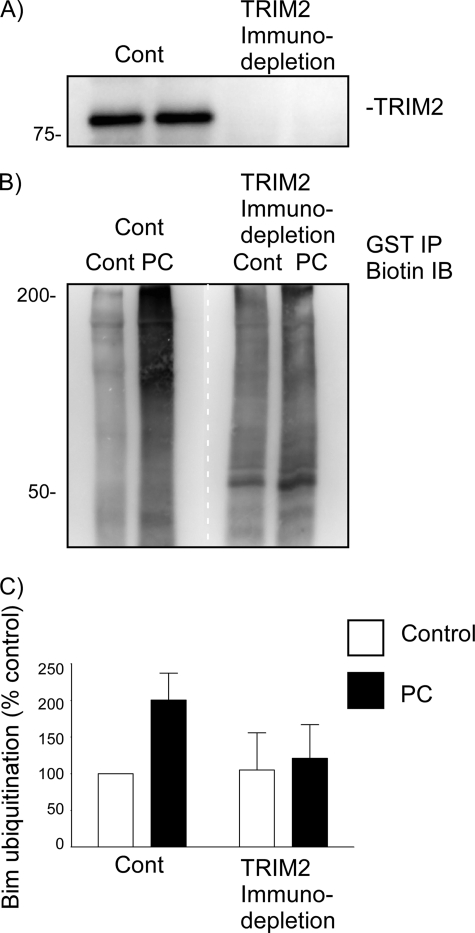
To determine further whether TRIM2 is the enzyme responsible for Bim ubiquitination following preconditioning ischemia, we investigated whether immunodepletion of TRIM2 from cell lysates prior to performing an ex vivo ubiquitination assay would prevent Bim ubiquitination. Cell lysates were subjected to either TRIM2 immunodepletion or control (CBP) immunodepletion prior to ubiquitination assay. TRIM2 immunodepletion was confirmed by immunoblotting (Fig. 4A). Bim ubiquitination was determined using an ex vivo ubiquitination assay (BioMol), whereby lysates were prepared incubated with biotinylated-ubiquitin, GST-Bim, E1 and E2 ligases, and ATP. Following termination of the reaction, the samples were subjected to GST immunoprecipitation and then immunoblotted for biotin. Following preconditioning there was an increase in Bim ubiquitination, as shown in Fig. 4B. Following immunodepletion of TRIM2 from the preconditioned cell lysates, Bim ubiquitination was reduced (Fig. 4, B and C). Taken together, these data confirm that under conditions where Bim ubiquitination increases, Bim binds to the E3 ligase TRIM2 in a p42/p44 MAPK-dependent manner. Furthermore, removal of TRIM2 from cell lysates prevents Bim ubiquitination following preconditioning ischemia.
FIGURE 4.
Immunodepletion of TRIM2 from preconditioned neuronal lysates reduces Bim ubiquitination. Cell lysates from control and preconditioned cells (30-min OGD, 1-h recovery) were prepared and subjected to TRIM2 or control (CBP) immunodepletion. Antibodies raised against CREB binding protein or TRIM2 were bound to protein A/G-agarose beads and incubated with prepared lysates. A, to confirm removal of TRIM2 from the lysate, a sample (5%) was subjected to immunoblotting with a TRIM2-specific antibody. B, control and TRIM2-immunodepleted lysates were then used to measure Bim ubiquitination. Cell lysates were incubated with GST-Bim, biotinylated ubiquitin, and E1 and E2 proteins. GST-Bim was precipitated by GST pulldown, and the ubiquitinated precipitated proteins were immunoblotted with a biotin-specific antibody. Preconditioning results in Bim ubiquitination in control (CBP) immunodepleted lysates, but not TRIM2 depleted lysates. The white dotted line denotes where the image was cut and reassembled to match the above lane loading. C, quantification of three independent experiments is shown.
Knockdown of TRIM2 Using shRNA and Its Effect on Rapid Ischemic Tolerance and Bim Levels
We next examined whether loss of TRIM2 will increase Bim protein stability and block rapid ischemic tolerance. We used shRNAmir to knock down TRIM2 expression. A lentivirus system delivered the shRNA plasmids to neurons. Cells were transduced for 48 h with a multiplicity of infection of 10 and transduction efficiency determined by GFP expression.
Incubation of cortical neurons with shRNA resulted in a 40% decrease in TRIM2 protein levels. We observed a corresponding 40% increase in Bim levels in the same cells (Fig. 5A). We determined the effect of TRIM2 shRNA on preconditioning-induced Bim degradation. Following preconditioning ischemia, Bim levels were reduced in control plasmid treated cells, but not TRIM2 shRNAmir-treated cells (Fig. 5B). As a control we show that the increase in phospho-p42/p44 MAPK levels following preconditioning ischemia are not affected by shRNAmir treatment (Fig. 5B).
