Abstract
Menaquinone (vitamin K2) serves as an electron carrier in the electron transport chain required for respiration in many pathogenic bacteria. Most bacteria utilize a common menaquinone biosynthetic pathway as exemplified by Escherichia coli. Recently, a novel biosynthetic pathway, the futalosine pathway, was discovered in Streptomyces. Bioinformatic analysis strongly suggests that this pathway is also operative in the human pathogens Campylobacter jejuni and Helicobacter pylori. Here, we provide compelling evidence that a modified futalosine pathway is operative in C. jejuni and that it utilizes 6-amino-6-deoxyfutalosine instead of futalosine. A key step in the Streptomyces pathway involves a nucleosidase called futalosine hydrolase. The closest homolog in C. jejuni has been annotated as a 5′-methylthioadenosine nucleosidase (MTAN). We have shown that this C. jejuni enzyme has MTAN activity but negligible futalosine hydrolase activity. However, the C. jejuni MTAN is able to hydrolyze 6-amino-6-deoxyfutalosine at a rate comparable with that of its known substrates. This suggests that the adenine-containing version of futalosine is the true biosynthetic intermediate in this organism. To demonstrate this in vivo, we constructed a C. jejuni mutant strain deleted for mqnA2, which is predicted to encode for the enzyme required to synthesize 6-amino-6-deoxyfutalosine. Growth of this mutant was readily rescued by the addition of 6-amino-6-deoxyfutalosine, but not futalosine. This provides the first direct evidence that a modified futalosine pathway is operative in C. jejuni. It also highlights the tremendous versatility of the C. jejuni MTAN, which plays key roles in S-adenosylmethionine recycling, the biosynthesis of autoinducer molecules, and the biosynthesis of menaquinone.
Keywords: Bacterial Genetics, Enzymes, Quinones, Respiration, Vitamin K, Campylobacter jejuni, Biosynthesis, Futalosine, Menaquinone, Nucleosidase
Introduction
Menaquinone (vitamin K2) is a lipid-soluble prenylated 2-methyl-1,4-naphthoquinone that plays a variety of roles in both eukaryotes and prokaryotes (see Fig. 1). In humans, it serves as a cofactor that is required for the post-translational generation of γ-carboxyglutamate residues in proteins involved in blood coagulation, bone metabolism, and vascular physiology (1). In bacteria, it plays a key role as an electron carrier in the electron transport chain required for respiration (2). Although most Gram-negative bacteria such as Escherichia coli use menaquinone under anaerobic conditions and ubiquinone under aerobic conditions, Gram-positive bacteria and many other Gram-negative bacteria rely on menaquinone as their sole electron carrier (3, 4). These include many pathogenic organisms such as Helicobacter pylori, Campylobacter jejuni, Staphylococcus aureus, and Mycobacterium tuberculosis (5–8). In these organisms, menaquinone is required for survival. Because humans are unable to synthesize menaquinone, the bacterial enzymes responsible for the biosynthesis of this vitamin serve as viable targets for the development of antibacterial compounds (2, 9).
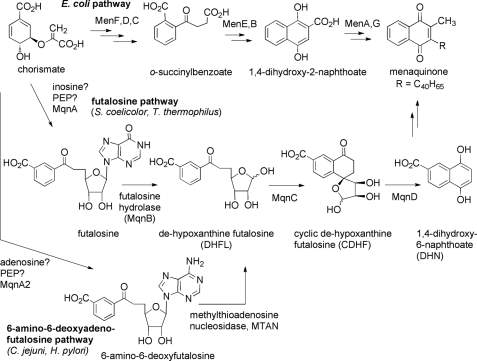
FIGURE 1.
Pathways for menaquinone biosynthesis in bacteria. The upper pathway shows the biosynthesis of menaquinone in E. coli. The middle pathway shows the futalosine pathway employed by S. coelicolor and T. thermophilus. The lower pathway shows the modified futalosine pathway employed by C. jejuni and H. pylori. PEP, phosphoenolpyruvate.
The biosynthesis of menaquinone in E. coli has been extensively studied (10, 11). It begins with the compound chorismate, which is an intermediate in the shikimate pathway for the biosynthesis of aromatic compounds (see Fig. 1). Five enzymes, MenB–MenF, generate the 1,4-dihydroxy-2-naphthoate core, and then MenA and MenG install the prenyl and methyl substituents to give menaquinone. In 2005, it was reported that various Streptomyces species lack orthologs of the menB--menF genes (12), suggesting that an entirely unique biosynthetic pathway is operative in these organisms. Interestingly, these genes are also absent in the pathogenic bacteria C. jejuni and H. pylori, even though these organisms are known to biosynthesize menaquinone (9). Isotopic feeding studies confirmed that a unique pathway is employed in Streptomyces, and 1,4-dihydroxy-6-naphthoate (DHN) was implicated as an intermediate (see Fig. 1) (13). This compound was demonstrated to recover the growth of a menaquinone auxotroph of Streptomyces coelicolor, providing strong evidence for this hypothesis. In 2008, Dairi and co-workers (14) reported the discovery of the alternate menaquinone biosynthetic pathway, or the futalosine pathway, and identified several of the genes in either S. coelicolor or Thermus thermophilus. They found that MqnA is involved in the early steps of futalosine biosynthesis, presumably utilizing chorismate, phosphoenolpyruvate, and inosine as precursors (Fig. 1). The enzyme futalosine hydrolase (MqnB or futalosine nucleosidase) then cleaves the hypoxanthine ring from futalosine to generate dehypoxanthinylfutalosine (DHFL).4 Finally, the enzymes MqnC and MqnD convert DHFL into 1,4-dihydroxy-6-naphthoate. Although the pathway was largely elucidated by generating S. coelicolor knock-out strains and isolating the intermediates that accumulated, in the case of futalosine hydrolase and MqnD, in vitro enzyme activity was demonstrated using recombinant T. thermophilus enzymes (15, 16).
