Abstract
Queuosine is a modified pyrrolopyrimidine nucleoside found in the anticodon loop of transfer RNA acceptors for the amino acids tyrosine, asparagine, aspartic acid, and histidine. Because it is exclusively synthesized by bacteria, higher eukaryotes must salvage queuosine or its nucleobase queuine from food and the gut microflora. Previously, animals made deficient in queuine died within 18 days of withdrawing tyrosine, a nonessential amino acid, from the diet (Marks, T., and Farkas, W. R. (1997) Biochem. Biophys. Res. Commun. 230, 233–237). Here, we show that human HepG2 cells deficient in queuine and mice made deficient in queuosine-modified transfer RNA, by disruption of the tRNA guanine transglycosylase enzyme, are compromised in their ability to produce tyrosine from phenylalanine. This has similarities to the disease phenylketonuria, which arises from mutation in the enzyme phenylalanine hydroxylase or from a decrease in the supply of its cofactor tetrahydrobiopterin (BH4). Immunoblot and kinetic analysis of liver from tRNA guanine transglycosylase-deficient animals indicates normal expression and activity of phenylalanine hydroxylase. By contrast, BH4 levels are significantly decreased in the plasma, and both plasma and urine show a clear elevation in dihydrobiopterin, an oxidation product of BH4, despite normal activity of the salvage enzyme dihydrofolate reductase. Our data suggest that queuosine modification limits BH4 oxidation in vivo and thereby potentially impacts on numerous physiological processes in eukaryotes.
Keywords: Gene Knockout; Metabolism; Pterin; RNA Modification; Transfer RNA (tRNA); 7,8-Dihydrobiopterin; Phenylalanine Hydroxylase; Queuine; Queuosine; Tetrahydrobiopterin
Introduction
Bacteria and humans have co-evolved for millennia, and many examples exist of how various symbiotic and commensal partnerships contribute to human health and nutrition ranging from the metabolism of complex carbohydrates to the provision of vital micronutrients (1). Queuosine is an example of a micronutrient, synthesized exclusively by bacteria but which, for poorly defined reasons, is utilized by almost all eukaryotic species with the exception of the baker's yeast, Saccharomyces cerevisiae (2).
Bacterial queuosine biosynthesis occurs in two stages. First, a series of five enzymatic steps convert guanosine triphosphate nucleoside (GTP) to the soluble 7-aminomethyl-7-deazaguanine molecule. Subsequently, 7-aminomethyl-7-deazaguanine is inserted into the wobble position of tRNA containing a GUN consensus sequence (Tyr, Asp, Asn, and His) by means of the single enzyme species, tRNA guanine transglycosylase (TGT), and is further remodeled in situ to queuosine (3). Eukaryotes must acquire queuosine or its free nucleobase, queuine, from food and the gut microflora. Curiously, both cytosolic and mitochondrial tRNA species are modified by queuosine (2). The eukaryotic enzyme that performs this reaction, queuine tRNA ribosyltransferase, has recently been identified as a heterodimeric complex, consisting of the eukaryotic homologue of the catalytic TGT subunit and a related protein called queuine tRNA ribosyltransferase domain containing 1 (QTRTD1), both of which localize to the mitochondria (4, 5).
Studies on germ-free (axenic) mice maintained on a chemically defined diet provided clear evidence that eukaryotes are nonautotrophic for queuosine biosynthesis (6). Unchallenged, these animals appear normal. However, withdrawal of tyrosine from the diet resulted in symptoms of squinting, stiffness, lethargy, convulsion, and ultimately death after 18 days (7). Re-administration of either chemically synthesized queuine or tyrosine alone prevented the symptoms; the latter result suggests that tyrosine uptake and utilization are unaffected by queuine status. It has been long established that tyrosine is a nonessential amino acid in higher eukaryotes as it can be synthesized from phenylalanine by the action of the phenylalanine hydroxylase (PAH)2 enzyme. It has therefore been suggested that the absence of queuine may affect the translation of the PAH enzyme leading to a dietary dependence on tyrosine supply (2, 8).
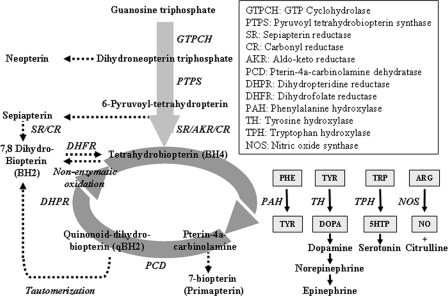
In humans, tyrosine production occurs principally in the liver and kidney correlating with the expression of the PAH enzyme (9). Deficiency in PAH leads to the disease phenylketonuria, characterized by increased blood levels of phenylalanine (referred to hyperphenylalaninemia) and reduced levels of tyrosine. In performing its reaction, PAH requires molecular oxygen and BH4 cofactor (Fig. 1). BH4 is produced from GTP by the enzymes GTP cyclohydrolase I, 6-pyruvoyltetrahydropterin synthase, and sepiapterin reductase. In generating BH4, other intermediary reactions at the sepiapterin reductase step are performed by carbonyl reductase and member proteins of the aldo-keto reductase family (10). The BH4 cofactor may also be recycled by the activity of two enzymes, pterin-4a-carbinolamine dehydratase and dihydropteridine reductase, which has particular importance for tyrosine biosynthesis in the liver. Deficiency in any of the BH4 enzymes, with the exception of sepiapterin reductase, causes hyperphenylalaninemia. Recent studies have shown that BH4 is also highly susceptible to auto-oxidation in vivo producing the metabolite 7,8-dihydrobiopterin (BH2), whose accumulation is limited by the enzyme dihydrofolate reductase (DHFR) through reduction of BH2 to BH4 (11).
FIGURE 1.
Schematic of tetrahydrobiopterin biosynthesis and recycling. Tetrahydrobiopterin cofactor (BH4) is synthesized from guanosine nucleoside triphosphate (GTP) via a series of reactions involving GTP cyclohydrolase (GTPCH), 6-pyruvoyltetrahydropterin synthase (PTPS), and a two-step reaction catalyzed by sepiapterin reductase (SR). The 6-pyruvoyltetrahydrobiopterin intermediate can also be converted to BH4 by members of the aldo-keto reductase family (AKR) and carbonyl reductase (CR), which are abundant in liver. Sepiapterin can be formed nonenzymatically from reaction intermediates produced by sepiapterin reductase, aldo-keto reductase family, or carbonyl reductase and may subsequently be converted to 7,8-dihydrobiopterin (BH2). Further sources of BH2 include nonenzymatic oxidation of BH4 and tautomerization of quinonoid dihydrobiopterin produced during BH4 recycling. The salvage reaction of DHFR limits the accumulation of oxidized biopterin in vivo by reducing BH2 back to BH4. Following catalysis by the aromatic amino acid hydroxylase enzymes (PAH, tyrosine hydroxylase (TH), and tryptophan hydroxylase (TPH)), BH4 cofactor is oxidized to its hydroxyl form and requires the activity of pterin-4a-carbinolamine dehydratase (PCD) and dihydropteridine reductase (DHPR) to regenerate reduced cofactor. Disorders that affect the biosynthesis or regeneration of BH4 may be detected by quantitative analysis of neopterin, sepiapterin, 7-biopterin, and BH2 as indicated. Adapted from Ref. 38.
