Abstract
Testicular cancer is highly curable with cisplatin-based therapy, and testicular cancer-derived human embryonal carcinoma (EC) cells undergo a p53-dominant transcriptional response to cisplatin. In this study, we have discovered that a poorly characterized member of the death-associated protein family of serine/threonine kinases, STK17A (also called DRAK1), is a novel p53 target gene. Cisplatin-mediated induction of STK17A in the EC cell line NT2/D1 was prevented with p53 siRNA. Furthermore, STK17A was induced with cisplatin in HCT116 and MCF10A cells but to a much lesser extent in isogenic p53-suppressed cells. A functional p53 response element that binds endogenous p53 in a cisplatin-dependent manner was identified 5 kb upstream of the first coding exon of STK17A. STK17A is not present in the mouse genome, but the closely related gene STK17B is induced with cisplatin in mouse NIH3T3 cells, although this induction is p53-independent. Interestingly, in human cells containing both STK17A and STK17B, only STK17A is induced with cisplatin. Knockdown of STK17A conferred resistance to cisplatin-induced growth suppression and apoptotic cell death in EC cells. This was associated with the up-regulation of detoxifying and antioxidant genes, including metallothioneins MT1H, MT1M, and MT1X that have previously been implicated in cisplatin resistance. In addition, knockdown of STK17A resulted in decreased cellular reactive oxygen species, whereas STK17A overexpression increased reactive oxygen species. In summary, we have identified STK17A as a novel direct target of p53 and a modulator of cisplatin toxicity and reactive oxygen species in testicular cancer cells.
Keywords: Antioxidant, General Transcription Factors, Metalloproteins, p53, Tumor Suppressor, DNA Damage, DRAK2, Testicular Cancer, Cisplatin, Reactive Oxygen Species
Introduction
The p53 tumor suppressor plays a pivotal role in preventing tumor formation and is induced in response to a variety of cellular stresses, including DNA damage leading mainly to apoptosis or cell cycle arrest (1). Other recognized actions of p53 involve regulation of genomic stability, DNA repair, senescence, autophagy, reactive oxygen generation, and metabolism (1, 2). The actions of p53 are mainly mediated by its potent transcriptional regulation of specific target genes. Because p53 is highly induced during the DNA damage response, it has a major, yet still not completely understood, role in determining whether cancer cells live or die with cytotoxic therapies. Furthermore, p53 regulation and downstream target gene activation may be cell type- and context-dependent, making it difficult to unravel the complete p53 transcriptional network (3). Identifying novel p53 target genes induced during DNA damage may reveal new mechanisms of p53 tumor suppression and provide new therapeutic opportunities.
Testicular germ cell tumor (TGCT)3 patients are cured at a rate approaching 90% with cisplatin-based combination therapy even when the disease is highly advanced (4). These responses have been linked to rapid and extensive apoptosis in patients and in xenografts and cultures of embryonal carcinoma, the putative stem cells of TGCTs (5, 6). However, the mechanisms responsible for the hypersensitivity to chemotherapy are not well understood. Because the p53 gene is rarely ever mutated in TGCT patients, it has been proposed that p53 may by uniquely latent during TGCT development and may have a specialized role in mediating the hypersensitivity of TGCTs to DNA-damaging agents (5–8). Using microarray analysis, we previously discovered that the transcriptional response to cisplatin in embryonal carcinoma cells is dominated by the induction of p53 target genes (9). In addition to many well known direct p53 target genes, there were several cisplatin-induced genes previously unassociated with p53, including the serine/threonine kinase 17A (STK17A).
STK17A is a member of the death-associated protein (DAP) family of serine/threonine protein kinases that includes the prototypic family member DAPK1 along with DAPK2 and DAPK3 (zipper-interacting protein kinase) (10–12). STK17A (DRAK1) and STK17B (DRAK2) are more distantly related and less characterized members of the DAPK family (13). DAPK1 has been linked to several cell death-related signaling pathways, including those resulting in autophagic and apoptotic cell death, and DAPK1 expression is repressed in a variety of cancers (10–12). Despite many proposed pathways and substrates, the precise mechanism(s) by which DAP family kinases induce cell death, including the key substrates that are phosphorylated, requires further elucidation.
In contrast to DAPK1, there is substantially less evidence to associate STK17A and STK17B with apoptosis or cell death. Although possessing moderate homology in the catalytic kinase domain, STK17A and STK17B both lack the death domain and the Ca2+/calmodulin regulatory domain of DAPK1 (10–13). One prior report demonstrated that overexpression of STK17A in NIH3T3 cells negatively impacted colony formation, implying that STK17A may also possess prodeath activity (13). Interestingly, STK17A is present near a chromosomal breakpoint region between human and rodents and is not present in the mouse or rat genome (14). Knock-out of STK17B in the mouse suggests that this protein negatively regulates T cell activation but appears not to directly regulate apoptosis in this setting (15, 16).
Here we demonstrate that STK17A is a novel, direct target gene of p53. STK17A was up-regulated by the DNA-damaging agent cisplatin in various cell lines in a p53-dependent manner, and p53 directly bound to an upstream element in the STK17A gene. Knockdown of STK17A in human embryonal carcinoma (EC) cells resulted in cisplatin resistance associated with increased expression of detoxifying and antioxidant genes and decreased levels of reactive oxygen species (ROS), demonstrating that STK17A plays a role in mediating cisplatin toxicity in TGCTs.
EXPERIMENTAL PROCEDURES
Cell Culture and Drug Treatments
NT2/D1, U2OS, 293T, and NIH3T3 (American Tissue Culture Collection (ATCC)) were cultured in DMEM with 10% fetal bovine serum supplemented with glutamine and antibiotics. The derivation of the NT2/D1 resistant cell line NT2/D1-R1 was described previously (17). HCT116 colon cancer cell lines p53+/+ and p53−/− were a gift from Dr. B. Vogelstein (The Johns Hopkins University) and cultured in DMEM. MCF10A immortalized breast epithelium cells and the MCF10A/Δp53 cell line stably transfected with p53 shRNA were described previously and were generously provided by Dr. A. Eastman (Dartmouth Medical School) and cultured in DMEM/F-12 supplemented with 10% FBS, 8 μg/ml insulin, 29 ng/ml epidermal growth factor, and 500 ng/ml hydrocortisone (18). ED1 cells are a mouse lung cancer cell line kindly provided by Dr. E. Dmitrovsky (Dartmouth Medical School) and were cultured in RPMI 1640 medium with 10% serum (19). The glioblastoma cell line A172 was cultured in DMEM and 5% serum and was a kind gift from Dr. M. Israel (Dartmouth Medical School). Cisplatin (Bristol Laboratories Ltd.) treatments were performed at the concentrations and time points indicated.