Bioinformatic analysis strongly implied that the futalosine pathway is also operative in the pathogenic organisms C. jejuni and H. pylori (9, 14). These bacteria lack homologs to the men genes of E. coli and possess homologs to the mqn genes of S. coelicolor. C. jejuni is the leading cause of bacterial gastroenteritis in the developed world and has been implicated as a causative agent of the debilitating paralysis associated with Guillain-Barré syndrome (17). H. pylori causes gastritis that can lead to peptic ulceration and gastric cancer (18). Because these bacteria require menaquinone biosynthesis for survival and because they use a biosynthetic pathway that differs from that employed by other beneficial intestinal microbiota such as lactobacilli, these enzymes represent attractive targets for the development of specific antibacterial compounds that may exhibit minimal adverse side effects (2, 9).
In this study, we describe our efforts in establishing that a modified futalosine pathway is operative in C. jejuni and in identifying the hydrolase/nucleosidase that is used by this organism. We have found that, unlike S. coelicolor, C. jejuni utilizes the adenine-containing version of futalosine, or 6-amino-6-deoxyfutalosine, as an intermediate in menaquinone biosynthesis. Furthermore, the enzyme responsible for the hydrolysis of the N-glycosidic bond in this organism is the 5′-methylthioadenosine nucleosidase (MTAN), which also plays roles in recycling by-products of S-adenosylmethionine-utilizing enzymes and in the biosynthesis of autoinducer molecules. Finally, we show that a C. jejuni deletion strain lacking an mqnA homolog (herein designated mqnA2) is auxotrophic for growth on 6-amino-6-deoxyfutalosine. This strongly supports the notion that the futalosine pathway for menaquinone biosynthesis is operative in C. jejuni and that the adenine-containing intermediate is utilized instead of the hypoxanthine-containing intermediate.
EXPERIMENTAL PROCEDURES
Materials and General Methods
5′-Methylthioadenosine (MTA) and xanthine oxidase (Grade III, from bovine milk) were purchased from Sigma. Protein concentration was determined by the Bradford method (19) using bovine serum albumin as the standard. 1H NMR spectra were acquired on a Bruker AV300 NMR spectrometer. Details regarding the synthetic procedures used to make 6-amino-6-deoxyfutalosine and the corresponding 1H NMR spectra are provided under supplemental “Materials and General Methods.”
Cloning of a Putative MTAN (cj0117)
The cj0117 gene was amplified from C. jejuni (strain NCTC 11168) genomic DNA by PCR. The oligonucleotide primers, including overhangs for ligation-independent cloning, were 5′-GGTATTGAGGGTCGCATGATGAAAATAGCAAT-3′ (sense) and 5′-AGAGGAGAGTTAGAGCCTCATAATTTCTCGCACAT-3′ (antisense). The PCR product was cloned into a pET-30Xa/LIC vector (Novagen) according to the manufacturer's instructions. The resulting recombinant plasmid, which encodes an N-terminal His6 tag on the target C. jejuni MTAN protein, was amplified in NovaBlue GigaSingles competent E. coli cells (Novagen).
Overexpression and Purification of C. jejuni MTAN
The recombinant plasmid was transformed into competent E. coli BL21(DE3) cells (Novagen), and these cells were grown on LB agar plates containing 30 mg/liter kanamycin at 37 °C. An individual colony was used to inoculate 10 ml of LB medium containing 30 mg/liter kanamycin, and this culture was incubated at 37 °C with shaking at 225 rpm for 12 h. The overnight culture was poured into 500 ml of LB medium containing 30 mg/liter kanamycin and incubated at 37 °C with shaking at 225 rpm until an A600 of 0.6–0.8 was reached. The culture was allowed to continue growing for 4 h after 70 mg/liter isopropyl β-d-thiogalactopyranoside was added to induce MTAN overexpression. Cells were harvested by centrifugation at 6000 rpm for 30 min, and the pellets were snap-frozen in liquid nitrogen and stored at −80 °C.