By extrapolation from the current data, the tyrosine dependence of queuine-deficient animals may relate to the loss of PAH activity or altered BH4 cofactor supply. In this study, we show that queuosine modification of tRNA, or hypothetically another unknown RNA substrate of the queuine tRNA ribosyltransferase enzyme, as opposed to free queuine base, is required for normal tyrosine production in eukaryotes. Decreased BH4 levels, concomitant with a marked accumulation of BH2 suggests that oxidation of BH4 cofactor underlies the defect.
EXPERIMENTAL PROCEDURES
Animals
Mice were bred and housed under specific pathogen-free conditions. Procedures were performed on mice at 6–8 weeks of age, unless otherwise stated, according to regulations and guidelines of the Ethics Committee, Trinity College Dublin, and the Irish Department of Health. Tyrosine-free chemically defined diet (AIN-76A-purified diet) was obtained from Harlan Teklad.
Cell Culture
HepG2 cells were purchased from the American Tissue Culture Collection and grown in an atmosphere of 5% CO2 at 37 °C in the following media: Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (DMEM/FBS) or 10% (v/v) horse serum (DMEM/HS), 2 mm l-glutamine, 1 mm sodium pyruvate, and penicillin/streptomycin (50 units/0.1 mg/ml) or in UltracultureTM serum-free medium (Lonza) supplemented with 2 mm l-glutamine and penicillin/streptomycin (10 units/20 μg/ml). When required, chemically synthesized queuine (a gift from Dr. Susumu Nishimura, Tsukuba University, Japan) was added at a final concentration of 300 nm.
Radiolabeled Phenylalanine Hydroxylase Assay
HepG2 cells were grown in serum-free (SF) medium, SF medium supplemented with queuine (300 nm), or in DMEM, FBS. Intracellular tyrosine and phenylalanine were depleted by incubation in SF medium not containing tyrosine or phenylalanine (Lonza; custom synthesis) and supplemented with cycloheximide (20 μg/ml) with constant shaking at 70 rpm for 1 h. Subsequently, cells were washed and incubated in KRHL buffer (120 mm NaCl, 4.8 mm KCl, 10 mm d-glucose, 2.5 mm CaCl2, 1.3 mm MgSO4, 2.5 mm Hepes, pH 7) containing 20, 60, and 100 μm l-[2,6-3H]phenylalanine (54 Ci/mmol; Amersham Biosciences) and 20 μg/ml cycloheximide for 30 min with constant shaking. Duplicate wells for each experimental point were used to determine cell protein content. Cells were solubilized with M-PER solution (Pierce), and the protein concentration was determined by Bradford assay (Bio-Rad). After 30 min, medium was removed, and cells were washed rapidly with ice-cold PBS and lysed in ice-cold 10% trichloroacetic acid (100 μl per well). Cells were scraped from the dish, and the lysate was centrifuged at 16,000 × g for 15 min. The supernatant was stored at −70 °C until analyzed. Intracellular phenylalanine and tyrosine were measured by HPLC on a Zorbax 300SB-C18 column (Agilent) pre-equilibrated with Mobile Phase Buffer (100 mm sodium phosphate, pH 1.9, 300 μm octyl sodium sulfate, 500 μm of EDTA and 6% HPLC grade methanol) and run at 1 ml/min. Cell extracts (50 μl) were spiked with cold tyrosine (0.1 mm final) and phenylalanine (0.5 mm final) and detected by fluorescence with an excitation of 258 nm and emission of 288 nm. Samples (1 ml) were collected, and radiolabeled amino acids were evaluated by scintillation counting.
Phenylalanine Metabolism in Mouse
l-Phenylalanine (40 mg/ml) was dissolved in 0.9% saline solution containing 7.5 mm NaOH and injected intraperitoneally at 1 mg/g of body weight. Blood was collected from the ventral caudal artery in K2-EDTA tubes (36 μg/ml of blood) and promptly centrifuged at 2,000 × g for 10 min at 4 °C. Samples were deproteinized by adding 9× volumes of 1.11 m HClO4, and the sample was allowed to stand for 10 min and then centrifuged at 16,000 × g for 6 min at 4 °C. An aliquot of the supernatant was treated with 0.091 volumes of 10 m KOH, incubated on ice for 10 min, and then centrifuged at 16,100 × g for 6 min at 4 °C. The supernatant was filtered through a 0.22-μm ultrafree-MC filter unit (Millipore) at 16,100 × g for 1 min at 4 °C. HPLC separations were performed at ambient temperature on a Zorbax 300SB-C18 column (Agilent) as described above. Samples (65 μl per run) had a pH value less than 7.0. Tyrosine and phenylalanine were detected by fluorescence with an excitation wavelength of 258 nm and an emission wavelength of 288 nm, and the levels were quantified by integration of the peak area using EZStart 7.3SPI software (Shimadzu).
PAH Antisera Production and Immunoblotting
PAH transcript from mouse liver was reverse-transcribed using the primer PAHR (5′-ccagtcgacgttctgtctatgacgtcactttctg-3′; SalI restriction site underlined) according to the Superscript III protocol (Invitrogen). First-strand products were separated using a nucleotide removal kit (Qiagen). PAH cDNA was amplified by PCR using the aforementioned reverse primer and the forward primer PAHF (5′-cacggatccatggcagctgttgtcctggagaacg-3′; BamHI restriction site underlined). The PAH cDNA was cloned into the BamHI and SalI restriction sites of pMal-c2 (New England Biolabs), which was transformed in Escherichia coli BL21 DE3 cells. Protein induction was performed by addition of 0.5 mm isopropyl 1-thio-β-d-galactopyranoside at 18 °C overnight. PAH-maltose-binding protein fusion protein was purified on amylose resin (New England Biolabs). Antisera were raised in New Zealand White rabbits (Harlan, UK) and counter-selected against purified maltose-binding protein bound to Ultralink Biosupport resin (Pierce) as described previously (4). PAH antisera was used at a dilution of 1:2,000.
PAH Activity Analysis
Liver was homogenized in ice-cold buffer (50 mm Tris-HCl, pH 7.5, 0.1 m KCl, 1 mm EDTA, 1 mm DTT, Sigma protease inhibitor mixture), and PAH activity was measured as described previously (12) using the natural cofactor (6R)-l-erythro-5,6,7,8-tetrahydrobiopterin ((6R)-BH4) at a concentration of 100 μm. Alternatively, production of 14C from [14C]phenylalanine was analyzed by thin layer chromatography according to published protocols using 100 μm (6R)-BH4 as cofactor (13).
Generation and Genotyping of Qtrt1 Genetap Mice
The AK7.1 ES cell line FHCRC-GT-S12–11A1 was obtained from and chimeric mice were generated by the mutant mouse regional resource center (MMRRC, University of California, Davis). Ear-punch samples from F1 offspring were analyzed by PCR for germ line transmission. Genotyping was performed for 35 cycles (10 s at 98 °C, 30 s at 66 °C, and 60 s at 72 °C) using the primer pair QWF (5′-gaggccggtgtgtggattcgatctg-3′) and QWR (5′-cagagcattctggatctccaccg-3′) to identify the wild-type allele (451 bp) and QWF in conjunction with the reverse primer QGR (5′-tctagcctcgaggtcgacggtatcg-3′) to identify the Qtrt1Gt(FHCRC-GT-S12–11A1)Sor allele (750 bp). PCRs to identify the promoter trap and poly(A) modules were performed for 35 cycles (10 s at 98 °C, 30 s at 67 °C, and 30 s at 72 °C) using the primers NeoF (5′-gcacgctgattgaagcagaagcctgc-3′) and NeoR (5′-ggtcagacgattcattggcaccatgc-3′) to confirm the presence of the β-Geo cassette (323 bp) and the primers HYGF (5′-aagttcgacagcgtctccgacctgatg-3′) and HYGR (5′-cgccatgtagtgtattgaccgattcc-3′) to detect the presence of the hygromycin cassette (389 bp).