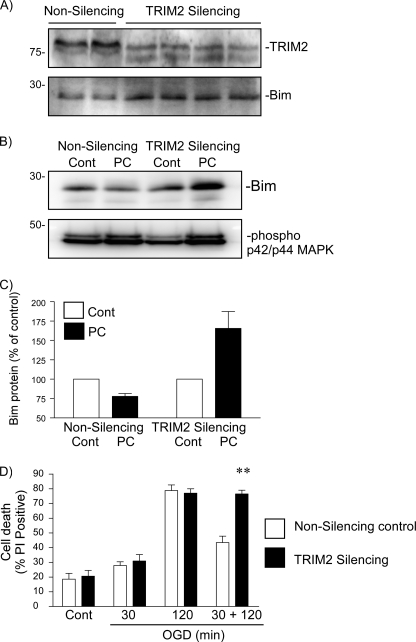
FIGURE 5.
Lentiviral-mediated delivery of anti-TRIM2 shRNAmir reveals a role of TRIM2 in rapid ischemic tolerance-induced neuroprotection. A, neuronal cells were incubated with control or anti-TRIM2 shRNAmir (multiplicity of infection of 10, 48 h). Cell lysates were prepared, and TRIM2 and Bim protein levels were determined by immunoblotting. B, cells were incubated with either nonsilencing control or TRIM2-silencing shRNAmir for 48 h. Cells were then preconditioned with 30-min OGD and recovered for 1 h. Bim protein levels and MAPK phosphorylation were determined by immunoblotting. Representative immunoblots are shown for Bim and phosphorylated p42/p44 MAPK in TRIM2 knockdown cells and control cells subjected to 30-min OGD and 1-h recovery. C, cells were incubated with TRIM2 shRNAmir or nonsilencing control for 48 h. Cells were then subjected to 30-min, 120-min or 30-min followed by 1-h recovery and a subsequent 120-min OGD. Cell death was assessed by propidium iodide assay 24 h later. Data shown are mean ± S.E. (error bars), n = 5. **, p < 0.001 versus nonsilencing treated cells (two-way ANOVA with Bonferroni post hoc test).
The effect of TRIM2 knockdown on neuronal sensitivity to ischemia was then examined. Cells were incubated with shRNAmir for 48 h and then subjected to 30- and 120-min OGD, followed by 24-h recovery. As can be seen in Fig. 5D, no significant differences were observed between silencing- and nonsilencing-treated cells following either 30-min or 120-min OGD. However, knockdown of TRIM2 resulted in a loss of neuroprotection in neuronal cultures subjected to preconditioning ischemia and a subsequent harmful ischemic challenge (Fig. 5D). This suggests that loss of TRIM2 blocks Bim degradation and rapid ischemic tolerance.
DISCUSSION
Here, we propose that TRIM2 functions as a Bim E3 ligase following preconditioning ischemia, mediating the ubiquitination of the cell death protein Bim. Our candidate satisfies previously established criteria that show that the interaction of Bim and TRIM2 is p42/p44 MAPK-dependent. We show the p42/p44 MAPK-dependent interaction between GST-Bim and exogenous TRIM2, synthesized in a cell-free expression system (Fig. 1). In addition, we show, in two different cell lines (MEF cells and cultured rat neurons), that TRIM2 associates with Bim under conditions known to promote Bim ubiquitination. We show that serum stimulation or TPA stimulation of MEF cells increased the binding of Bim to TRIM2 in a MAPK-dependent manner (Fig. 2). In cultured neurons subjected to brief ischemia, Bim binding to TRIM2 increased consistent with Bim degradation (Fig. 3). The binding of Bim to TRIM2 was reduced when a nonphosphorylatable mutant Bim was used in the reactions. Finally, we show that knockdown of Bim by shRNAmir reduced Bim degradation and rapid ischemic tolerance (Fig. 5). Taken together, these data support the role of TRIM2 as mediating p42/p44 MAPK-dependent Bim degradation following preconditioning ischemia.