To purify MTAN, a frozen cell pellet was thawed and resuspended in 10 ml of potassium phosphate buffer (20 mm, pH 7.0). The cells were lysed twice at 20,000 p.s.i. using a French pressure cell. The cell lysate was subsequently centrifuged at 6500 rpm for 30 min and passed through a 0.22-μm filter before affinity chromatography. A column containing 10 ml of chelating Sepharose Fast Flow resin (GE Healthcare) was charged with 20 ml of 100 mm NiSO4 and then washed with 20 ml of distilled H2O and 30 ml of sodium phosphate buffer (20 mm, pH 7.0) containing 0.5 m NaCl and 5 mm imidazole. The filtered lysate was loaded onto the column and eluted with the same buffer containing increasing amounts of imidazole in a stepwise fashion (5, 125, and 500 mm). Fractions that eluted with 500 mm imidazole and showed absorbance at 280 nm were collected. These fractions were concentrated and buffer-exchanged into an appropriate reaction buffer using an Amicon Ultra-4 membrane filtration device (Mr 10,000 cutoff, Millipore) at 5000 rpm. Enzyme samples were stored at 5 °C and used within 24 h of purification.
Monitoring C. jejuni MTAN Activity with Futalosine, 5′-Methylthioadenosine, and 6-Amino-6-deoxyfutalosine Using 1H NMR Spectroscopy
Samples of freshly purified C. jejuni MTAN (250 μg each) that had been subjected to buffer exchange into 50 mm NaH2PO4/D2O buffer (pD 7.0, 200 μl each) were added to solutions of MTA, futalosine, and 6-amino-6-deoxyfutalosine (3.4 mm each) in 800 μl of D2O (1-ml final volume). These samples were incubated at room temperature, and 1H NMR spectra were acquired at timed intervals to monitor the progress of the enzymatic reaction. Spectra taken of control reactions lacking enzyme showed no change over the course of several days at room temperature.
Continuous Coupled Assay for C. jejuni MTAN Activity
The activity of C. jejuni MTAN was quantified by measuring the amount of adenine or hypoxanthine produced during the enzymatic reactions using a coupled spectrophotometric assay that employs xanthine oxidase (20–22). All kinetic assays were performed in 50 mm potassium phosphate buffer (final volume of 1.0 ml, pH 7.0) containing 0.28 units of xanthine oxidase and a variable concentration of MTA (0.5–5 μm), 6-amino-6-deoxyfutalosine (0.5–5 and 150 μm), or futalosine (150 μm). Assay mixtures were incubated at 25 °C for 10 min before enzymatic reactions were initiated by the addition of a fixed amount of C. jejuni MTAN (50 ng for MTA and 200 ng for 6-amino-6-deoxyfutalosine or futalosine). Rates were measured by monitoring the increased absorbance at 305 nm (MTA and 6-amino-6-deoxyfutalosine) or at 290 nm (futalosine). Changes in absorbance were converted to changes in concentration using molar absorption coefficients of 15,400 m−1 cm−1 for the adenine assay and 12,200 m−1 cm−1 for the hypoxanthine assay. Kinetic parameters were determined by fitting initial velocities to the Michaelis-Menten equation using GraFit 7.0.
Construction and Testing of a C. jejuni ΔmqnA2 Deletion Strain
cjj81176_1302, encoding a mqnA homolog we have designated mqnA2, was PCR-amplified from strain 81-176 genomic DNA using primers mqnA2KO-1 (5′-TCA TTG TAT CAA TCA TCC ATT GAT CG-3′) and mqnA2KO-2 (5′-TTG GCT CAG TTG TAG CAG ATG AAC-3′) and cloning the PCR product into the commercial vector pGEM-T (Promega). Inverse PCR was performed on the generated plasmid construct using primers mqnA2KO-3 (5′-AAA GAA TTC TTA AGA TAT ATA TGT AAA GG-3′) and mqnA2KO-4 (5′-AAA GGA TCC TTA AAG CGT TTT GTA AAG-3′). The resulting amplicon and plasmid pUC18K-2, carrying a non-polar kanamycin resistance cassette (aphA-3), were each digested with BamHI and EcoRI restriction enzymes. (Restriction site locations are underlined in the corresponding primers.) aphA-3 was ligated to the PCR amplicon to form plasmid pGEM-mqnA2::aphA-3. The E. coli-derived plasmid was delivered to strain 81-176 by natural transformation. Colonies were isolated on Mueller-Hinton agar (Oxoid, Hampshire, England) containing 5 μg/ml trimethoprim and 10 μg/ml vancomycin (MH-TV) and supplemented with 50 μg/ml kanamycin and 100 μg/ml 6-amino-6-deoxyfutalosine. C. jejuni isolates harboring mqnA2 disrupted by the kanamycin resistance cassette were confirmed via PCR and sequencing analysis. Growth analyses were performed on plates with MH-TV containing 50 μg/ml kanamycin or supplemented with 100 μg/ml 6-amino-6-deoxyfutalosine, 100 μg/ml futalosine, or 100 μg/ml menaquinone-4 (Sigma).