Southern Blotting
Southern blotting probes 5′ and 3′ to the ROSAFARY insertion site were generated by PCR and cloned into the EcoRI and XhoI restriction sites of pBluescript II SK(+). To do this, genomic DNA was isolated from wild-type liver using the RecoverEase DNA isolation kit (Stratagene) according to the manufacturer's instructions. The 5′-probe (399 bp) was amplified by PCR for 35 cycles (20 s at 94 °C, 30 s at 66 °C, and 45 s at 72 °C) using the primers 5SF (5′-cgactcgaggctgccgcatctgcttggg-3′; XhoI site underlined) and 5SR (5′-gcggaattcctcctccgtcacctcggac-3′; EcoRI site underlined), and the 3′-probe (285 bp) was amplified by PCR for 35 cycles (20 s at 94 °C, 30 s at 65 °C, and 70 s at 72 °C) using the primers 3SF (5′-cgactcgagcttccgctcgccctacgat-3′; XhoI site underlined) and 3SR (5′-gcggaattcgagacagggtttctctgtg-3′; EcoRI site underlined). The neomycin probe (323 bp) was generated using the NeoF and NeoR primers used previously to confirm the presence of the β-Geo cassette. Southern blotting was performed as described previously (14).
Sequencing of the Gene-trap Insertion Site
Nested PCRs were used to amplify and clone the regions of Qtrt1 exon 3 flanking the ROSAFARY integration site. The 5′- and 3′-junction site of exon 3 with the ROSAFARY cassette was amplified by PCR for 35 cycles (20 s at 94 °C, 30 s at 61 °C, and 120 s at 72 °C) using the primers Q/LF (5′-tccgaactcgtcagttccaccacg-3′) and Q/LR (5′-ctactgccagctttgacatccaa-3′) to amplify the 5′-junction site and L/QF (5′-tccgaactcgtcagttccaccacgg-3′) and L/QR (5′-ttggatgtcaaagctggcagtag-3′) to amplify the 3′-junction site. Subsequently, nested PCR was carried out for 35 cycles (20 s at 94 °C, 60 s at 63 °C, and 60 s at 72 °C) using the NQ/LF (5′-cgagaattcgacaatcggacagacacag-3′) and NQ/LR (5′-gcggaattcgctgggattaaaggcatg-3′) primers to re-amplify the 5′-junction site and NL/QF (5′-ctcgaggacaatcggacagacacagataag-3′) and NL/QR (5′-gaattccgcatgcctttaatcccagcactcg-3′) to re-amplify the 3′-junction site. The primer set for the nested re-amplification of the 5′-junction contained EcoRI restriction sites, and the primers to re-amplify the 3′-junction contained an EcoRI and XhoI restriction site (underlined). These amplicons were digested with restriction enzymes, cloned into the E. coli plasmid pBluescript II SK(+) (Stratagene), and sequenced (MWG, Ebersberg, Germany) using the M13 (−21) forward and M13 reverse primers.
Tetrahydrobiopterin Measurement in Plasma and Urine
Blood (150 μl) was collected from the tail ventral caudal artery in tubes containing K2EDTA (36 μg/ml of blood) and 0.1% (w/v) DTT, and plasma was separated by centrifugation at 2,000 × g for 10 min at room temperature. Urine samples were collected and kept at 4 °C and out of sunlight. Total protein was determined by the Bradford assay (Bio-Rad). Prior to biopterin measurements, urine samples were diluted to 30 μg/ml and plasma samples to 3 mg/ml. BH4 levels were measured by HPLC analysis after iodine oxidation in acidic or alkaline conditions as described previously (15) on a Spherisorb ODS1 C18 column (particle size 5 μm; 250 × 4.6 mm; Waters) by an isocratic gradient in water with 5% (v/v) methanol at a flow rate of 0.6 ml/min.
LC-MS Analysis of tRNA
Queuosine nucleoside content of bulk tRNA (16) was determined by liquid chromatography-tandem mass spectrometry according to published protocols (17).
DHFR Activity Assay
Liver (1 g) was homogenized in 3 ml of ice-cold homogenization buffer (0.2 m Tris-HCl, pH 7.6, at 4 °C containing 10 mm DTT and protease inhibitors) and centrifuged at 100,000 × g for 60 min at 4 °C. The standard spectrophotometric assay was used to measure DHFR activity (18).
RESULTS
Queuine Deficiency in HepG2 Cells Compromises Phenylalanine to Tyrosine Conversion
The original queuine and tyrosine depletion study was performed on an outbred Swiss mouse strain, raising the possibility that the effect is unique to mice or to the genetic background of the animals used. To rule out this possibility, we examined the effect of queuine deficiency on tyrosine production in HepG2 cells, a human hepatoma cell line functionally capable of synthesizing tyrosine from phenylalanine and containing the necessary enzymes for BH4 synthesis and recycling (19, 20).
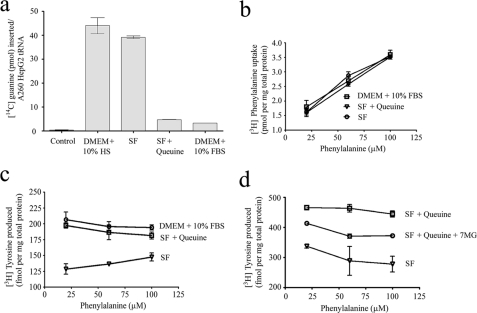
Cell growth medium supplemented with 10% fetal bovine serum (FBS) contains ∼1–2 × 10−8 m queuine (21), allowing for full modification of tRNA. By contrast, HS is essentially queuine-free (22) providing a means to deplete queuine from cells. Unfortunately, FBS and HS influence PAH activity in ways unrelated to queuine (23). Therefore, for the purpose of this study, HepG2 cells were also grown in SF medium in the absence or presence of chemically synthesized queuine. The guanine incorporation assay (4) was used to evaluate the queuosine status of tRNA (Fig. 2a). High levels of guanine incorporation occurred in tRNA extracted from HepG2 cells grown in SF- or HS-containing medium (signifying a depletion of the queuosine modification in tRNA), whereas guanine incorporation was low for tRNA extracted from cells grown in SF medium supplemented with queuine or in FBS-containing medium. The results indicate that SF medium can deplete queuosine-modified tRNA (Q-tRNA) and, by inference, queuine levels in cells. The results of the enzymatic assay were confirmed by LC-MS analysis (supplemental Fig. 1).
FIGURE 2.