Real Time PCR and Immunoblot Analysis
Reverse transcription was performed on 1 μg of RNA using the TaqMan RT kit (Applied Biosystems). The cDNA (20 ng) was used with SYBR Green (Applied Biosystems) for quantitative real time PCR assays using the ΔΔCt method normalized to GAPDH and the ABI Prism Sequence Detection System 7700. Primers are provided in supplemental Table S1. For Western analysis, cells were lysed in radioimmune precipitation buffer and separated by SDS-PAGE as described previously (9). Antibodies to STK17A (IMG-157-1, Imgenex), p53 (DO-1, sc0126, Santa Cruz Biotechnology, Inc.), and actin (C-11, sc01615, Santa Cruz Biotechnology, Inc.) were used.
Chromatin Immunoprecipitation (ChIP) Assays
Briefly, 1 × 107 cells/15-cm dish were treated for 6 h with 2.0 μm cisplatin and 10 h later fixed with 1% formaldehyde for 10 min at 37 °C. Cells were lysed in ChIP lysis buffer in the presence of protease inhibitors and sonicated on ice for 11 × 15-s pulses with 20% amplitude on a Vibra Cell sonicator (Danbury, CT). Diluted samples were incubated overnight with p53 DO-1 or mouse IgG control antibodies (sc2025, Santa Cruz Biotechnology, Inc.) prebound to G protein-coupled Dynabeads (Invitrogen). Following wash steps, antibody complexes were eluted and incubated for 65 °C overnight, and DNA was then purified using the QIAquick PCR amplification kit (Qiagen). Enrichment for target sequences was performed using real time PCR normalized to 10% input. Primers are provided in supplemental Table S1.
Reporter Assays
A 350-bp fragment containing STK17Ap53RE1 and located 5.0 kb upstream of the transcriptional start site of human STK17A was generated by PCR using genomic DNA from NT2/D1 cells and primers 5′-TTGCTCCTTATCTAGGCTCCTTA and 5′-GATGGAGTCTACTCCTGTTGAGA. The product was cloned into pCR2.1-TOPO (Invitrogen) and then inserted into the BamHI and XhoI sites of TK-Luc (kindly provided by Dr. J. DiRenzo, Dartmouth Medical School) to generate STK-TK-Luc. A version of STK-TK-Luc in which p53RE1 was mutated from GGGCATGCTCAGGCAAGTCC to GGGaATaCTCAGGaAAaTCC (where lowercase letters indicate the mutations) was constructed using QuikChange site-directed mutagenesis (Stratagene). All constructs were sequence-confirmed. Cells were plated at 1.5 × 105 cells/well of a 6-well plate. FuGENE transfections were performed with 0.6 μg of reporter, 0.2 μg of Renilla TK and 0.5 μg of either p53 expression plasmid, dominant-negative p53 (DN-p53) (17), or empty vector control. Cells were harvested 24 h following transfection, and Dual-Luciferase reporter analyses were performed (Promega). Luciferase activity was normalized against Renilla activity.
RNAi Knockdown
Lentiviral silencing shRNAs targeting human STK17A (TRCN0976, STK17Ash1 and TRCN0975, STK17Ash2) were purchased from Open Biosystems along with pLKO.1 control. Lentiviral stocks were generated from 293T cells as described previously (20). NT2/D1 cells were cultured with lentiviral stocks for 24 h, and stable pools were selected with 1.0 μg/ml puromycin. Two independent siRNAs were designed that target sequences AAGACUCCAGUGGUAAUCUAC and CGGCAUGAACCGGAGGCCCAU of human p53 in NT2/D1 cells and GAAUGAGGCCUUAGAGUUAUU and UAACUCUAAGGCCUCAUUCUU of mouse p53 in NIH3T3 cells (Dharmacon). A final concentration of 120 nm siRNA was transfected using Oligofectamine reagent (Invitrogen) as described previously (21). The siRNA duplex control used was the Scramble II sequence (Dharmacon).
Cell Proliferation, Apoptosis, and ROS Assays
Cell proliferation and survival were assessed with Cell-Titer Glo (Promega). Cells were plated at 5 × 104/well of a 6-well plate and the next day treated with the indicated dosages of cisplatin for 3 days. For acute treatments, cells were treated with cisplatin for 6 h and then assayed 3 days later. For cell cycle analysis, 7 × 105 cells/10-cm dish were treated the next day with cisplatin for 6 h and assayed 24 h later. For continuous treatment, 5 × 105 cells were treated with cisplatin for 3 days. Adherent and floating cells were fixed in 70% ethanol and stained for 4 h with propidium iodide prior to analysis on a BD Biosciences FACScan flow cytometer. The percentage of cells with sub-G1 DNA was determined using CellQuest software.
For ROS detection, cells were incubated with 10 μm 2′,7′-dichlorodihydrofluorescein diacetate (Molecular Probes) for 30 min at 37 °C in Opti-MEM (Invitrogen) supplemented with 10% serum. After incubation, cells were washed with PBS, trypsinized, and resuspended in PBS; fluorescence was measured using a BD Biosciences FACScan; and data were analyzed with CellQuest software. The human STK17A cDNA was purchased from Origene (SC323411) and subcloned into pcDNA3.1. 293T cells were transfected with expression vectors using Lipofectamine 2000 (Invitrogen) and 24 h later assayed for ROS generation as described above.
Microarray Analysis
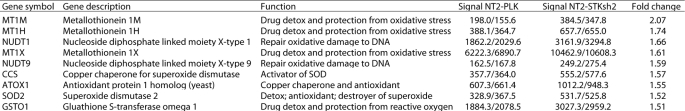
Total RNA was isolated with TRI Reagent. Hybridization was performed according to Illumina guidelines using the Illumina HumanHT-12 v4 Expression BeadChip array, which provides coverage for more than 47,000 transcripts and known splice variants across the human transcriptome. NT2-PLK and NT2-STK17Ash2 cells were hybridized in biological duplicate. Raw data were normalized using the quantile method utilizing GenomeStudio software. Comparing the average of duplicate normalized intensity values yielded 126 transcripts up-regulated 1.5-fold or greater (range, 1.5–3.7-fold) and 209 transcripts down-regulated 1.5 or greater (range, 1.5–3.4-fold). The genes in Table 1 were selected from the 126 up-regulated genes. The lists of genes can be provided upon request.
TABLE 1.
Genes induced with STK17A knockdown in NT2/D1 cells
Statistics
When a value for statistical significance is provided, a two-sample, two-tailed t test assuming unequal variance was performed.
RESULTS
STK17A Is Induced in Response to Cisplatin in a p53-dependent Manner
Testicular cancer patients can be cured with cisplatin-based chemotherapy even when the disease is highly metastatic (4). In prior studies, it was shown that acute treatment of the testicular cancer-derived human EC cell line NT2/D1 with cisplatin mediates a transcriptional response dominated by the up-regulation of known p53 target genes (9). One gene up-regulated in the screen but not previously known to be a p53 target gene was the poorly characterized DAP family kinase STK17A.