We have previously reported that Bim ubiquitination is increased following preconditioning ischemia (1). We therefore determined whether TRIM2 may function as a Bim E3 ligase in this model. The binding of exogenous and endogenous Bim to TRIM2 was increased following preconditioning ischemia. Both exogenous (GST-Bim) and endogenous Bim interactions with TRIM2 were investigated to take advantage of mutant Bim proteins with nonphosphorylatable residues. We observed a decrease in Bim binding to TRIM2 when p42/p44 MAPK activation was blocked. This observation is consistent with reports that Bim ubiquitination is p42/p44 MAPK-dependent (11). Furthermore, TRIM2 did not precipitate the nonphosphorylatable mutant 3ABim, which further supports a role of MAPK in regulating the interaction of Bim and TRIM2. When TRIM2 was immunodepleted from neuronal lysates we did not observe an increase in Bim ubiquitination following preconditioning ischemia. Therefore, our identification of TRIM2 as a Bim E3 ligase is consistent with the known biology of Bim ubiquitination as reported by Cook's group (11, 14).
Our studies support the identification of TRIM2 as a Bim E3 ligase. TRIM2 is a member of the TRIM superfamily of E3 ligase proteins containing at least 37 members (19). The proteins are so named because they contain a consensus RING domain, B-Box, and coiled-coil structures in their amino terminus. The RING domain is the molecular signature of the E3 ligase. Unlike other TRIM family proteins that contain two B-box domains, TRIM2 contains only one B-box domain, although the significance of this is as yet unclear (19). Little functional data are available for TRIM2 or neural activity regulated ring finger protein (NARF) since its identification as being induced following seizures in a kainate model of epilepsy (20). Interestingly, Bim levels are regulated by seizures (10, 21). It was shown that TRIM2 was associated with microtubule associated motor protein myosin V (20). TRIM2 was also identified in a cancer genomic paper as being down-regulated in cervical squamous cell carcinoma using gene chip microarrays, although further analysis with real time RT-PCR shows that TRIM2 changes were not consistent (22).
Recently, a TRIM2 knock-out transgenic mouse was described (23) with features consistent with our hypothesis of TRIM2 regulating Bim degradation. The authors show that TRIM2 regulates neurofilament light chain ubiquitination, and loss of TRIM2 promotes a neurodegenerative phenotype in mouse brain. Interestingly, the authors suggest that UbcH5a regulates neurofilament light chain ubiquitination and TRIM2 autoubiquitination (23). In contrast, our own preliminary studies suggest that UbcH5b regulates TRIM2-mediated ubiquitination of Bim.4 We therefore agree with the author's conclusion that multiple E2 proteins may regulate TRIM2-mediated protein ubiquitination. The observation of the neurodegeneration phenotype would be predicted if TRIM2 regulates ubiquitination of Bim, a regulator of neuronal cell death. The role of neurofilament light chain in this mechanism is not yet clear, especially following preconditioning ischemia. Although the authors did not measure Bim levels in their knockdown animals (23), our experiments show that knockdown of TRIM2 increases endogenous Bim protein levels in neurons and blocks the preconditioning-induced decrease in Bim (Fig. 5).
Two additional Bim E3 ligases have been proposed, CIS and c-Cbl (12, 13). However, other studies fail to support their identification, for example Bim protein levels do not change in the testis of c-Cbl knock-out mice, which would be expected if the E3 ligase were absent (24). Bim levels increase following knockdown of TRIM2 (Fig. 5). c-Cbl is a tyrosine kinase, but mutation of all four tyrosine residues in Bim had no effect on Bim ubiquitination (14), whereas mutation of serine residues (Ser65) prevents Bim ubiquitination (11, 25) Serum stimulation of MEFs, which increases Bim ubiquitination, did not increase the interaction between Bim and c-Cbl (14). Finally, p42/p44 MAPK-mediated Bim ubiquitination occurred in MEF cells obtained from c-Cbl knock-out mice (14). A more recent candidate for the Bim E3 ligase is CIS, a member of the SKIP-Cullen E3 ligase family (13). However, the authors did not show a p42/p44 MAPK dependence of the interaction. Neither CIS nor c-Cbl interacts with Bim in a manner consistent with Bim degradation following preconditioning ischemia (supplemental Fig. 1). Following preconditioning ischemia, we did not observe a significant increase in immunoprecipitation of CIS or c-Cbl with Bim. These data suggest that neither CIS, nor c-Cbl, mediates Bim ubiquitination following preconditioning, but they do not rule out the potential role of these E3 ligases under alternative biological conditions. Interestingly, in our experiments we did observe an increase in c-Cbl binding to Bim in the presence of U0126, which is under further investigation (supplemental Fig. 1).