Queuine deficiency impairs phenylalanine to tyrosine conversion in HepG2 cells. a, Q-tRNA levels in HepG2 cells grown in DMEM supplemented with 10% horse serum (DMEM + 10% HS), serum-free medium, serum-free medium plus queuine (SF + Queuine), and DMEM supplemented with 10% FBS (DMEM + 10% FBS). tRNA that has been modified by queuosine is unable to accept [14C]guanine into the anticodon loop by the E. coli TGT enzyme reaction. The control reaction contained tRNA from SF grown cells but did not contain TGT enzyme. b, influence of queuine on phenylalanine uptake; c, tyrosine production by HepG2 cells. Cells, grown in the indicated medium, were placed in a phenylalanine- and tyrosine-deficient medium for 1 h. Subsequently, [14C]phenylalanine was added for 30 min. Cells were lysed and radiolabeled amino acids analyzed by separation on a C18 HPLC column followed by scintillation counting. d, effect of 7-methylguanine on tyrosine biosynthesis by HepG2 cells. Cells were cultivated in either SF medium alone (SF), SF medium supplemented with queuine (SF + Queuine), or SF medium pretreated with 7-methylguanine (7MG) for 24 h prior to and after the addition of queuine (SF + Queuine + 7MG). Cells were incubated with increasing concentrations of [14C]phenylalanine for 1 h (rather than the 30 min used earlier) and intracellular phenylalanine and tyrosine levels measured as described above.
To measure tyrosine production in vivo, HepG2 cells were depleted of tyrosine and phenylalanine and subsequently incubated with increasing concentrations of [14C]phenylalanine. Intracellular phenylalanine (Fig. 2b) and tyrosine (Fig. 2c) were analyzed by reversed phase HPLC. Our results show that queuine status does not influence phenylalanine uptake, which is linear with respect to the concentration supplied. However, the ability to produce tyrosine is reduced by 15–40% depending on the amount of phenylalanine administered. The lack of a dose response for tyrosine synthesis in the experiment is considered to arise from a low capacity of HepG2 cells for phenylalanine hydroxylation, which is saturated even at the lowest concentration of [14C]phenylalanine used.
In an attempt to distinguish whether the effect on tyrosine formation arises from the depletion of queuine base or queuosine modification of (t)RNA, a known inhibitor of the queuine tRNA ribosyltransferase activity, 7-methylguanine (24), was used to treat HepG2 cells (Fig. 2d). Taking the queuosine status of tRNA of cells grown in serum-free medium as being fully unmodified (39.15 pmol of [14C]guanine insertion/A260 tRNA) and those grown in serum-free medium in the presence of queuine as being fully modified (4.54 pmol of [14C]guanine insertion/A260 tRNA), the administration of 7-methylguanine resulted in tRNA being ∼19% unmodified with respect to queuosine (11.16 pmol of [14C]guanine insertion/A260 tRNA). This was concomitant with a significant reduction in tyrosine formation compared with queuine-sufficient controls. These results suggest that the status of queuosine-modified (t)RNA, as opposed to the levels of queuine nucleobase, influences the ability of HepG2 cells to produce tyrosine.
Qtrt1 Gene-trap Mice Are Deficient in Queuosine-modified tRNA
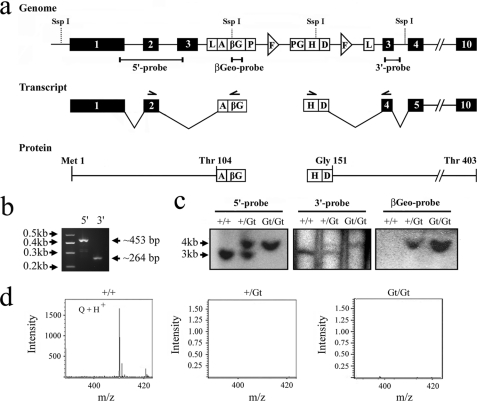
To create mice lacking the TGT enzyme, a gene-trap insertion strategy was employed. The ES cell line FHCRC-GT-S12-11A1 contains an integrated copy of the ROSAFARY vector in the Qtrt1 gene, which encodes the TGT subunit. The ROSAFARY insert was putatively mapped to intron 2 creating the allele Qtrt1Gt(FHCRC-GT-S12–11A1)Sor subsequently abbreviated as Qtrt1Gt.
Attempts to confirm the suspected position of the gene-trap cassette were unsuccessful, and a nested PCR approach was used to map the region spanning exons 1–4. Our results placed the cassette in exon 3 of the Qtrt1 gene (Fig. 3a). Subsequent cloning and sequencing of the 5′- and 3′-flanking regions of the cassette unequivocally mapped the cassette to position 21,216,900 on chromosome 9, dividing exon 3 of Qtrt1 in approximately two parts. ES cells were microinjected into blastocysts, giving rise to 12 male mice exhibiting 50–98% chimerism, 2 of which achieved germ line transmission on multiple occasions.
FIGURE 3.
Gene-trap disruption of Qtrt1 results in loss of Q-tRNA. a, disruption of the Qtrt1 gene by the ROSAFARY gene-trap. The insertion site was mapped by DNA sequencing. Exons are shown numbered in black boxes and introns are depicted as intervening lines. The positions of restriction sites for the SspI DNA endonuclease and probes for Southern blotting are shown. The ROSARARY vector contains 5′- and 3′-long term repeats (L), a splice acceptor site (A), coding sequence for the β-galactosidase-neomycin fusion protein (βG), a polyadenylation signal sequence (P), a selectable marker for hygromycin resistance (H), expressed under the PGK promoter (PG), a splice donor site (D), and two frt elements (F). The spliced transcripts from the Qtrt1Gt allele and the encoded fusion protein products are shown. b, RT-PCR analysis of the Qtrt1Gt transcripts demonstrating that exon 3 is not transcribed. Total RNA was isolated from the liver of homozygous gene-trap animals and reverse-transcribed using reverse primers located in the β-Geo cassette and exon 4. Subsequently, forward primers in exon 2 and the hygromycin cassette were used in combination with the reverse primers yielding amplicons of 453 and 264 bp, consistent with the loss of exon 3 during splicing. c, Southern blotting of the Qtrt1 gene locus of wild-type (+/+), heterozygous (+/Gt), and homozygous (Gt/Gt) animals. Genomic DNA was digested with SspI and hybridized to probes complementary to regions 5′ and 3′ to the ROSAFARY insert and a probe internal to the βGeo cassette. d, bulk tRNA was isolated from the liver of 6-week-old animals and enzymatically hydrolyzed before being analyzed by LC-MS. The queuosine peak is identifiable at 410 m/z.
The ROSAFARY vector is designed such that a promoter trap module (SAβgeo*pA) with an artificial adenoviral splice acceptor acts as the 3′-terminal exon to create a β-galactosidase-neomycin (β-geo) fusion marker with any upstream exons. In addition, a poly(A) trap module containing the hygromycin resistance gene (PGKhygSD) with a downstream splice donor site forms a fusion transcript with any downstream exons (25). The placement of the cassette in exon 3 of Qtrt1 would necessitate that this exon is skipped during splicing as a readthrough transcript would encounter multiple stop codons precluding the production of the β-geo protein. This was not considered possible because β-geo was used to select for G418 antibiotic resistance during ES cell screening (25). Analysis of the transcript from the Qtrt1Gt allele by RT-PCR demonstrated that, as expected, exon 3 is not produced (Fig. 3b), and the TGT protein is made in two fragments, a Met1–Thr104::β-geo fusion and a hygromycin::Gly151–Thr403 fusion (Fig. 3a). Importantly, the loss of exon 3 would remove an essential active site aspartate (Asp102; according to mouse TGT numbering) and a serine residue involved in substrate recognition (Ser103) negating the possibility of catalytic activity (4, 5).