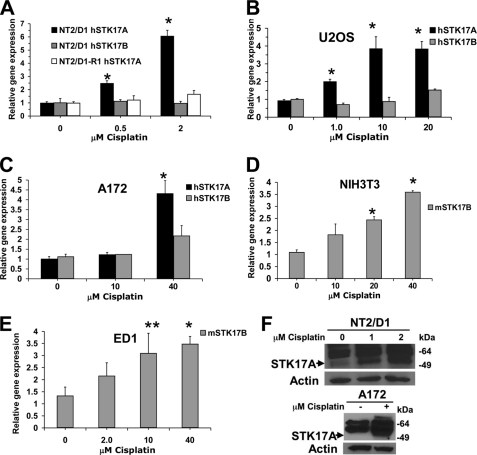
Because it has been suggested that p53 may be an important mediator of chemotherapeutic toxicity in testicular cancer and other tumor types and because DAPK family members have previously been associate with apoptosis, it was investigated whether STK17A is a direct target of p53 in EC and other tumor cells. Cisplatin induced STK17A in a dose-dependent manner in NT2/D1 cells but not in a cisplatin-resistant derivative, NT2/D1-R1, previously shown to have a defect in p53 signaling (Fig. 1A) (17). Furthermore, the STK17A homolog STK17B was not induced with cisplatin in NT2/D1 cells (Fig. 1A). Importantly, STK17A but not STK17B was also induced in human osteosarcoma U20S and human glioblastoma A172 cells (Fig. 1, B and 1C). STK17A is missing from the mouse and rat genomes likely due to a consequence of it residing at an evolutionary chromosome rearrangement (14). Interestingly, in mouse NIH3T3 and mouse ED1 cells, STK17B was significantly induced with cisplatin (Fig. 1, D and E). Importantly, cisplatin induction of STK17A protein also occurred in NT2/D1 and A172 cells (Fig. 1F). Furthermore, STK17A was also induced with the DNA-damaging agents vinblastine, doxorubicin, and etoposide (supplemental Fig. S1). These data indicate that STK17A is inducible upon genotoxic stress in multiple cell contexts and suggests that DNA damage-mediated induction of STK17B may compensate for the absence of STK17A in mouse cells.
FIGURE 1.
STK17A is induced with cisplatin in a variety of cell lines. A, STK17A is induced with cisplatin in cisplatin-sensitive human NT2/D1 cells but not in cisplatin-resistant NT2/D1-R1 cells. Real time PCR analysis was performed on RNA harvested 24 h following a 6-h cisplatin treatment. B and C, STK17A but not STK17B is induced with cisplatin in human U2OS osteosarcoma and A172 human glioblastoma cells treated with cisplatin as in A. Expression was measured by real time PCR. D and E, STK17B is induced with cisplatin in mouse NIH3T3 and mouse lung cancer ED1 cells. Real time PCR analysis was performed on RNA harvested 24 h following a 6-h cisplatin treatment. F, STK17A protein is induced with cisplatin in NT2/D1 and A172 cells as determined by Western analysis. The band representing STK17A is directly below the two nonspecific bands. All data points represent the average of biologic triplicates. Error bars are S.D. *, p < 0.01; **, p < 0.05. Data are representative of at least two independent experiments.
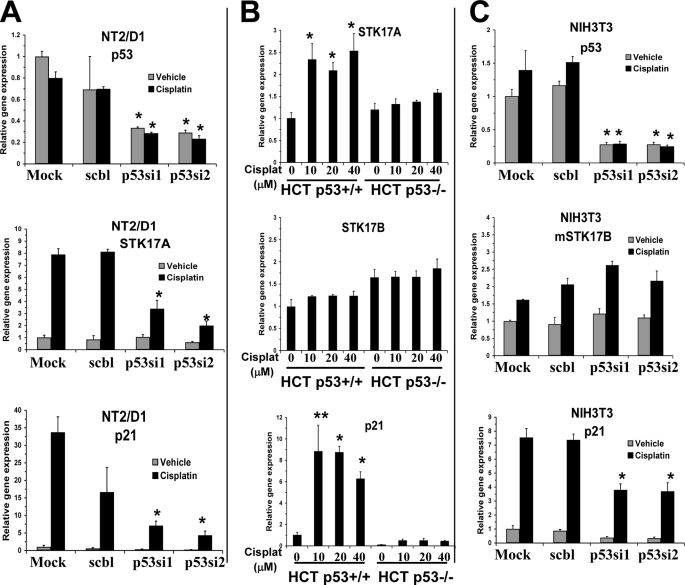
To investigate the p53 dependence of cisplatin-mediated STK17A and STK17B induction, several isogenic systems of p53 repression were used. Cisplatin induction of STK17A was substantially repressed in NT2/D1 cells treated with two independent p53 siRNAs to a degree similar to that of the known p53 target gene p21 (Fig. 2A). STK17A was induced with cisplatin in HCT116p53+/+ cells; however, induction was greatly diminished in the p53-deleted cell line HCT116p53−/−, again similar to the pattern of p21 expression (Fig. 2B). Similar results suggesting that STK17A induction is p53-dependent were obtained in MCF10A cells either expressing wild-type p53 or stably expressing shRNA to p53 (MCF10A/Δp53) (supplemental Fig. S2). In contrast to the situation with STK17A, STK17B expression was unaffected by cisplatin in both HCT116p53+/+ and HCT116p53−/− cells (Fig. 2B). Furthermore, the cisplatin induction of mouse STK17B in NIH3T3 cells was unaffected by p53 siRNA treatment that efficiently repressed p21 induction (Fig. 2C). Thus, cisplatin induction of STK17A is p53-dependent in a variety of cell contexts in contrast to the induction of mouse STK17B in NIH3T3 cells.
FIGURE 2.
STK17A is induced with cisplatin in a p53-dependent manner. A, cisplatin induction of STK17A in NT2/D1 cells is repressed with p53 siRNA knockdown. Expression analysis of p53 (top panel), STK17A (middle panel), or p21 (bottom panel) in NT2/D1 cells mock-transfected or transfected with either control siRNA (scbl) or two independent siRNAs targeting p53 and then treated with cisplatin for 6 h followed by real time PCR analysis 24 h later. *, p < 0.01 compared with identically treated control cells. B, STK17A is up-regulated following cisplatin (Cisplat) treatment of wild-type HCT116 cells (HCT116p53+/+) but to a much lesser extent in the isogenic p53-deleted line (HCT116p53−/−), whereas STK17B is not induced with cisplatin and not repressed in HCT116p53−/− cells. Cells were treated with cisplatin and harvested as in A for real time PCR analysis of STK17A (top), STK17B (middle), and p21 (bottom). *, p < 0.02; **, p < 0.05 compared with no cisplatin treatment. C, STK17B is up-regulated with cisplatin in a p53-independent manner in NIH3T3 cells. Expression of p53 (top panel), STK17B (middle panel), or p21 (bottom panel) in NIH3T3 cells mock-transfected or transfected with either control siRNA (scbl) or two independent siRNA targeting mouse p53 and then treated with cisplatin for 6 h followed by real time PCR analysis 24 h later. *, p < 0.02 compared with the identically treated mock control. All data points represent the average of biological triplicates. Error bars are S.D. Data are representative of at least two independent experiments.