RESULTS
Studies with Futalosine, 5′-Methylthioadenosine, and C. jejuni MTAN (Cj0117)
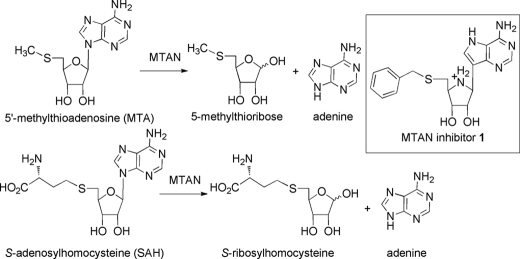
Our first approach toward identifying the futalosine pathway in C. jejuni involved cloning the putative futalosine hydrolase and determining whether it shows activity for synthetic futalosine. BLAST searches against the established futalosine hydrolases from S. coelicolor (KEGG entry SCO4327) and T. thermophilus (entry TTHA0556) showed only one reasonable candidate: a C. jejuni protein that was encoded by the cj0117 gene in C. jejuni strain 11168 (31 and 26% amino acid sequence identities, respectively). This protein had been annotated as a 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase (MTAN). MTAN is a nucleosidase that hydrolyzes the N-glycosidic bond in either MTA or S-adenosylhomocysteine and releases free adenine and 5′-methylthioribose or S-ribosylhomocysteine, respectively (Fig. 2) (23–25). MTA is a by-product in the S-adenosylmethionine-dependent biosynthesis of polyamines such as spermidine, and MTAN serves to recycle it back into basic metabolic pathways. S-Adenosylhomocysteine is a by-product of S-adenosylmethionine-dependent methyltransferases and is converted into S-ribosylhomocysteine by MTAN. S-Ribosylhomocysteine then serves as a precursor to the quorum-sensing molecule autoinducer-2 (26). Because the cj0117 gene product shares sequence homology with this known nucleosidase, it seemed reasonable that it may function as a futalosine hydrolase, an MTAN, or both. The latter possibility seemed reasonable because MTAN is known to show substrate promiscuity and accept either MTA or S-adenosylhomocysteine, and the carboxylate of futalosine could occupy a position in the active site of MTAN that is analogous to that occupied by the carboxylate of S-adenosylhomocysteine. Furthermore, very potent inhibitors of MTAN are known that contain a phenyl ring positioned in a very similar manner to that of futalosine (e.g. inhibitor 1), indicating that the active site can accommodate a substrate with this functionality (20).
FIGURE 2.
Reactions catalyzed by MTAN. The inset shows the structure of the potent MTAN inhibitor 1.
We therefore cloned cj0117, expressed its gene product as a His-tagged protein, and purified the protein via ion affinity chromatography. We prepared futalosine using our previously described synthetic route from inosine (27). Using 1H NMR spectral analysis, we found that Cj0117 was active in catalyzing the hydrolysis of MTA but showed negligible activity (<5%) with futalosine under identical conditions. Thus, it appears that Cj0117 is a functional MTAN, but not a futalosine hydrolase.
Studies with 6-Amino-6-deoxyfutalosine and C. jejuni MTAN
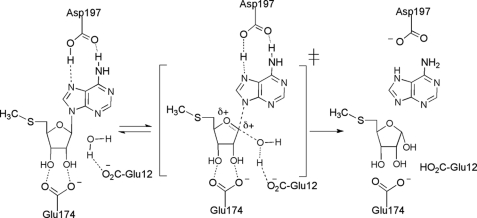
The absence of an alternate candidate to serve as a futalosine hydrolase in C. jejuni led us to suspect that futalosine is not an actual intermediate in menaquinone biosynthesis in this organism. Despite this uncertainty, it still appeared that the latter steps of the futalosine pathway are operative in C. jejuni, as homologs to MqnC and MqnD are present in the C. jejuni proteome. Notably, the putative MqnC (encoded by cj0462 in C. jejuni strain 11168) shares 44 and 41% amino acid sequence identities with the S. coelicolor and T. thermophilus enzymes, respectively. Therefore, we began to suspect that only the first steps in the futalosine pathway in C. jejuni involve an alternate intermediate. A clue to the nature of the true intermediate was found from previous mechanistic and structural studies on the E. coli MTAN. A kinetic isotope effect analysis showed that the enzyme utilizes a highly dissociative mechanism with a transition state that has considerable oxocarbenium ion character (Fig. 3) (28). Mutagenesis and structural studies have shown that there are three conserved residues that are absolutely essential for the activity of this enzyme: Glu-12, Glu-174, and Asp-197 (29–32). Glu-12 is thought to serve as a general base and to deprotonate the water that ultimately attacks the oxocarbenium ion. Glu-174 forms key H-bonds with the ribose C-2′ and C-3′ hydroxyl groups and likely plays important roles in controlling electron density in the ribose ring and substrate positioning. Finally, Asp-197 forms two H-bonds with the departing adenine and likely plays the role of a general acid catalyst. To analyze the potential role of these residues in futalosine hydrolysis, we performed a sequence alignment between the E. coli and C. jejuni MTANs and the S. coelicolor and T. thermophilus futalosine hydrolases (Fig. 4) (33). All three of the conserved E. coli MTAN residues aligned with their counterparts in the C. jejuni MTAN, consistent with its observed catalytic activity on MTA. In the case of the futalosine hydrolases, the residues corresponding to Glu-174 were present as anticipated, and it was not clear which residues played the role of Glu-12 as the N-terminal sequences differed significantly. Most interestingly, however, was the observation that an asparagine residue was present in place of Asp-197 in both of the futalosine hydrolases. This is significant because this residue interacts with the purine leaving group, and a key difference between futalosine and MTA is that the former possesses a hypoxanthine base and the latter possesses an adenine base. It is even more compelling when one considers that hypoxanthine exists primarily as the keto tautomer, whereas adenine exists as the enamine tautomer (34). This affects the H-bonding properties of the exocyclic heteroatom in the purine of futalosine so that it can function only as an H-bond acceptor. Thus, the interaction with an aspartate residue could not form the same bifunctional acceptor/donor H-bonding pattern described for MTAN catalysis. It therefore seems likely that MTAN-related nucleosidases that act on adenine-containing substrates retain the key aspartate; however, those that act on hypoxanthine-containing substrates replace it with an asparagine. An analysis of all MTAN homologs present in all sequenced genomes of Helicobacter or Campylobacter species indicated that the residue corresponding to Asp-197 is always conserved. This suggests that a modified futalosine pathway is operative in C. jejuni and H. pylori in which 6-amino-6-deoxyfutalosine is used in place of futalosine.