To obtain mice lacking active TGT protein, heterozygous animals (Qtrt1Gt/+) were intercrossed. Southern blotting analyses of the 5′- and 3′-ROSAFARY insertion sites and the β-geo cassette region are consistent with a single gene-trap insertion in exon 3 of the Qtrt1 locus (Fig. 3c). As explained above, disruption of the Qtrt1 gene, which encodes the catalytic subunit of the eukaryotic queuine tRNA ribosyltransferase complex, would be expected to be functionally incapable of Q-tRNA formation. Confirmation of the Q-tRNA status of animals was made by LC-MS analysis of bulk tRNA extracted from the liver of 6-week-old Qtrt1+/+, Qtrt1Gt/+, and Qtrt1Gt/Gt mice (Fig. 3d). As expected, wild-type animals contained Q-tRNA. However, Q-tRNA could not be detected in either heterozygous or homozygous animals. That heterozygous mice failed to produce detectable levels of Q-tRNA may be explained by the fact animals are born germ-free and without Q-tRNA leading to the possibility that the single normal Qtrt1 allele is haploinsufficient or that the fusion constructs are acting in a dominant negative manner to sequester away limited amounts of QTRTD1 from active TGT protein. Irrespectively, analysis of tRNA from the liver of older heterozygous animals, at 16 weeks of age, revealed that appreciable levels of Q-tRNA had been produced (supplemental Fig. 2).
Genotype analysis of 36 litters (229 pups) from heterozygous intercrossing (supplemental Fig. 3) showed that TGT deficiency does not influence viability or sex bias (supplemental Table 1). In addition, breeding of homozygous animals revealed that both males and females have normal fecundity (supplemental Table 2), concurring with the lack of an obvious phenotype in queuine-deficient fly (26), worm (27), and mouse (28).
TGT Disruption in Mouse Decreases Tyrosine Production from Phenylalanine
The studies on HepG2 cells suggested that queuine tRNA ribosyltransferase inactivation negatively impacts tyrosine biosynthesis. The generation of TGT-deficient mice provided a means to explore this effect in whole animals. Phenylalanine was injected into the peritoneum of 6–8-week-old mice (1 mg/g of body weight) that had been maintained on a normal diet. At various time points, blood samples were collected from the ventral caudal artery and plasma tyrosine analyzed by HPLC (Fig. 4a). Mice of all three genotypes were found to produce equivalent amounts of tyrosine within the 1st h. However, subsequent to this, a sharp decline in tyrosine production by Qtrt1Gt/+ and Qtrt1Gt/Gt mice was observed. This contrasts with the sustained production of tyrosine in wild-type animals, which only began to decline from 2 h onward as serum phenylalanine from the initial peritoneal bolus became exhausted. The increased ability of wild-type mice to produce tyrosine relative to hetero- and homozygous gene-trap mice is readily apparent from the conversion ratio of phenylalanine to tyrosine in plasma (Fig. 4b). Similar results were obtained using animals that had been fasted for 24 h prior to phenylalanine loading (supplemental Fig. 4).
FIGURE 4.
TGT deficiency in mouse impairs phenylalanine to tyrosine conversion. a, phenylalanine (1 mg/g body weight) was administered intraperitoneally to eight individual wild-type (+/+), heterozygous (+/Gt), and homozygous (Gt/Gt) animals, and at the times indicated blood was withdrawn from the ventral caudal artery. Plasma phenylalanine and tyrosine were separated by HPLC on a reversed-phase C18 column and detected by their absorbance at 206 nm. Each point was measured in triplicate. b, ratio of plasma tyrosine to phenylalanine (conversion ratio) in animals at various times after the administration of phenylalanine.
To determine whether TGT deficiency phenocopies the tyrosine dependence of axenic, queuine-deficient mice, two male animals of each genotype Qtrt1Gt/+, Qtrt1Gt/+ and Qtrt1Gt/Gt were maintained on a chemically defined tyrosine-free diet (AIN-76A based) for 2 months. No lethality occurred, and no overt physical or behavioral abnormalities could be visually ascribed. This result suggests that either the germfree status of the original study presented additional compounding factors, that it was the lack of free queuine, which was responsible for the phenotype, and/or that bacteria in the gut of TGT deficient mice can supply sufficient amino acid to permit survival. In this regard, studies show that the intestinal microbiota of adult humans may provide 1–20% of circulating plasma lysine and threonine (1).
TGT Inactivation Does Not Affect PAH Expression or Activity in Mouse Liver
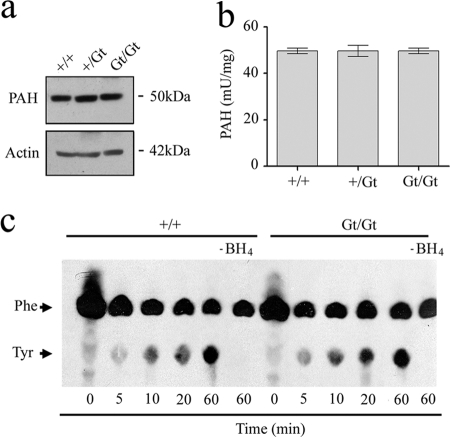
A decrease in the ability to metabolize phenylalanine to tyrosine is a characteristic of the disease phenylketonuria that can arise from defects in the expression or activity of the PAH enzyme. The position of queuosine within the wobble position of the anticodon could potentially affect PAH translation as the absence of queuosine was shown to drastically decrease translation of virF mRNA in the pathogen Shigella flexneri (29) and to induce +1 frameshifting in bacteria (30). To explore a possible effect on PAH translation, antisera were raised to recombinant PAH, and liver samples analyzed by immunoblot assay (Fig. 5a). No observable difference in PAH expression was detected across wild-type and gene-trap animals ruling out a defect in translation.
FIGURE 5.
TGT status does not influence PAH expression or activity. a, liver cytosolic extract was prepared from 6- to 8-week-old wild-type and gene-trap mice and analyzed by immunoblot using antisera against PAH and actin, the latter serving as an internal control. b, analysis of PAH activity in mouse liver extracts. Values are given per mg of total protein (n = 3 measurements from three different animals per group). c, representative figure (of three experiments) showing the measurement of [14C]phenylalanine hydroxylation in liver cytosol from wild-type and homozygous gene-trap animals that had been diluted only marginally (1:20) by the addition of phenylalanine (0.2 mm cold phenylalanine; 33 nCi [14C]phenylalanine), catalase (3.4 μg), and BH4 (100 μm). The [14C]phenylalanine and [14C]tyrosine were resolved by thin layer chromatography and visualized by autoradiography.
A variety of other mechanisms are known to regulate PAH activity in vivo, including its activation by phenylalanine and phosphorylation, in addition to its allosteric inhibition by BH4 cofactor (31, 32). Previous reports suggest that queuine can enhance the phosphorylation of unspecified cytosolic proteins (22), whereas in other cases a decrease in phosphorylation was observed (33–35). In addition, notable similarities exist between the structure of queuine and tetrahydrobiopterin, both being derived from GTP. Indeed, biopterins are known inhibitors of the queuine ribosyltransferase activity in vitro (36) and in vivo (24). As such, a reciprocal relationship between the queuosine modification and PAH could be envisaged by, for example, counterbalancing the allosteric inhibition of PAH by BH4.
Liver cytosolic fractions were examined for PAH activity in all three genotypes revealing that queuine has no impact on the specific activity of tyrosine formation; all were ∼50 milliunits·mg−1 across each of the genotypes (Fig. 5b). The results clearly show that the ex vivo activity of the PAH enzyme is not affected by the queuosine status of (t)RNA.