STK17A Is a Direct Transcriptional Target of p53
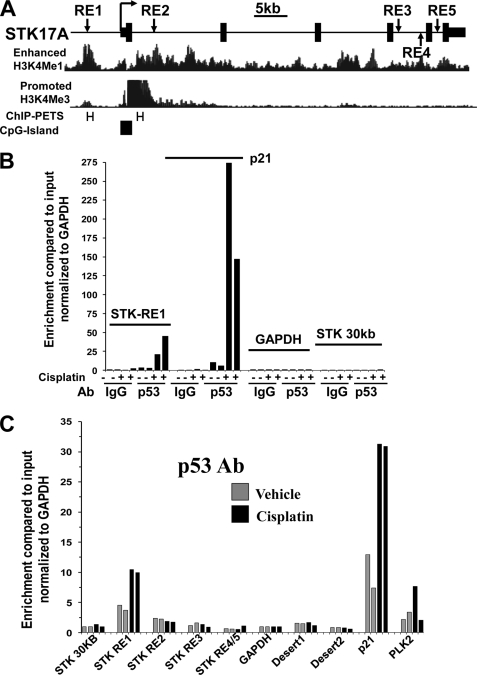
The p53MH algorithm was used to search for p53 binding sites within the human STK17A gene (22). In total, 62 kb of sequence was interrogated, including all STK17A exons and introns and 12 and 5 kb of 5′- and 3′-untranslated sequence, respectively. A 100% consensus matching p53-responsive element (STK17A-p53RE1) was found 5.0 kb upstream of the transcriptional start site of STK17A (Figs. 3A and 4B). Importantly, this site has no gap between the two p53 binding half-sites, which is the situation for the vast majority of p53 sites found with genome-wide p53 ChIP sequencing analysis (23). In addition, four weaker intronic consensus sites were found (Fig. 3A and supplemental Table S2). Interestingly STK17A-p53RE1 is within a region predicted to be an enhancer based on the presence of the H3K4Me1 histone mark (Fig. 3A). Furthermore, STK17A-p53RE1 was within a site identified by Wei et al. (23) using p53 ChIP followed by sequencing (Fig. 3A). We could not find a p53 consensus binding sequence within the only other predicted p53 ChIP-PET from that study, although this ChIP-PET is ∼2 kb from STK17A-p53RE2 (Fig. 3A).
FIGURE 3.
p53 binds in vivo to endogenous p53RE in STK17A. A, genomic organization of the human STK17A gene. Depicted are the locations of predicted p53-responsive elements (RE1–RE5) in relation to the transcriptional start site (arrow), exons (solid boxes), and introns (solid line) of STK17A. Positional enrichment of H3K4Me1 and H3K4Me3 modifications, p53 ChIP-PETs (PET regions are labeled with H), and CpG islands were downloaded from the UCSC Genome Browser assembly March 2006 (NCBI36/hg18). B, ChIP analysis in biological duplicate of NT2/D1 cells treated with 2.0 μm cisplatin for 6 h and harvested 10 h later. A p53 antibody, but not IgG, enriched in a cisplatin-dependent manner DNA fragments containing STK17A-p53RE1 and the well characterized p53 binding site of p21 but not for fragments of the GAPDH promoter or a region of the STK17A gene 30 kb upstream of the transcriptional start site. Real time PCR amplifications were performed for each precipitation with primers surrounding each site normalized to the signal from input DNA. C, an independent ChIP experiment under identical conditions as B with primer sets to STK17A-p53RE1 to STK17A-p53RE5 demonstrates that p53 only binds efficiently to STK17A-p53RE1 and that this was comparable with p53 binding to the previously characterized p53RE in the PLK2 gene. Note that the same primer set was used to detect STK17A-p53RE4 and STK17A-p53RE5 due to their close proximity. Two additional ChIP experiments were performed in biological duplicate and demonstrated greater than 20-fold-enrichment of p53 binding to STK17A-p53RE1.
FIGURE 4.
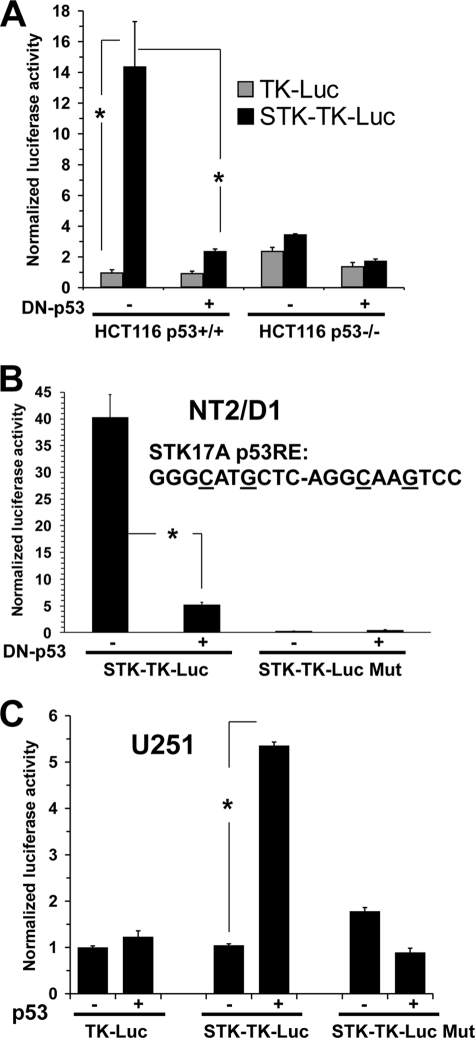
STK17A contains a functional p53-responsive element. A, reporter assay of HCT116p53+/+ and HCT116p53−/− cells transfected with either TK-Luc control reporter or STK17A reporter (STK-TK-Luc; TK-Luc containing a 350-bp fragment surrounding STK17A-p53RE1) and either a control vector or DN-p53. B, reporter activity of NT2/D1 cells transfected with STK-TK-Luc or STK-TK-Luc-Mut (STK-TK-Luc where the core C and G in each half-site is mutated) and either control vector or DN-p53. C, the U251 cell line containing mutated p53 was transfected with TK-Luc control reporter or STK-TK-Luc or STK-TK-Luc-Mut reporter and either a control vector or a p53 expression vector. All data points are the average of biologic triplicate transfections. Error bars are S.D. *, p < 0.02. Data are representative of at least two independent experiments.
ChIP analysis utilizing a p53 antibody demonstrated a 3-fold enrichment of endogenous p53 binding to STK17A-p53RE1 compared with the non-p53 target gene GAPDH. This -fold enrichment in binding increased to up to 45-fold in the presence of cisplatin (Fig. 3B). No enrichment of the STK17A-p53RE1 fragment was obtained with a control IgG antibody, and there was no binding of p53 detected in a region 30 kb upstream of the STK17A gene (Fig. 3B). The known p53 target gene p21 was used as a positive control and showed considerably more p53 binding compared with STK17A-p53RE1. However, this difference in ChIP signal is also seen with many other p53 target genes as a result of the very tight binding of p53 to the p21 enhancer (23). In a direct comparison of STK17A-p53REs, p53 bound only to STK17A-p53RE1, and this binding was similar in extent to the binding of p53 to the previously characterized p53RE of the direct p53 target gene PLK2 (Fig. 3C) (23). Two gene desert regions were used as additional negative controls.