FIGURE 3.
Proposed mechanism for the reaction catalyzed by the E. coli MTAN and the roles of key active site residues.
FIGURE 4.
Alignment of the E. coli MTAN (EcMTAN), the C. jejuni MTAN (CjMTAN), the T. thermophilus futalosine hydrolase (TtMqnB), and the S. coelicolor futalosine hydrolase (ScMqnB). Accession numbers are P0AF14, Q0PC20, Q5SKT7, and Q9KXN0, respectively. Numbered residues correspond to key active site residues identified in the E. coli MTAN. Boldface residues with a gray background correspond to key active site residues identified in the E. coli MTAN and their corresponding conserved homologs. White residues with a black background indicate the replacement of the E. coli MTAN Glu-197 with Gln. Asterisks identify residues conserved in all four sequences. The alignment was performed using COBALT (33).
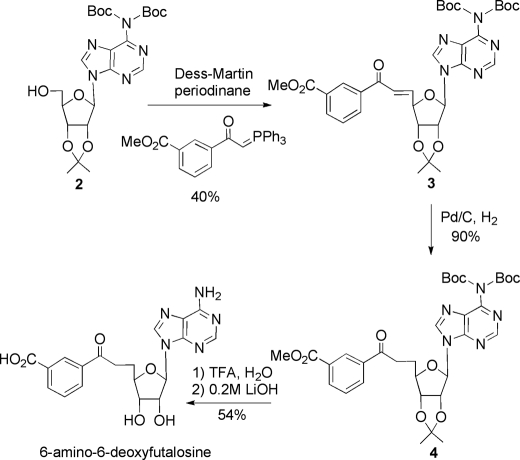
To test this hypothesis, it was necessary to synthesize authentic 6-amino-6-deoxyfutalosine. This was done using a procedure similar to that described for the synthesis of futalosine with the exception that the adenosine was used as the starting material (27). The 6-amino group was protected as a bis-t-butoxycarbonyl derivative, and the 2′,3′-hydroxyl groups were protected with an acetonide moiety to give the known compound 2 (Fig. 5). A one-pot oxidation/Wittig procedure was then used to produce alkene 3 as the trans-isomer (35). Hydrogenation then provided a single isomer of compound 4. Removal of the acetonide and t-butoxycarbonyl protecting groups was achieved by treatment with trifluoroacetic acid, and hydrolysis of the methyl ester using LiOH generated 6-amino-6-deoxyfutalosine.
FIGURE 5.
Synthetic route used to prepare 6-amino-6-deoxyfutalosine. Boc, t-butoxycarbonyl.
With 6-amino-6-deoxyfutalosine in hand, it was possible to test whether it serves as a substrate for the C. jejuni MTAN. Monitoring an enzymatic incubation by 1H NMR spectroscopy (see supplemental “Materials and General Methods”) confirmed that 6-amino-6-deoxyfutalosine was converted into adenine and DHFL by MTAN. This suggests that the DHFL required for menaquinone biosynthesis in C. jejuni could be provided by the action of MTAN on 6-amino-6-deoxyfutalosine. To assess the catalytic efficiency of this process, we employed a coupled kinetic assay for adenine release that had previously been used in monitoring MTAN kinetics with MTA (20). The nucleosidase activity with 6-amino-6-deoxyfutalosine was found to follow Michaelis-Menten kinetics, and the kinetic constants obtained were kcat = 0.53 ± 0.03 s−1 and Km = 1.03 ± 0.12 μm (see supplemental “Materials and General Methods”). We also determined the activity of the C. jejuni MTAN against MTA and obtained kinetic constants of kcat = 2.7 ± 0.1 s−1 and Km = 0.93 ± 0.10 μm. The similarity of these values indicates that 6-amino-6-deoxyfutalosine is a good substrate for the C. jejuni MTAN and supports the notion that this activity may be biologically relevant. The same assay can be used to analyze for hypoxanthine release, so we reinvestigated the ability of MTAN to catalyze the hydrolysis of futalosine. Using 150 μm concentrations of substrates, we found that a low level of activity could be observed with futalosine; however, it was only 2% of that observed with 6-amino-6-deoxyfutalosine.