Conceivably, the absence of TGT could lead to the production of an inhibitory metabolite for the PAH reaction that would be diluted out in the standard spectrophotometric assay. It is known for example that 7-biopterin, formed by spontaneous re-arrangement of 4a-hydroxytetrahydrobiopterin during BH4 recycling, can competitively inhibit the PAH enzyme (37). To rule out this possibility, tyrosine production by cytosolic liver extracts diluted only marginally (1:20) by the addition of buffer, catalase (170 ng/μl), and (6R)-BH4 cofactor (100 μm) was performed (Fig. 5c). The results show that tyrosine production remained unchanged between wild-type and homozygous gene-trap mice over a 1-h period. It can therefore be concluded that neither the expression of PAH protein nor its ex vivo activity is affected by the queuosine status of (t)RNA, and further that queuosine deficiency does not lead to the accumulation of inhibitory metabolites in the liver.
TGT Disruption Results in Decreased Plasma BH4 and Elevated BH2 in Plasma and Urine
Given that loss of TGT has no impact on PAH, an analysis of pterins in plasma and urine was carried out following differential oxidation with iodine under acidic and basic conditions (representative figures of these results are presented in supplemental Fig. 5).
In the plasma of heterozygous and homozygous gene-trap animals, no significant changes occurred in the total biopterin levels relative to wild-type animals (Fig. 6a). However, the levels of BH4 were decreased by ∼30% (Fig. 6b) concomitant with an increase in the oxidized biopterin, dihydrobiopterin (Fig. 6c). In urine, there was an increase of ∼20% in total biopterin in gene-trap mice relative to wild-type animals (Fig. 6d). Although no significant changes were seen in BH4 levels (Fig. 6e), an increase of ∼40% in BH2 was observed (Fig. 6f). These results could be explained either by increased auto-oxidation of BH4, changes in the activity of the sepiapterin reductase and dihydropterin reductase enzymes, or alternatively that the activity of the salvage pathway for BH2, through the dihydrofolate reductase enzyme, is suboptimal.
FIGURE 6.
TGT-deficient animals have decreased levels of BH4 in plasma and elevated BH2 in plasma and urine. a, total biopterin; (b) BH4; (c) BH2 levels in the plasma; (d) total biopterin; (e) BH4; and (f) BH2 levels in the urine of wild-type (+/+), heterozygous (+/Gt), and homozygous (Gt/Gt) Qtrt1 gene-trap mice. Pterin levels were determined from concentration curves made using authentic standards. Each point represents the average value from triplicate measurements from 12 individual mice. ***, p < 0.001, analysis of variance, Dunnett's test.
TGT Inactivation Does Not Affect the Activity of Dihydrofolate Reductase in Liver
Previously, reports have shown that mice treated with methotrexate, a potent inhibitor of DHFR, experience increased levels of BH2 coupled to a decrease in endogenous BH4 in liver, kidney, and blood (15). Interestingly, studies show that exogenously administered BH4 is primarily oxidized to BH2 in the body through an ill-defined mechanism before being taken up by tissue and reduced back to BH4 (38), underlining the importance of DHFR in limiting the accumulation of oxidized biopterin.
In light of the clear accumulation of BH2 in the plasma and urine of Qtrt1 gene-trap mice, the levels of DHFR were examined. DHFR activity was measured spectrophotometrically in liver homogenate using 7,8-dihydrofolate as substrate (Fig. 7a). There was no detectable difference in activity across each of the three genotypes ruling out the possibility of a defect in the BH2 salvage pathway and instead pointing to increased production of oxidized BH4 as being a principal defect in Qtrt1 gene-trap mice (Fig. 7b).
FIGURE 7.
DHFR activity in liver is not affected by TGT status suggesting an increase in BH2 production accounts for the elevated levels seen in plasma and urine. a, activity of DHFR in liver was determined spectrophotometrically using 7,8-dihydrofolate as substrate. (n = 3 measurements from three individual animals per group.) b, model showing possible reasons for BH2 accumulation in TGT-deficient animals. Under conditions of full queuosine modification of (t)RNA, only basal levels of BH2 are produced and are cleared by DHFR. However, when (t)RNA is unmodified by queuosine, BH2 production is enhanced, despite normal DHFR activity, either through (i) increased production of BH2 by sepiapterin reductase (SR)/carbonyl reductase (CR) at the pyruvoyl tetrahydrobiopterin to BH4 reaction step, (ii) increased production and tautomerization of quinonoid dihydrobiopterin caused by a defect at the DHPR recycling step, (iii) nonenzymatic oxidation of BH4, or (iv) competition between BH2 and dihydrofolate for reduction by the DHFR enzyme.
DISCUSSION
Previously, it has been shown that animals deficient in the bacterially derived queuine molecule require dietary tyrosine for their survival (7). In this study, we demonstrate that both human HepG2 cells made deficient in queuine and transgenic animals incapable of forming queuosine-modified (t)RNA (Qtrt1 gene-trap mice) exhibit a decreased ability to produce tyrosine from phenylalanine; the former result suggests that the defect is cell autonomous and relevant to humans, and the latter result indicates that the defect arises from the absence of the queuosine modification in RNA, either tRNA or hypothetically another unknown RNA substrate of the queuine tRNA ribosyltransferase complex, as opposed to the free queuine nucleobase. The relatively mild defect seen in the catabolism of phenylalanine to tyrosine would not be expected to present as hyperphenylalanemia except under circumstances of unusually high phenylalanine intake. Therefore, that mutations to the queuine pathway may be of relevance to phenylketonuria in humans is doubtful.
Qtrt1 gene-trap animals, similar to queuine-deficient mice (28), appeared normal, displaying similar viability and fecundity to wild-type littermates. This observation is in agreement with earlier studies on various other queuine-deprived eukaryotic species, including Dictyostelium (39), fly (26), and worm (27). It may be concluded that queuine or queuosine-modified (t)RNA does not impact on the development, growth, or reproduction of eukaryotic organisms under laboratory conditions.
At variance with the lack of an overt phenotype in queuine-deficient animals, the physiological manifestation of animals co-deficient in queuine and tyrosine was dramatic and included symptoms of lethargy, labored breathing, convulsion, and death after only 18 days (7). Although TGT deficient animals had decreased ability to produce tyrosine, none of the aforementioned symptoms presented when these animals were placed on a tyrosine-free diet. A number of explanations can be envisaged. First, the germ-free status of the animals in the queuine depletion study coupled to the administration of a synthetic liquid diet may have compounded the severity of tyrosine deprivation. This situation contrasts with our study where animals were fed a chow-based chemically defined diet and would be expected to have normal gut flora. Second, and related to the above point, queuine deficiency and TGT disruption may not be equivalent. Queuine-deficient mice have no queuine or queuosine-modified (t)RNA, whereas Qtrt1 gene-trap mice are almost certainly unabated in their ability to harvest queuine from the gut and transport it into the cell. Third, it is conceivable that Qtrt1 gene-trap animals acquire sufficient tyrosine from the gut microflora as studies using 15N-labeled microbial amino acids determined that up to 20% of circulating lysine and threonine can be derived from the intestinal microbiota (1).