We cloned a 350-bp region containing STK17A-p53RE1 in front of the basal thymidine kinase promoter driving luciferase expression. HCT116p53+/+ and HCT116p53−/− cells were transfected with either control reporter (TK-Luc) or the STK17A reporter construct (STK-TK-Luc). Only in HCT116p53+/+ cells did STK-TK-Luc have appreciable activity above that of the control reporter, and the activity was substantially repressed with addition of DN-p53 (Fig. 4A). STK17A-TK-Luc also had activity in p53 wild type-containing NT2/D1 cells that was again inhibited by DN-p53. Mutating four bases in the p53 binding motif of STK17A-RE1 abolished this activity (Fig. 4B). Furthermore, STK17A-Luc demonstrated activity similar to that of the TK-Luc control and STK17A-Luc-Mut reporters in human glioblastoma cells harboring mutant p53, and transfection of wild-type p53 increased activity only in cells transfected with the intact STK17-p53RE1 reporter. The reporter data, together with the above ChIP results, establish that STK17A-p53RE1 is a functional p53-responsive element in the STK17A gene, thereby strongly indicating that STK17A is a novel and direct p53 target gene.
STK17A Knockdown Results in Decreased Sensitivity to Cisplatin Associated with Decreased Apoptosis in NT2/D1 Cells
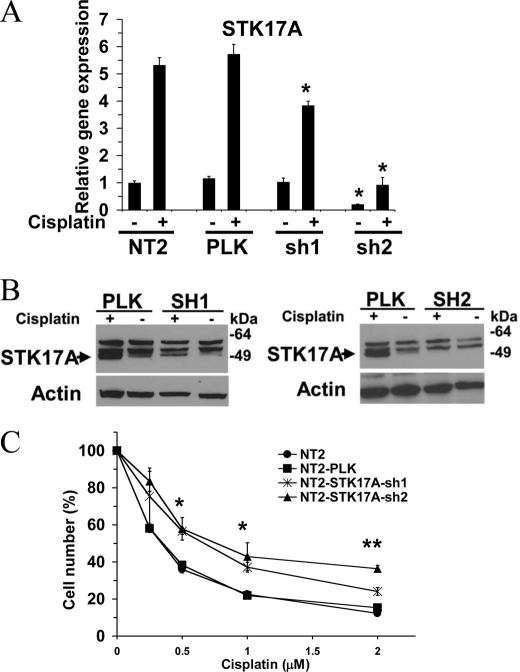
EC cells are hypersensitive to DNA-damaging agents that are associated with curability of testicular cancer (24). To assess the importance of STK17A in the response of EC cells to cisplatin, NT2/D1 cells with stable shRNA knockdown of STK17A were generated. STK17Ash2 efficiently knocked down both basal and cisplatin-induced expression of STK17A, whereas STK17Ash1 gave an intermediate effect that was more pronounced at the level of protein expression (Fig. 5, A and B). STK17A knockdown resulted in decreased sensitivity to cisplatin in NT2/D1 cells as demonstrated by increased proliferation and survival of STK17Ash2 cells compared with control cells with STK17Ash1 cells demonstrating an intermediate phenotype (Fig. 5C). The increase in cell number in STK17Ash2 cells was seen with both continuous (3-day) (Fig. 5C) and acute (6-h) cisplatin treatments (data not shown) and was reproducibly demonstrated utilizing three separately derived selections of STK17Ash lentiviral cells.
FIGURE 5.
STK17A knockdown results in decreased sensitivity to cisplatin in NT2/D1 cells. A, real time PCR analysis of STK17A expression in control NT2/D1 cells and cells transduced with two independent STK17A shRNA lentiviruses. NT2 represents mock-transduced cells, and PLK represents cells transduced with empty pLKO.1 vector. *, p < 0.01 compared with identically treated NT2/D1 control cells. B, Western analysis of STK17A expression in NT2/D1 cells stably transduced with pLKO.1 and STK17A shRNA lentiviruses. The band for STK17A is directly below the two nonspecific bands. C, dose response after 3-day cisplatin treatment of mock-transduced NT2/D1 cells or cells transfected with control or STK17A shRNA lentiviruses. Cell proliferation and survival were measured by the Cell-Titer Glo assay. *, p < 0.05; **, p < 0.01 compared with identically treated NT2/D1 control cells. Data points are the average of biological triplicates. Error bars are S.D. Data are representative of three independent experiments.
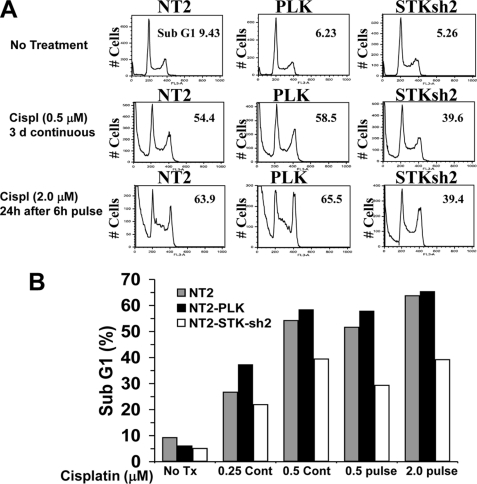
It was apparent that the increase in cell number with cisplatin in STK17Ash2 cells was in large part due to a decrease in cell death by the observation of fewer floating cells in the medium. There was a substantial decrease in apoptosis in STK17Ash2 cells after cisplatin treatment compared with controls as assessed by the level of cells with sub-G1 DNA content (Fig. 6, A and B). The decrease in apoptosis in STK17Ash2 cells was seen under both continuous (3-day) and acute (6-h) cisplatin treatments (Fig. 6, A and B). Furthermore, the G2 arrest seen with cisplatin treatment was similar in STK17Ash2 and control cells, indicating that STK17A knockdown does not greatly alter DNA damage-activated cell cycle checkpoints in NT2/D1 cells (Fig. 6B).
FIGURE 6.
STK17A knockdown results in decreased cisplatin-mediated apoptotic cell death. A, cell cycle analysis of NT2/D1 control, NT2-PLK, and NT2-STK17A-sh2 cells indicates fewer apoptotic sub-G1 cells upon cisplatin (Cispl) treatment of STK17A knockdown cells compared with control cells, whereas cell cycle phase distributions are similar. B, graph of data in A along with additional cisplatin dosages. Tx, treatment; Cont, control.