Studies with a C. jejuni Strain Deleted for mqnA2
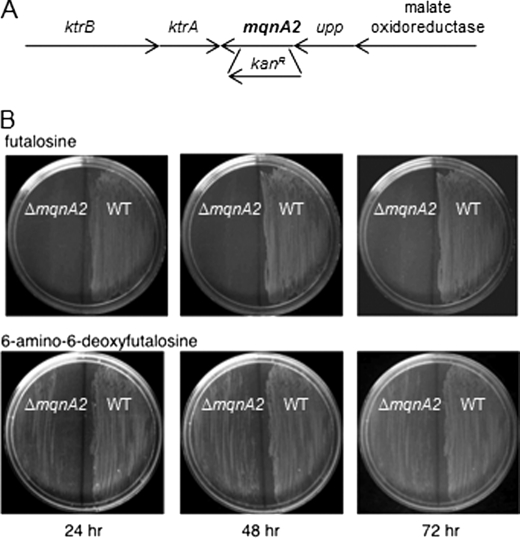
We next wished to provide in vivo evidence that the modified futalosine pathway for menaquinone biosynthesis is operative in C. jejuni and that 6-amino-6-deoxyfutalosine is used in place of futalosine in this organism. We anticipated that a MqnA-like enzyme in C. jejuni would be involved in the biosynthesis of 6-amino-6-deoxyfutalosine, and therefore, a mutant lacking the gene encoding for this protein would grow only when supplemented with 6-amino-6-deoxyfutalosine, but not with futalosine. A search for homologs in the C. jejuni genome indicated that the gene products of cj1285c and cjj81176_1302, in C. jejuni strains 11168 and 81-176, respectively, are 99.6% identical to each other and share 26% sequence identity with MqnA from S. coelicolor. On the basis of our prediction that the C. jejuni MqnA-like enzyme would synthesize 6-amino-6-deoxyfutalosine instead of futalosine, we designated this gene mqnA2 and constructed a deletion mutant in C. jejuni 81-176 by replacing most of the mqnA2 open reading frame with a non-polar kanamycin resistance cassette (aphA-3) using double crossover homologous recombination (Fig. 6A). Mutant clones were selected on standard rich medium (Mueller-Hinton) plates supplemented with kanamycin and 6-amino-6-deoxyfutalosine. Mueller-Hinton plates containing either futalosine (Fig. 6B, upper) or 6-amino-6-deoxyfutalosine (Fig. 6B, lower) were streaked with both wild-type C. jejuni 81-176 and 81-176 ΔmqnA2. After 24 and 48 h, the wild type grew well on either plate, whereas growth of the ΔmqnA2 mutant was observed only in the presence of 6-amino-6-deoxyfutalosine. No growth was observed on unsupplemented plates (data not shown). This supports the hypothesis that cj1285c/cjj81176_1302 encodes MqnA2, which is required for the biosynthesis of 6-amino-6-deoxyfutalosine, and that an essential modified futalosine pathway is operative in C. jejuni. Very weak growth of the mutant strain on the futalosine plates was observed after 72 h of incubation. This is likely due to the slow hydrolysis of futalosine catalyzed by the C. jejuni MTAN (2% of the activity observed with either MTA or 6-amino-6-deoxyfutalosine). In contrast to the work with S. coelicolor, we could not restore growth of the mutant strain with commercially available menaquinone-4 (bearing a C20 prenyl side chain) (data not shown). This may be due to the insolubility of the compound and the resulting limited penetration of a very non-polar molecule across the outer membrane of the Gram-negative organism. It could also be due to a more stringent requirement for menaquinone bearing a C30 prenyl side chain, menaquinone-6, which is normally utilized by C. jejuni (36).
FIGURE 6.
Genomic context of mqnA2 and growth of wild-type C. jejuni 81-176 and ΔmqnA2 on plates supplemented with either futalosine or 6-amino-6-deoxyfutalosine. A, the genomic organization surrounding mqnA2 showing that the gene is positioned at the end of an operon. The position of insertion of the kanamycin resistance cassette is indicated. B, growth of ΔmqnA2 (left) and the wild type (right) C. jejuni 81-176 is shown on plates supplemented with either futalosine (upper) or 6-amino-6-deoxyfutalosine (lower) as a function of time.
DISCUSSION
This study provides the first direct evidence that a modified futalosine pathway is operative in C. jejuni. A search for the gene encoding the putative futalosine hydrolase indicated that the C. jejuni MTAN was the most reasonable candidate to play this role. After finding that the C. jejuni MTAN catalyzed the hydrolysis of MTA, but not of futalosine, we began to suspect that the pathway in C. jejuni differed from that in S. coelicolor. An analysis of the active site architecture of the E. coli MTAN showed that a key aspartate residue, Asp-197, is required to act as an acid catalyst in promoting the departure of the adenine leaving group. This residue is present in all gene products annotated as MTANs in Helicobacter and Campylobacter species but was found to be an asparagine in the two identified futalosine hydrolases from S. coelicolor and T. thermophilus (14, 15). This suggested that an adenine-containing version of futalosine is the true intermediate in Helicobacter and Campylobacter species. Ultimately, we found that the C. jejuni MTAN could hydrolyze 6-amino-6-deoxyfutalosine, implying that this is the source of the DHFL used in menaquinone biosynthesis in this organism. The amino acid change implies that there are key differences in the mechanisms employed by the MTANs and the futalosine hydrolases. Because hypoxanthine is a better leaving group than adenine due to the more electronegative oxygen, it may not require an acid catalyst to depart; the H-bonding with an asparagine residue may be sufficient to promote catalysis. Alternatively, an as yet unidentified residue may serve as an acid catalyst in the futalosine hydrolases.