At the outset of the study, it was considered that queuine status may influence the activity of the PAH protein, which is required for the catabolism of phenylalanine to tyrosine. Our results show that neither the expression nor ex vivo activity of the PAH protein is affected by TGT disruption. Rather, the levels of the essential PAH cofactor BH4 is significantly decreased in plasma concomitant with an accumulation of BH2, the inactive oxidized product of BH4. It is important to stress that the Qtrt1 gene-trap mice are capable of producing significant amounts of tyrosine, but this ability is lost over time following a phenylalanine challenge, presumably because of diminished BH4 levels and accumulating BH2, a known competitive inhibitor of PAH from in vitro studies (40). That a 30% decrease in plasma BH4 levels and a 2-fold increase in BH2, as observed in our study, could impinge on tyrosine formation may be appreciated from the fact that the concentration of BH4 in mouse liver (21 pmol/mg protein) is only half that of PAH (40 pmol of PAH subunit/mg of protein) (41) and that under normal conditions the levels of BH4 are subsaturating (5–10 μm) with respect to the PAH Km value for BH4 cofactor (25 μm) (42). Thus, even small changes in cofactor supply could negatively impact the production of tyrosine from phenylalanine.
Despite the increased levels of BH2 seen in Qtrt1 gene-trap mice, the activity of the DHFR enzyme, responsible for salvaging oxidized cofactor, remained unaffected. This result suggests that DHFR is incapable of maintaining tetrahydrobiopterin cofactor in a reduced state under queuosine-deficient conditions. The ineffectiveness of the DHFR enzyme in this regard may be due to the low levels of BH2 generated in liver, given that we observe only a concentration of 1 μm in plasma, and the fact that the Km value of the DHFR enzyme for BH2, at 6.42 μm, is very much higher than the other principal DHFR substrate 7,8-dihydrofolate, which has a Km of 0.17 μm (43). Although this study has not considered how queuosine deficiency may relate to changes in folate metabolism, one could envisage based on the arguments above that an elevation in 7,8-dihydrofolate could directly compete with BH2 for reduction by DHFR, and as such, the problems seen with tetrahydrobiopterin metabolism in the current study could be secondary to changes in folate metabolism.
The impact of Qtrt1 disruption on brain function may differ significantly from that of liver, where the concentration of DHFR is low (44). Should a similar accumulation of BH2 occur, it could potentially limit the production of numerous biogenic amino neurotransmitters whose production is also under the control of BH4 through the activity of tyrosine hydroxylase and tryptophan hydroxylase (Fig. 1).
BH4 is also required for the activity of each of the nitric-oxide synthase isoforms, inducible, neural, and endothelial NOS and participates in several steps in nitric oxide generation through stabilizing the active dimeric form of the enzyme, acting as an electron donor during oxygen activation and functioning in electron recapture prior to nitric oxide release (45). Numerous studies indicate that BH4 is highly susceptible to auto-oxidation to BH2 in vivo (15, 37) and that a low BH4/BH2 ratio, as seen in Qtrt1 gene-trap mice, results in the uncoupling of endothelial NOS leading to superoxide formation and endothelial dysfunction (11, 46). Indeed, increased BH4 has proven to be protective in several experimental disease models by its ability to reduce blood pressure (47), decrease atherosclerosis (48), and prevent diabetic complications (49). It remains to be determined whether queuine has a protective role in any of these disease processes.
The underlying cause of BH4 depletion and BH2 accumulation in TGT deficient mice is uncertain at present. In vitro, oxygen and peroxynitrite can oxidize BH4 to quinonoid dihydrobiopterin, which readily rearranges to BH2 (50, 51), and in vivo it has been shown that BH2 rapidly forms in the circulation following administration of BH4 (15). The authors consider changes to intracellular homeostasis as the most probable explanation for the increased oxidation of tetrahydrobiopterin as queuine deficiency has previously been suggested to affect the activity of a number of antioxidant systems (52, 53) and to influence the metabolic state of the cell (33, 52, 54). Other explanations for the altered BH4/BH2 ratio include inadequate BH4 recycling at the dihydropterin reductase step, responsible for quinonoid BH2 reduction to BH4, or increased production of BH2 through the sepiapterin reductase/carbonyl reductase step. A further cause for elevated BH2 may relate to elevated intracellular dihydrofolate because DHFR has a Km value for dihydrofolate that is more than an order of magnitude lower than that of dihydrobiopterin, as described earlier (43).
It is reasonable to assume that the queuosine modification of tRNA may subtly affect a number of biological processes through broad changes in the protein translation profile. In this regard, it has previously been shown using molecular simulations that queuosine helps to confine the dynamic movement of the anticodon (55), and studies using two histidine isoacceptors from Drosophila showed that queuosine can limit the bias that exists among synonymous codon usage (56). It is hoped that greater physiological analysis of TGT-deficient mice will provide greater insight into the role of this intriguing bacterially derived micronutrient.
Supplementary Material
Acknowledgments
We thank Dr. Tim Mantle, Dr. Paul Voorheis, and Prof. John Scott for helpful advice and discussion.
This work was supported by Programme Grant 05-IN3-I761 from Science Foundation Ireland.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5 and Tables 1 and 2.
- PAH
- phenylalanine hydroxylase
- TGT
- tRNA guanine transglycosylase
- BH4
- tetrahydrobiopterin
- BH2
- dihydrobiopterin
- DHFR
- dihydrofolate reductase
- BH2
- 7,8-dihydrobiopterin
- HS
- horse serum
- SF
- serum-free.
REFERENCES
- 1. Hooper L. V., Midtvedt T., Gordon J. I. (2002) Annu. Rev. Nutr. 22, 283–307 [DOI] [PubMed] [Google Scholar]
- 2. Nishimura S. (1983) Prog. Nucleic Acids Res. Mol. Biol. 28, 49–73 [DOI] [PubMed] [Google Scholar]
- 3. Iwata-Reuyl D., de Crécy-Lagard V. (2009) in DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution (Grosjean H. ed) Landes Bioscience, Austin, TX [Google Scholar]