STK17A Regulates ROS
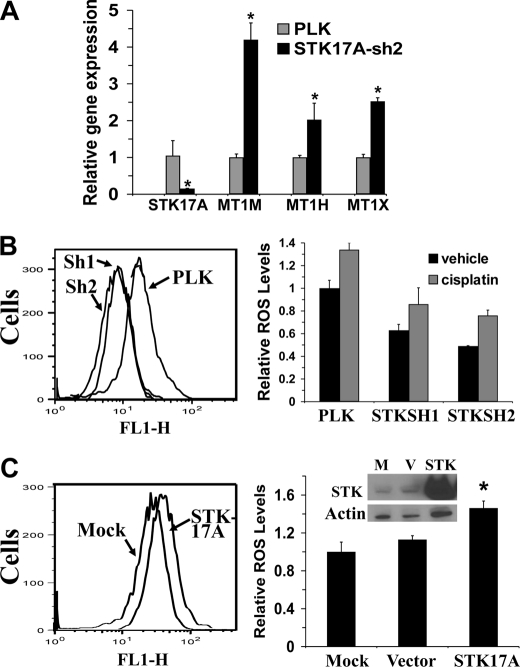
Little is known concerning the biological function of STK17A apart from its association with decreased colony formation when overexpressed in NIH3T3 cells (13). To further understand the cisplatin resistance in STK17A knockdown cells, genome-wide gene expression in NT2/D1-PLK control and NT2/D1-STK17Ash2 cells was compared. Taking the average of biological duplicate samples resulted in 126 transcripts with increased expression in NT2/D1-STK17Ash2 cells compared with control cells and 209 transcripts with lower expression. Many of the 126 transcripts up-regulated with STK17A knockdown have known roles in protective detoxification and antioxidation responses to cancer therapeutics (Table 1), including three members of the metallothionein gene family. Increased metallothionein expression is a commonly proposed mechanism of cisplatin resistance, and three metallothionein species were confirmed to be overexpressed in STK17A knockdown cells (Fig. 7A) (25). This is in contrast to levels of members of the BCL2 family of pro- and antiapoptotic mediators that were unchanged upon STK17A knockdown (supplemental Fig. S3).
FIGURE 7.
STK17A modulates ROS. A, NT2-STK17Ash2 cells express higher levels of metallothionein genes MT1M, MT1H, and MT1X compared with NT2-PLK control cells as determined by real time PCR. Data points are the average of biological triplicates. Error bars are S.D. *, p < 0.02. B, NT2/D1 cells stably expressing STK17A shRNAs (Sh1 and Sh2) have lower basal and cisplatin-induced ROS levels than control cells (PLK) as measured by the 2′,7′-dichlorodihydrofluorescein diacetate assay. Cells were incubated with or without 1 μm cisplatin for 18 h prior to ROS determination as described under “Experimental Procedures.” Tracings of representative basal ROS determinations in NT2-STK17A-sh1, NT2-STK17A-sh2, and NT2-PLK cells are shown. Bars are the average of two biological replicates, and error bars are the ranges of the two values. The experiment was repeated with similar results. C, 293T cells overexpressing STK17A have higher basal ROS levels. Cells were mock-transfected or transfected with empty vector or an expression plasmid for STK17A, and ROS levels were determined 24 h later. Representative tracings are at left. Bars represent the average of biological triplicate transfections, and error bars are S.D. *, p < 0.005 compared with mock or vector controls. The inset is a Western blot of STK17A overexpression. The experiment was repeated with similar results. M, mock; V, vector.
Furthermore, the antioxidant gene expression profile associated with STK17A knockdown correlated with a decrease in both basal and cisplatin-induced intracellular ROS levels (Fig. 7B), whereas transient overexpression of STK17A in 293T cells resulted in an increase in ROS levels (Fig. 7C). The extent of the alterations in ROS levels with STK17A modulation is comparable with similar shifts in ROS levels in other systems (26–29). These data indicate that STK17A may play a role in enhancing ROS by repressing the expression of genes involved in detoxification and antioxidation. In conclusion, this study identifies STK17A as a novel p53 target gene and provides molecular and biologic evidence that it is involved in apoptotic responses mediated by cisplatin in EC cells.
DISCUSSION
TGCTs are one of the few solid tumors cured with conventional chemotherapy in advanced stages (4). The underlying mechanisms are unclear, although one proposed mechanism is linked to the characteristically high levels of wild-type p53 in TGCTs (5–8). Here we have identified STK17A as a novel p53 target gene in TGCT-derived EC cells and show that STK17A is a modulator of the cytotoxic cisplatin response in EC cells. Evidence that STK17A is a direct p53 target gene includes demonstration of p53-dependent up-regulation of STK17A in response to cisplatin in a variety of isogenic cell models and demonstration that endogenous p53 binds to an upstream p53RE within the STK17A gene. In addition, STK17A-p53RE1 was able to drive p53-dependent transcription when fused to a heterologous reporter gene.
We previously showed through microarray analysis that the transcriptional response to cisplatin in EC cells was dominated by p53 target genes (9). In that original screen, stringent cutoffs were used to compile a list of 46 genes up-regulated with cisplatin in EC cells. Of these genes, 20 were known direct target genes of p53, and evidence was provided via gene-specific and global p53 siRNA analyses that many of the remaining genes were likely new p53 target genes (9). Remarkably, since this publication, nine additional genes of the original 46, DRAM1 (FLJ11259), GPR87, PHLDA3, GLS2, TIGAR (C12orf5), RPS27L, PAI1, CAV1, and now STK17A have been validated as novel direct p53 target genes (21, 28–37). These findings suggest a hyperactivity of p53 in EC cells that may reflect their germ cell origins. It is tempting to speculate that hyperactivity of p53 in EC and by extension the curability of TGCTs could be a by-product of safeguard mechanisms designed to protect the germ line from deleterious mutations.
Our data indicate that STK17A has a role in mediating cisplatin-induced apoptosis in NT2/D1 cells. STK17A knockdown conferred reproducible but modest (∼2-fold) decreased sensitivity to cisplatin in NT2/D1 cells. Several previous studies have shown that TGCT lines have a rather modest 2–4-fold greater cisplatin sensitivity compared with other cancer types, which has been suggested to account for the difference between cure and failure in the clinical setting of large tumor burden, suggesting that even small changes in sensitivity may have clinical relevance (for a review, see Ref. 38). Furthermore, the level of resistance seen in STK17A knockdown NT2/D1 cells is similar to what we have reported previously when p53 itself is knocked down in NT2/D1 cells (∼3-fold resistance) (9). The response to DNA-damaging agents and mechanisms of anticancer drug resistance are complex and multifactorial. It is likely that STK17A is one of several mediators of the cisplatin response in TGCT cells. We were unable to generate stable STK17A-overexpressing NT2/D1 cells, which may be related to the previous report of toxicity when STK17A was overexpressed in NIH3T3 cells (13). However, it is noteworthy that the cisplatin-resistant NT2/D1 line, NT2/D1-R1, failed to up-regulate STK17A in response to cisplatin (Fig. 1A). In addition, down-regulation of STK17A was closely associated with oxaliplatin resistance in colon cancer cells and etoposide resistance in melanoma cells (39, 40).