We have recently described the role of the ubiquitin-proteasome system in regulating rapid ischemic tolerance. The identification of the Bim E3 ligase in this system shows that the investigation of molecular mechanisms of rapid ischemic tolerance may yield novel therapeutic targets, which may help to reduce brain injury following ischemia. In addition, these targets may enable the activation of a neuroprotective phenotype when ischemia may be predicted, such as occurs with heart bypass surgery. It is not yet clear whether TRIM2 acts on its own or in conjunction with other proteins to regulate Bim ubiquitination. Although Bim phosphorylation of Ser69 is deemed necessary for the interaction between Bim and TRIM2, it is not yet clear whether this represents the binding sequence on Bim or whether the phosphorylation of this residue results in a conformational change in the structure of Bim, enabling its interaction with TRIM2. Some E3 ligase may also form dimers, which may be important for their function, but it is not yet clear whether TRIM2 acts in a similar manner. Finally, it is not yet clear which species of polyubiquitination TRIM2 modulates on the Bim protein. The ubiquitinated Bim is degraded by the proteasome, hence one would presume a Lys48 linkage polyubiquitin modification. Interestingly when TRIM2 is immunodepleted from cell lysates, we still observe a small increase in a lower molecular mass Bim (approximately GST-Bim +1 ubiquitin), which may be indicative of monoubiquitination. It has been suggested that separate E3 ligases mediate mono- and polyubiquitination of a substrate (26). On the basis of our results TRIM2, may regulate polyubiquitination and function as an E4 ligase. However, the distinction between E4 and E3 ligases is not clear, and TRIM2 contains a RING domain that is associated with E3 ligases, rather than the U-box domain of the E4 ligase. The identification of E3 and E4 ligases for a specific substrate may reveal additional targets for drug manipulation of the target protein.
The previous studies of Bim E3 ligases determined several criteria that they must satisfy to be considered a candidate Bim E3 ligase (14). TRIM2 satisfies these criteria for consideration as a Bim E3 ligase in multiple cell lines, using expressed proteins and following shRNAmir knockdown. Given the potential role of Bim in regulating apoptosis in many cells and pathological conditions, further knowledge of this enzyme offers the exciting potential of regulating Bim protein levels in cells as a therapeutic strategy.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants NS50669 and NS59588 (to R. M.) and NS24728 and NS39016 (to R. P. S.). This work was also supported by National Institutes of Health/National Center for Research Resources/Research Centers at Minority Institutes Grants G12-RR03034 and U54 NS060659 (to Morehouse School of Medicine).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
R. Meller, unpublished observations.
- HECT
- homologous to the E6-AP carboxyl terminus
- CIS
- cytokine-inducible Src homology 2 domain-containing protein
- Bim
- Bcl-2-interacting mediator of cell death
- MEF
- mouse embryonic fibroblast
- OGD
- oxygen and glucose deprivation
- RING
- really interesting new gene
- TPA
- 12-O-tetradecanoylphorbol-13-acetate
- TRIM
- tripartite motif-containing protein.
REFERENCES
- 1. Meller R., Cameron J. A., Torrey D. J., Clayton C. E., Ordonez A. N., Henshall D. C., Minami M., Schindler C. K., Saugstad J. A., Simon R. P. (2006) J. Biol. Chem. 281, 7429–7436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nakamura M., Nakakimura K., Matsumoto M., Sakabe T. (2002) J. Cereb. Blood Flow Metab. 22, 161–170 [DOI] [PubMed] [Google Scholar]
- 3. Reshef A., Sperling O., Zoref-Shani E. (2000) Adv. Exp. Med. Biol. 486, 217–221 [PubMed] [Google Scholar]
- 4. Reshef A., Sperling O., Zoref-Shani E. (2000) Neuroreport 11, 463–465 [DOI] [PubMed] [Google Scholar]
- 5. Pérez-Pinzón M. A., Born J. G. (1999) Neuroscience 89, 453–459 [DOI] [PubMed] [Google Scholar]