A very recent report has also demonstrated that the H. pylori MTAN catalyzes the hydrolysis of 6-amino-6-deoxyfutalosine, but not of futalosine (37). Although in vivo studies and kinetics were not reported, the authors proposed that this is the true biosynthetic intermediate in that organism. Our in vivo studies in C. jejuni have demonstrated that the mqnA2 gene is required for the biosynthesis of 6-amino-6-deoxyfutalosine and that this activity is essential for growth of the organism. Together with the studies on S. coelicolor, this strongly suggests that menaquinone is biosynthesized in C. jejuni and H. pylori via a modified futalosine pathway. This also implicates menaquinone biosynthetic enzymes as viable targets for the development of antibacterials directed against C. jejuni and H. pylori.
This work points out the truly remarkable number of roles that MTAN plays in these pathogenic bacteria. The enzyme displays a great deal of flexibility in regard to the nature of the group attached to the 5′-position of the substrate ribose moiety yet appears to be quite specific in regard to the nature of the nucleoside base. It hydrolyzes MTA in its role in recycling the by-products of the S-adenosylmethionine-dependent biosynthesis of spermidine (38). It also hydrolyzes S-adenosylhomocysteine in its role in the biosynthesis of the quorum-sensing molecule autoinducer-2, which is generated in both C. jejuni and H. pylori (39–42). Finally, as we have shown in this work, MTAN plays an essential role in the hydrolysis of 6-amino-6-deoxyfutalosine, an activity that has been implicated as a requirement for menaquinone biosynthesis. Thus, this multifunctional enzyme may serve as the Achilles' heel of these pathogens, and the very potent MTAN inhibitors that are already available from work with the E. coli enzyme may serve as effective antibacterial agents. Given the multiple important biological roles of MTAN, it may be difficult for resistance mechanisms to develop.
Supplementary Material
This work was supported in part by a grant from the Natural Sciences and Engineering Research Council of Canada (NSERC; to M. E. T.) and by a Burroughs Wellcome Fund Career Development Award in the Biomedical Sciences, a Canada Research Chair Award, and Canadian Institutes of Health Research Operating Grant MOP-68981 (to E. C. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Materials and General Methods,” Figs. S1–S7, and additional references.
- DHFL
- dehypoxanthinylfutalosine
- MTAN
- 5′-methylthioadenosine nucleosidase
- MTA
- 5′-methylthioadenosine.
REFERENCES
- 1. Furie B., Bouchard B. A., Furie B. C. (1999) Blood 93, 1798–1808 [PubMed] [Google Scholar]
- 2. Kurosu M., Begari E. (2010) Molecules 15, 1531–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Unden G., Bongaerts J. (1997) Biochim. Biophys. Acta 1320, 217–234 [DOI] [PubMed] [Google Scholar]
- 4. Collins M. D., Jones D. (1981) Microbiol. Rev. 45, 316–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dhiman R. K., Mahapatra S., Slayden R. A., Boyne M. E., Lenaerts A., Hinshaw J. C., Angala S. K., Chatterjee D., Biswas K., Narayanasamy P., Kurosu M., Crick D. C. (2009) Mol. Microbiol. 72, 85–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Marcelli S. W., Chang H. T., Chapman T., Chalk P. A., Miles R. J., Poole R. K. (1996) FEMS Microbiol. Lett. 138, 59–64 [DOI] [PubMed] [Google Scholar]
- 7. Collins M. D., Costas M., Owen R. J. (1984) Arch. Microbiol. 137, 168–170 [DOI] [PubMed] [Google Scholar]
- 8. Moss C. W., Lambert-Fair M. A., Nicholson M. A., Guerrant G. O. (1990) J. Clin. Microbiol. 28, 395–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dairi T. (2009) J. Antibiot. 62, 347–352 [DOI] [PubMed] [Google Scholar]
- 10. Meganathan R. (2001) Vitam. Horm. 61, 173–218 [DOI] [PubMed] [Google Scholar]
- 11. Bentley R., Meganathan R. (1982) Microbiol. Rev. 46, 241–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Borodina I., Krabben P., Nielsen J. (2005) Genome Res. 15, 820–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Seto H., Jinnai Y., Hiratsuka T., Fukawa M., Furihata K., Itoh N., Dairi T. (2008) J. Am. Chem. Soc. 130, 5614–5615 [DOI] [PubMed] [Google Scholar]