Little information is available concerning the function of STK17A. STK17A is only distantly related to the prototype DAP kinase DAPK1 (41–45). DAPK1 is a cytosolic actin filament-associated, calcium/calmodulin-dependent, serine/threonine kinase that promotes apoptosis in response to various stimuli, including FAS, γ interferon, and TNF-α (10). DAPK1 can repress transformation due to activation of p53, and DAPK1 expression is repressed by promoter methylation in a variety of cancers (41–45). Interestingly, a prior study has indicated that, like STK17A, DAPK1 is a p53 target gene (46). The DAP family shares homology in their catalytic domains (10). However, their extracatalytic domains and biologic properties differ markedly (10). For example, STK17A lacks the death and calcium/calmodulin regulatory domains of DAPK1 and in contrast to DAPK1 resides in the nucleus (13). Furthermore, the key immediate upstream and downstream effectors of the DAP kinase family are largely unknown. Although our data strongly indicate that inhibition of ROS production is involved in decreased sensitivity to cisplatin in STK17A knockdown cells, precisely how this occurs and whether other mechanisms play a role will require future investigation. STK17A does not regulate BCL2 family expression (see supplemental Fig. S3). Furthermore, STK17A appears not to have a role in feedback regulation of the p53 pathway, as proposed for DAPK1 (42), because knockdown and overexpression of STK17A did not appreciably alter p53 induction, p53 target gene activation, or p53-based reporter activity (data not shown).
Interestingly, STK17A is not present in the mouse or rat genomes (14). However, the closely related gene STK17B is induced with cisplatin in mouse cell lines, although it is not induced in several human lines that contain both STK17A and STK17B. These data imply an importance for either STK17A or STK17B induction upon DNA damage and suggest that STK17A and STK17B may have overlapping species- and/or context-specific roles downstream of the DNA damage response. The mechanisms that account for STK17B induction with cisplatin in mouse cells and the potential role of p53 in this induction require further study. In mice, STK17B is enriched in lymphocytes and raises the threshold for T cell activation while maintaining T cell survival but appears not to have a direct role in apoptosis in this setting (15, 16). However, other studies suggest a role for STK17B in apoptosis, including UV radiation-induced apoptosis in rat kidney and rat colon cancer cells (47, 48). Together, these and the present findings suggest a possible role for STK17B in apoptosis and cell survival during cancer therapy that warrants further investigation.
Our data demonstrate that knockdown of the nuclear kinase STK17A induces the expression of several antioxidant and detoxification genes in NT2/D1 cells, including several of the metallothioneins. This response could readily explain the decreased sensitivity to cisplatin in STK17A knockdown cells because ROS generation has been proposed as a major mechanism of cisplatin antitumor actions, and up-regulation of metallothioneins is a commonly invoked mechanism of cisplatin resistance (25, 49). p53 has been shown to have both pro-oxidant and antioxidant activity due to activation of specific downstream targets, including TIGAR, GLS2, PUMA, FDXR, and PIG3 (28–30, 50, 51). The decision as to which program is activated may be related to cell context and the extent of genotoxic stress (2). Interestingly, p53 has been shown previously to inhibit the expression of canonical antioxidant genes through inhibition of NRF2, the master transcriptional regulator that binds to antioxidant response elements to activate antioxidant genes (52). In contrast, the DAPK family has not been implicated in ROS regulation in prior studies. Our data demonstrate that knockdown or overexpression of STK17A results in decreased and increased ROS levels, respectively, and thus for the first time associates a member of the DAPK family with ROS regulation. It will be of interest in the future to determine whether the regulation of the antioxidant genes and ROS is a direct or indirect consequence of STK17A knockdown and whether these actions are shared by other DAPK family members.
In summary, we have shown for the first time that STK17A is a DNA damage-inducible, direct p53 target gene with a role in modulating cell survival, apoptosis, and ROS accumulation. The data indicate that deregulation of STK17A may have clinical implications for the efficacy of cancer therapies. It will be of interest to investigate, similarly to the case with p53, whether or not STK17A has cell context-dependent effects on cell survival and apoptosis during cancer therapy.
Supplementary Material
Acknowledgments
We thank Dr. Alan Eastman, Dr. Ethan Dmitrovsky, Dr. Mark Israel (Dartmouth Medical School), and Dr. Bert Vogelstein (The Johns Hopkins University) for cell lines and Dr. James Direnzo (Dartmouth Medical School) for the TK-luciferase reporter construct.
This work was supported, in whole or in part, by National Institutes of Health Grant R21-CA124817. This work was also supported by United States Department of Defense Investigator-Initiated Research Award PR093629 and a Prouty development award from the Norris Cotton Cancer Center.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S3.
- TGCT
- testicular germ cell tumor
- DAP
- death-associated protein
- EC
- embryonal carcinoma
- RE
- response element
- ROS
- reactive oxygen species
- STK
- serine/threonine kinase
- DAPK
- DAP kinase
- Luc
- luciferase
- TK
- thymidine kinase
- DN
- dominant-negative
- PET
- paired end tag.
REFERENCES
- 1. Zilfou J. T., Lowe S. W. (2009) Cold Spring Harb. Perspect. Biol. 1, a001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gottlieb E., Vousden K. H. (2010) Cold Spring Harb. Perspect. Biol. 2, a001040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beckerman R., Prives C. (2010) Cold Spring Harb. Perspect. Biol. 2, a000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Einhorn L. H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 4592–4595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Houldsworth J., Korkola J. E., Bosl G. J., Chaganti R. S. (2006) J. Clin. Oncol. 24, 5512–5518 [DOI] [PubMed] [Google Scholar]
- 6. Giuliano C. J., Freemantle S. J., Spinella M. J. (2006) Curr. Cancer Ther. Rev. 2, 255–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Peng H. Q., Hogg D., Malkin D., Bailey D., Gallie B. L., Bulbul M., Jewett M., Buchanan J., Goss P. E. (1993) Cancer Res. 53, 3574–3578 [PubMed] [Google Scholar]