- 6. Meller R., Thompson S. J., Lusardi T. A., Ordonez A. N., Ashley M. D., Jessick V., Wang W., Torrey D. J., Henshall D. C., Gafken P. R., Saugstad J. A., Xiong Z. G., Simon R. P. (2008) J. Neurosci. 28, 50–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ordonez A. N., Jessick V. J., Clayton C. E., Ashley M. D., Thompson S. J., Simon R. P., Meller R. (2010) Int. J. Physiol. Pathophysiol. Pharmacol. 2, 36–44 [PMC free article] [PubMed] [Google Scholar]
- 8. Hershko A., Heller H., Elias S., Ciechanover A. (1983) J. Biol. Chem. 258, 8206–8214 [PubMed] [Google Scholar]
- 9. Ardley H. C., Robinson P. A. (2005) Essays Biochem. 41, 15–30 [DOI] [PubMed] [Google Scholar]
- 10. Murphy B. M., Engel T., Paucard A., Hatazaki S., Mouri G., Tanaka K., Tuffy L. P., Jimenez-Mateos E. M., Woods I., Dunleavy M., Bonner H. P., Meller R., Simon R. P., Strasser A., Prehn J. H., Henshall D. C. (2010) Cell Death Differ. 17, 459–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ley R., Balmanno K., Hadfield K., Weston C., Cook S. J. (2003) J. Biol. Chem. 278, 18811–18816 [DOI] [PubMed] [Google Scholar]
- 12. Akiyama T., Bouillet P., Miyazaki T., Kadono Y., Chikuda H., Chung U. I., Fukuda A., Hikita A., Seto H., Okada T., Inaba T., Sanjay A., Baron R., Kawaguchi H., Oda H., Nakamura K., Strasser A., Tanaka S. (2003) EMBO J. 22, 6653–6664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang W., Cheng G. Z., Gong J., Hermanto U., Zong C. S., Chan J., Cheng J. Q., Wang L. H. (2008) J. Biol. Chem. 283, 16416–16426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wiggins C. M., Band H., Cook S. J. (2007) Cell. Signal. 19, 2605–2611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Putcha G. V., Le S., Frank S., Besirli C. G., Clark K., Chu B., Alix S., Youle R. J., LaMarche A., Maroney A. C., Johnson E. M., Jr. (2003) Neuron 38, 899–914 [DOI] [PubMed] [Google Scholar]
- 16. Qi X. J., Wildey G. M., Howe P. H. (2006) J. Biol. Chem. 281, 813–823 [DOI] [PubMed] [Google Scholar]
- 17. Meroni G., Diez-Roux G. (2005) Bioessays 27, 1147–1157 [DOI] [PubMed] [Google Scholar]
- 18. Ley R., Hadfield K., Howes E., Cook S. J. (2005) J. Biol. Chem. 280, 17657–17663 [DOI] [PubMed] [Google Scholar]
- 19. Reymond A., Meroni G., Fantozzi A., Merla G., Cairo S., Luzi L., Riganelli D., Zanaria E., Messali S., Cainarca S., Guffanti A., Minucci S., Pelicci P. G., Ballabio A. (2001) EMBO J. 20, 2140–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ohkawa N., Kokura K., Matsu-Ura T., Obinata T., Konishi Y., Tamura T. A. (2001) J. Neurochem. 78, 75–87 [DOI] [PubMed] [Google Scholar]
- 21. Shinoda S., Schindler C. K., Meller R., So N. K., Araki T., Yamamoto A., Lan J. Q., Taki W., Simon R. P., Henshall D. C. (2004) J. Clin. Invest. 113, 1059–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Miyatake T., Ueda Y., Nakashima R., Yoshino K., Kimura T., Murata T., Nomura T., Fujita M., Buzard G. S., Enomoto T. (2007) Int. J. Cancer 120, 2068–2077 [DOI] [PubMed] [Google Scholar]
- 23. Balastik M., Ferraguti F., Pires-da Silva A., Lee T. H., Alvarez-Bolado G., Lu K. P., Gruss P. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12016–12021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. El Chami N., Ikhlef F., Kaszas K., Yakoub S., Tabone E., Siddeek B., Cunha S., Beaudoin C., Morel L., Benahmed M., Régnier D. C. (2005) J. Cell Biol. 171, 651–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ley R., Ewings K. E., Hadfield K., Howes E., Balmanno K., Cook S. J. (2004) J. Biol. Chem. 279, 8837–8847 [DOI] [PubMed] [Google Scholar]
- 26. Koegl M., Hoppe T., Schlenker S., Ulrich H. D., Mayer T. U., Jentsch S. (1999) Cell 96, 635–644 [DOI] [PubMed] [Google Scholar]
- 27. Meller R., Minami M., Cameron J. A., Impey S., Chen D., Lan J. Q., Henshall D. C., Simon R. P. (2005) J. Cereb. Blood Flow Metab. 25, 234–246 [DOI] [PubMed] [Google Scholar]
- 28. Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 29. Gatlin C. L., Kleemann G. R., Hays L. G., Link A. J., Yates J. R., 3rd (1998) Anal. Biochem. 263, 93–101 [DOI] [PubMed] [Google Scholar]
- 30. Hemsley A., Arnheim N., Toney M. D., Cortopassi G., Galas D. J. (1989) Nucleic Acids Res. 17, 6545–6551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cleary S. P., Tan F. C., Nakrieko K. A., Thompson S. J., Mullineaux P. M., Creissen G. P., von Stedingk E., Glaser E., Smith A. G., Robinson C. (2002) J. Biol. Chem. 277, 5562–5569 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.