- 14. Hiratsuka T., Furihata K., Ishikawa J., Yamashita H., Itoh N., Seto H., Dairi T. (2008) Science 321, 1670–1673 [DOI] [PubMed] [Google Scholar]
- 15. Hiratsuka T., Itoh N., Seto H., Dairi T. (2009) Biosci. Biotechnol. Biochem. 73, 1137–1141 [DOI] [PubMed] [Google Scholar]
- 16. Arai R., Murayama K., Uchikubo-Kamo T., Nishimoto M., Toyama M., Kuramitsu S., Terada T., Shirouzu M., Yokoyama S. (2009) J. Struct. Biol. 168, 575–581 [DOI] [PubMed] [Google Scholar]
- 17. Butzler J. P. (2004) Clin. Microbiol. Infect. 10, 868–876 [DOI] [PubMed] [Google Scholar]
- 18. van Amsterdam K., van Vliet A. H., Kusters J. G., van der Ende A. (2006) FEMS Microbiol. Rev. 30, 131–156 [DOI] [PubMed] [Google Scholar]
- 19. Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 20. Singh V., Evans G. B., Lenz D. H., Mason J. M., Clinch K., Mee S., Painter G. F., Tyler P. C., Furneaux R. H., Lee J. E., Howell P. L., Schramm V. L. (2005) J. Biol. Chem. 280, 18265–18273 [DOI] [PubMed] [Google Scholar]
- 21. Klenow H. (1952) Biochem. J. 50, 404–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kalckar H. M. (1947) J. Biol. Chem. 167, 429–443 [PubMed] [Google Scholar]
- 23. Duerre J. A. (1962) J. Biol. Chem. 237, 3737–3741 [Google Scholar]
- 24. Cornell K. A., Riscoe M. K. (1998) Biochim. Biophys. Acta 1396, 8–14 [DOI] [PubMed] [Google Scholar]
- 25. Cornell K. A., Swarts W. E., Barry R. D., Riscoe M. K. (1996) Biochem. Biophys. Res. Commun. 228, 724–732 [DOI] [PubMed] [Google Scholar]
- 26. Schauder S., Shokat K., Surette M. G., Bassler B. L. (2001) Mol. Microbiol. 41, 463–476 [DOI] [PubMed] [Google Scholar]
- 27. Li X., Tanner M. E. (2010) Tetrahedron Lett. 51, 6463–6465 [Google Scholar]
- 28. Singh V., Lee J. E., Núñez S., Howell P. L., Schramm V. L. (2005) Biochemistry 44, 11647–11659 [DOI] [PubMed] [Google Scholar]
- 29. Lee J. E., Luong W., Huang D. J., Cornell K. A., Riscoe M. K., Howell P. L. (2005) Biochemistry 44, 11049–11057 [DOI] [PubMed] [Google Scholar]
- 30. Lee J. E., Singh V., Evans G. B., Tyler P. C., Furneaux R. H., Cornell K. A., Riscoe M. K., Schramm V. L., Howell P. L. (2005) J. Biol. Chem. 280, 18274–18282 [DOI] [PubMed] [Google Scholar]
- 31. Lee J. E., Smith G. D., Horvatin C., Huang D. J., Cornell K. A., Riscoe M. K., Howell P. L. (2005) J. Mol. Biol. 352, 559–574 [DOI] [PubMed] [Google Scholar]
- 32. Lee J. E., Cornell K. A., Riscoe M. K., Howell P. L. (2001) Structure 9, 941–953 [DOI] [PubMed] [Google Scholar]
- 33. Papadopoulos J. S., Agarwala R. (2007) Bioinformatics 23, 1073–1079 [DOI] [PubMed] [Google Scholar]
- 34. Medeiros G. C., Thomas G. J., Jr. (1971) Biochim. Biophys. Acta 238, 1–4 [DOI] [PubMed] [Google Scholar]
- 35. Ikejiri M., Ohshima T., Fukushima A., Shimotohno K., Maruyama T. (2008) Bioorg. Med. Chem. Lett. 18, 4638–4641 [DOI] [PubMed] [Google Scholar]
- 36. Carlone G. M., Anet F. A. L. (1983) J. Gen. Microbiol. 129, 3385–3393 [DOI] [PubMed] [Google Scholar]
- 37. Arakawa C., Kuratsu M., Furihata K., Hiratsuka T., Itoh N., Seto H., Dairi T. (2011) Antimicrob. Agents Chemother. 55, 913–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Evans G. B., Furneaux R. H., Schramm V. L., Singh V., Tyler P. C. (2004) J. Med. Chem. 47, 3275–3281 [DOI] [PubMed] [Google Scholar]
- 39. Quiñones B., Miller W. G., Bates A. H., Mandrell R. E. (2009) Appl. Environ. Microbiol. 75, 281–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rader B. A., Campagna S. R., Semmelhack M. F., Bassler B. L., Guillemin K. (2007) J. Bacteriol. 189, 6109–6117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cloak O. M., Solow B. T., Briggs C. E., Chen C. Y., Fratamico P. M. (2002) Appl. Environ. Microbiol. 68, 4666–4671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Elvers K. T., Park S. F. (2002) Microbiology 148, 1475–1481 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.