- 4. Boland C., Hayes P., Santa-Maria I., Nishimura S., Kelly V. P. (2009) J. Biol. Chem. 284, 18218–18227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen Y. C., Kelly V. P., Stachura S. V., Garcia G. A. (2010) RNA 16, 958–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Farkas W. R. (1980) J. Biol. Chem. 255, 6832–6835 [PubMed] [Google Scholar]
- 7. Marks T., Farkas W. R. (1997) Biochem. Biophys. Res. Commun. 230, 233–237 [DOI] [PubMed] [Google Scholar]
- 8. Iwata-Reuyl D. (2003) Bioorg. Chem. 31, 24–43 [DOI] [PubMed] [Google Scholar]
- 9. Lichter-Konecki U., Hipke C. M., Konecki D. S. (1999) Mol. Genet. Metab. 67, 308–316 [DOI] [PubMed] [Google Scholar]
- 10. Iino T., Tabata M., Takikawa S., Sawada H., Shintaku H., Ishikura S., Hara A. (2003) Arch. Biochem. Biophys. 416, 180–187 [DOI] [PubMed] [Google Scholar]
- 11. Crabtree M. J., Tatham A. L., Hale A. B., Alp N. J., Channon K. M. (2009) J. Biol. Chem. 284, 28128–28136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shiman R., Gray D. W. (1980) J. Biol. Chem. 255, 4793–4800 [PubMed] [Google Scholar]
- 13. Christensen R., Güttler F., Jensen T. G. (2002) Mol. Genet. Metab. 76, 313–318 [DOI] [PubMed] [Google Scholar]
- 14. Kelly V. P., Suzuki T., Nakajima O., Arai T., Tamai Y., Takahashi S., Nishimura S., Yamamoto M. (2005) Mol. Cell. Biol. 25, 3658–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sawabe K., Wakasugi K. O., Hasegawa H. (2004) J. Pharmacol. Sci. 96, 124–133 [DOI] [PubMed] [Google Scholar]
- 16. Yang W. K., Novelli G. D. (1971) Methods Enzymol. 20, 44–55 [Google Scholar]
- 17. Phillips G., Chikwana V. M., Maxwell A., El-Yacoubi B., Swairjo M. A., Iwata-Reuyl D., de Crécy-Lagard V. (2010) J. Biol. Chem. 285, 12706–12713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bailey S. W., Ayling J. E. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 15424–15429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Darlington G. J. (1987) Methods Enzymol. 151, 19–38 [DOI] [PubMed] [Google Scholar]
- 20. Nicosia A., Tafi R., Monaci P. (1992) Nucleic Acids Res. 20, 5321–5328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Katze J. R., Basile B., McCloskey J. A. (1982) Science 216, 55–56 [DOI] [PubMed] [Google Scholar]
- 22. Langgut W., Kersten H. (1990) FEBS Lett. 265, 33–36 [DOI] [PubMed] [Google Scholar]
- 23. Haggerty D. F., Young P. L., Buese J. V., Popják G. (1975) J. Biol. Chem. 250, 8428–8437 [PubMed] [Google Scholar]
- 24. Muralidhar G., Utz E. D., Elliott M. S., Katze J. R., Trewyn R. W. (1988) Anal. Biochem. 171, 346–351 [DOI] [PubMed] [Google Scholar]
- 25. Chen W. V., Delrow J., Corrin P. D., Frazier J. P., Soriano P. (2004) Nat. Genet. 36, 304–312 [DOI] [PubMed] [Google Scholar]
- 26. Siard T. J., Jacobson K. B., Farkas W. R. (1991) Biofactors 3, 41–47 [PubMed] [Google Scholar]
- 27. Gaur R., Björk G. R., Tuck S., Varshney U. (2007) J. Biosci. 32, 747–754 [DOI] [PubMed] [Google Scholar]
- 28. Reyniers J. P., Pleasants J. R., Wostmann B. S., Katze J. R., Farkas W. R. (1981) J. Biol. Chem. 256, 11591–11594 [PubMed] [Google Scholar]
- 29. Durand J. M., Dagberg B., Uhlin B. E., Björk G. R. (2000) Mol. Microbiol. 35, 924–935 [DOI] [PubMed] [Google Scholar]
- 30. Urbonavicius J., Qian Q., Durand J. M., Hagervall T. G., Björk G. R. (2001) EMBO J. 20, 4863–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kobe B., Jennings I. G., House C. M., Michell B. J., Goodwill K. E., Santarsiero B. D., Stevens R. C., Cotton R. G., Kemp B. E. (1999) Nat. Struct. Biol. 6, 442–448 [DOI] [PubMed] [Google Scholar]
- 32. Gersting S. W., Staudigl M., Truger M. S., Messing D. D., Danecka M. K., Sommerhoff C. P., Kemter K. F., Muntau A. C. (2010) J. Biol. Chem. 285, 30686–30697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mahr U., Böhm P., Kersten H. (1990) Biofactors. 2, 185–192 [PubMed] [Google Scholar]
- 34. Langgut W. (1995) Biochem. Biophys. Res. Commun. 207, 306–311 [DOI] [PubMed] [Google Scholar]
- 35. Pathak C., Jaiswal Y. K., Vinayak M. (2008) Biosci. Rep. 28, 73–81 [DOI] [PubMed] [Google Scholar]
- 36. Farkas W. R., Jacobson K. B., Katze J. R. (1984) Biochim. Biophys. Acta 781, 64–75 [DOI] [PubMed] [Google Scholar]
- 37. Pey A. L., Martinez A., Charubala R., Maitland D. J., Teigen K., Calvo A., Pfleiderer W., Wood J. M., Schallreuter K. U. (2006) FASEB J. 20, 2130–2132 [DOI] [PubMed] [Google Scholar]
- 38. Hasegawa H., Sawabe K., Nakanishi N., Wakasugi O. K. (2005) Mol. Genet. Metab. 86, S2–S10 [DOI] [PubMed] [Google Scholar]
- 39. Ott G., Kersten H., Nishimura S. (1982) FEBS Lett. 146, 311–314 [DOI] [PubMed] [Google Scholar]
- 40. Xia T., Gray D. W., Shiman R. (1994) J. Biol. Chem. 269, 24657–24665 [PubMed] [Google Scholar]
- 41. Thöny B., Ding Z., Martínez A. (2004) FEBS Lett. 577, 507–511 [DOI] [PubMed] [Google Scholar]
- 42. Kure S., Sato K., Fujii K., Aoki Y., Suzuki Y., Kato S., Matsubara Y. (2004) Mol. Genet. Metab. 83, 150–156 [DOI] [PubMed] [Google Scholar]
- 43. Webber S., Whiteley J. M. (1985) Arch. Biochem. Biophys. 236, 681–690 [DOI] [PubMed] [Google Scholar]
- 44. Kaufman S. (1991) Neurochem. Res. 16, 1031–1036 [DOI] [PubMed] [Google Scholar]
- 45. Daff S. (2010) Nitric Oxide 23, 1–11 [DOI] [PubMed] [Google Scholar]
- 46. Takeda M., Yamashita T., Shinohara M., Sasaki N., Takaya T., Nakajima K., Inoue N., Masano T., Tawa H., Satomi-Kobayashi S., Toh R., Sugiyama D., Nishimura K., Yokoyama M., Hirata K., Kawashima S. (2009) Circ. J. 73, 955–962 [DOI] [PubMed] [Google Scholar]
- 47. Du Y. H., Guan Y. Y., Alp N. J., Channon K. M., Chen A. F. (2008) Circulation 117, 1045–1054 [DOI] [PubMed] [Google Scholar]
- 48. Alp N. J., McAteer M. A., Khoo J., Choudhury R. P., Channon K. M. (2004) Arterioscler. Thromb. Vasc. Biol. 24, 445–450 [DOI] [PubMed] [Google Scholar]
- 49. Shinozaki K., Nishio Y., Okamura T., Yoshida Y., Maegawa H., Kojima H., Masada M., Toda N., Kikkawa R., Kashiwagi A. (2000) Circ. Res. 87, 566–573 [DOI] [PubMed] [Google Scholar]
- 50. Milstien S., Katusic Z. (1999) Biochem. Biophys. Res. Commun. 263, 681–684 [DOI] [PubMed] [Google Scholar]
- 51. Kirsch M., Korth H. G., Stenert V., Sustmann R., de Groot H. (2003) J. Biol. Chem. 278, 24481–24490 [DOI] [PubMed] [Google Scholar]
- 52. Szabo L., Nishimura S., Farkas W. R. (1988) Biofactors 1, 241–244 [PubMed] [Google Scholar]
- 53. Pathak C., Vinayak M. (2005) Mol. Biol. Rep. 32, 191–196 [DOI] [PubMed] [Google Scholar]
- 54. Reisser T., Langgut W., Kersten H. (1994) Eur. J. Biochem. 221, 979–986 [DOI] [PubMed] [Google Scholar]
- 55. Morris R. C., Brown K. G., Elliott M. S. (1999) J. Biomol. Struct. Dyn. 16, 757–774 [DOI] [PubMed] [Google Scholar]
- 56. Meier F., Suter B., Grosjean H., Keith G., Kubli E. (1985) EMBO J. 4, 823–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.