- 8. Lutzker S. G., Levine A. J. (1996) Nat. Med. 2, 804–810 [DOI] [PubMed] [Google Scholar]
- 9. Kerley-Hamilton J. S., Pike A. M., Li N., DiRenzo J., Spinella M. J. (2005) Oncogene 24, 6090–6100 [DOI] [PubMed] [Google Scholar]
- 10. Bialik S., Kimchi A. (2006) Annu. Rev. Biochem. 75, 189–210 [DOI] [PubMed] [Google Scholar]
- 11. Lin Y., Hupp T. R., Stevens C. (2010) FEBS J. 277, 48–57 [DOI] [PubMed] [Google Scholar]
- 12. Michie A. M., McCaig A. M., Nakagawa R., Vukovic M. (2010) FASEB J. 277, 74–80 [DOI] [PubMed] [Google Scholar]
- 13. Sanjo H., Kawai T., Akira S. (1998) J. Biol. Chem. 273, 29066–29071 [DOI] [PubMed] [Google Scholar]
- 14. Fitzgerald J., Bateman J. F. (2004) Trends Genet. 20, 408–412 [DOI] [PubMed] [Google Scholar]
- 15. McGargill M. A., Wen B. G., Walsh C. M., Hedrick S. M. (2004) Immunity 21, 781–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gatzka M., Newton R. H., Walsh C. M. (2009) J. Immunol. 183, 285–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Curtin J. C., Dragnev K. H., Sekula D., Christie A. J., Dmitrovsky E., Spinella M. J. (2001) Oncogene 20, 2559–2569 [DOI] [PubMed] [Google Scholar]
- 18. Levesque A. A., Kohn E. A., Bresnick E., Eastman A. (2005) Oncogene 24, 3786–3796 [DOI] [PubMed] [Google Scholar]
- 19. Liu X., Sempere L. F., Galimberti F., Freemantle S. J., Black C., Dragnev K. H., Ma Y., Fiering S., Memoli V., Li H., DiRenzo J., Korc M., Cole C. N., Bak M., Kauppinen S., Dmitrovsky E. (2009) Clin. Cancer Res. 15, 1177–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Beyrouthy M. J., Garner K. M., Hever M. P., Freemantle S. J., Eastman A., Dmitrovsky E., Spinella M. J. (2009) Cancer Res. 69, 9360–9366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kerley-Hamilton J. S., Pike A. M., Hutchinson J. A., Freemantle S. J., Spinella M. J. (2007) Biochim. Biophys. Acta 1769, 209–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hoh J., Jin S., Parrado T., Edington J., Levine A. J., Ott J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 8467–8472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wei C. L., Wu Q., Vega V. B., Chiu K. P., Ng P., Zhang T., Shahab A., Yong H. C., Fu Y., Weng Z., Liu J., Zhao X. D., Chew J. L., Lee Y. L., Kuznetsov V. A., Sung W. K., Miller L. D., Lim B., Liu E. T., Yu Q., Ng H. H., Ruan Y. (2006) Cell 124, 207–219 [DOI] [PubMed] [Google Scholar]
- 24. Spierings D. C., de Vries E. G., Vellenga E., de Jong S. (2003) J. Pathol. 200, 137–148 [DOI] [PubMed] [Google Scholar]
- 25. Knipp M. (2009) Curr. Med. Chem. 16, 522–537 [DOI] [PubMed] [Google Scholar]
- 26. Chang T. S., Cho C. S., Park S., Yu S., Kang S. W., Rhee S. G. (2004) J. Biol. Chem. 279, 41975–41984 [DOI] [PubMed] [Google Scholar]
- 27. Boswell S. A., Ongusaha P. P., Nghiem P., Lee S. W. (2007) J. Biol. Chem. 282, 4850–4858 [DOI] [PubMed] [Google Scholar]
- 28. Hu W., Zhang C., Wu R., Sun Y., Levine A., Feng Z. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 7455–7460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Suzuki S., Tanaka T., Poyurovsky M. V., Nagano H., Mayama T., Ohkubo S., Lokshin M., Hosokawa H., Nakayama T., Suzuki Y., Sugano S., Sato E., Nagao T., Yokote K., Tatsuno I., Prives C. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 7461–7466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bensaad K., Tsuruta A., Selak M. A., Vidal M. N., Nakano K., Bartrons R., Gottlieb E., Vousden K. H. (2006) Cell 126, 107–120 [DOI] [PubMed] [Google Scholar]
- 31. Crighton D., Wilkinson S., O'Prey J., Syed N., Smith P., Harrison P. R., Gasco M., Garrone O., Crook T., Ryan K. M. (2006) Cell 126, 121–134 [DOI] [PubMed] [Google Scholar]
- 32. Zhang Y., Qian Y., Lu W., Chen X. (2009) Cancer Res. 69, 6049–6056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kawase T., Ohki R., Shibata T., Tsutsumi S., Kamimura N., Inazawa J., Ohta T., Ichikawa H., Aburatani H., Tashiro F., Taya Y. (2009) Cell 136, 535–550 [DOI] [PubMed] [Google Scholar]
- 34. Li J., Tan J., Zhuang L., Banerjee B., Yang X., Chau J. F., Lee P. L., Hande M. P., Li B., Yu Q. (2007) Cancer Res. 67, 11317–11326 [DOI] [PubMed] [Google Scholar]
- 35. He H., Sun Y. (2007) Oncogene 26, 2707–2716 [DOI] [PubMed] [Google Scholar]
- 36. Kortlever R. M., Higgins P. J., Bernards R. (2006) Nat. Cell Biol. 8, 877–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yu X., Riley T., Levine A. J. (2009) FEBS J. 276, 2201–2212 [DOI] [PubMed] [Google Scholar]
- 38. Masters J. R., Köberle B. (2003) Nat. Rev. Cancer 3, 517–525 [DOI] [PubMed] [Google Scholar]
- 39. Tang H., Liu Y. J., Liu M., Li X. (2007) Anticancer Drugs 18, 633–639 [DOI] [PubMed] [Google Scholar]
- 40. Wittig R., Nessling M., Will R. D., Mollenhauer J., Salowsky R., Münstermann E., Schick M., Helmbach H., Gschwendt B., Korn B., Kioschis P., Lichter P., Schadendorf D., Poustka A. (2002) Cancer Res. 62, 6698–6705 [PubMed] [Google Scholar]
- 41. Inbal B., Cohen O., Polak-Charcon S., Kopolovic J., Vadai E., Eisenbach L., Kimchi A. (1997) Nature 390, 180–184 [DOI] [PubMed] [Google Scholar]
- 42. Raveh T., Droguett G., Horwitz M. S., DePinho R. A., Kimchi A. (2001) Nat. Cell Biol. 3, 1–7 [DOI] [PubMed] [Google Scholar]
- 43. Gozuacik D., Bialik S., Raveh T., Mitou G., Shohat G., Sabanay H., Mizushima N., Yoshimori T., Kimchi A. (2008) Cell Death Differ. 15, 1875–1886 [DOI] [PubMed] [Google Scholar]
- 44. Harrison B., Kraus M., Burch L., Stevens C., Craig A., Gordon-Weeks P., Hupp T. R. (2008) J. Biol. Chem. 283, 9999–10014 [DOI] [PubMed] [Google Scholar]
- 45. Gozuacik D., Kimchi A. (2006) Autophagy 2, 74–79 [DOI] [PubMed] [Google Scholar]
- 46. Martoriati A., Doumont G., Alcalay M., Bellefroid E., Pelicci P. G., Marine J. C. (2005) Oncogene 24, 1461–1466 [DOI] [PubMed] [Google Scholar]
- 47. Kuwahara H., Nakamura N., Kanazawa H. (2006) Biol. Pharm. Bull. 29, 225–233 [DOI] [PubMed] [Google Scholar]
- 48. Mao J., Qiao X., Luo H., Wu J. (2006) J. Biol. Chem. 281, 12587–12595 [DOI] [PubMed] [Google Scholar]
- 49. Brozovic A., Ambrioviæ-Ristov A., Osmak M. (2010) Crit. Rev. Toxicol. 40, 347–359 [DOI] [PubMed] [Google Scholar]
- 50. Polyak K., Xia Y., Zweier J. L., Kinzler K. W., Vogelstein B. (1997) Nature 389, 300–305 [DOI] [PubMed] [Google Scholar]
- 51. Hwang P. M., Bunz F., Yu J., Rago C., Chan T. A., Murphy M. P., Kelso G. F., Smith R. A., Kinzler K. W., Vogelstein B. (2001) Nat. Med. 7, 1111–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Faraonio R., Vergara P., Di Marzo D., Pierantoni M. G., Napolitano M., Russo T., Cimino F. (2006) J. Biol. Chem. 281, 39776–39784 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.