Abstract
Sphingolipid metabolites, such as ceramide (Cer), sphingosine (SPH), and sphingosine 1-phosphate (S1P), contribute to multiple aspects of carcinogenesis including cell proliferation, migration, angiogenesis, and tumor resistance. The cellular balance between Cer and S1P levels, for example, is an important determinant of cell fate, with the former inducing apoptosis and the later mitogenesis. Acid ceramidase (ASAH1) plays a pivotal role in regulating the intracellular concentration of these two metabolites by hydrolyzing Cer into SPH, which is rapidly phosphorylated to form S1P. Genistein is a phytoestrogen isoflavone that exerts agonist and antagonist effects on the proliferation of estrogen-dependent MCF-7 cells in a dose-dependent manner, primarily as a ligand for estrogen receptors. Genistein can also activate signaling through GPR30, a G-protein-coupled cell surface receptor. Based on the relationship between bioactive sphingolipids and tumorigenesis, we sought to determine the effect of genistein on ASAH1 transcription in MCF-7 breast cancer cells. We show herein that nanomolar concentrations of genistein induce ASAH1 transcription through a GPR30-dependent, pertussis toxin-sensitive pathway that requires the activation of c-Src and extracellular signal regulated kinase 1/2 (ERK1/2). Activation of this pathway promotes histone acetylation and recruitment of phospho-estrogen receptor α and specificity protein-1 to the ASAH1 promoter, ultimately culminating in increased ceramidase activity. Finally, we show that genistein stimulates cyclin B2 expression and cell proliferation in an ASAH1-dependent manner. Collectively, these data identify a mechanism through which genistein promotes sphingolipid metabolism and support a role for ASAH1 in breast cancer cell growth.
Keywords: Breast Cancer, Gene Regulation, Signal Transduction, Sphingolipid, Steroid Hormone Receptor, GPR-30, Acid Ceramidase, Estrogen Receptor, Genistein
Introduction
The soybean isoflavone genistein (4,5,7-trihydroxyisoflavone) exerts many cellular effects including activating apoptosis, inhibiting protein-tyrosine kinase activity, and suppressing angiogenesis (1, 2). Genistein also acts in a dose-dependent manner to both positively (3–7) and negatively (8–14) regulate tumorigenesis. In estrogen receptor (ER)2-positive cells, high doses of genistein (>10 μm) are associated with tumor suppression, whereas low doses (0.01–1 μm) have a mitogenic effect (10, 15, 16). Genistein promotes proliferation of estrogen-dependent breast and thyroid cancer cells (11, 17, 18), and induces the expression of vascular endothelial growth factor (19), c-FOS (20), peroxisome proliferator-activated receptor γ (21), and proteinase inhibitor 9 (22). The proliferative properties of genistein are mostly due to its ability to activate multiple genomic and non-genomic estrogenic pathways by binding to ERα and -β (12). Furthermore, genistein activates GPR30 (also called GPER-1) (17, 20, 23), a transmembrane G-protein-coupled receptor that binds most ER ligands and mediates rapid estrogenic signaling (24, 25). The binding of 17β-estradiol (E2) to GPR30 stimulates the cAMP pathway (26), increases intracellular Ca2+ (24, 25), and induces epidermal growth factor receptor transactivation in ER-negative breast cancer cells (27, 28). Signaling through GPR30 promotes the proliferation of multiple carcinomas (29, 30), stimulates cell migration through induction of the connective tissue growth factor gene (31), and promotes c-FOS (20), BCL-2 (32, 33), cyclin D2 (34), and estrogen-related receptor α (35) gene expression.
Sphingolipids are a large family of lipids involved in many aspects of cell regulation (reviewed in Ref. 36–38). Notably, ceramide (Cer) and sphingosine 1-phosphate (S1P) have been extensively studied for their opposing roles in the regulation of various aspects of cancer pathogenesis and therapy (39). Cer inhibits cell growth and promotes apoptosis and senescence, whereas S1P induces cell proliferation and migration by signaling through five S1P receptors (40–42). In this manner, the relative concentrations of these two molecules determine whether the cell undergoes apoptosis or proliferates (39). Consequently, targeting pathways that elevate Cer or decrease S1P accumulation has been a promising therapeutic approach in cancer treatment (43).
Acid ceramidase (encoded by ASAH1) is a lipid hydrolase that directly regulates Cer metabolism by catalyzing its degradation to sphingosine (SPH) and a free fatty acid (44). SPH is phosphorylated by sphingosine kinases (SPHK) to form S1P. Because Cer hydrolysis is the major pathway for SPH generation (45), ASAH1 plays a key role in regulating cellular homeostasis by controlling the Cer/SPH/S1P balance within the cell. In addition, the aberrant expression of ASAH1 in various human cancers (46–49), including breast cancer (50), prompted the emergence of this enzyme as a potential target for chemotherapy (48, 51, 52). Inhibitors of ASAH1 such as B13 and LCL464 were shown to cause Cer accumulation and prevent tumor growth (51, 53). At the transcriptional level, functional characterization of ASAH1 has been previously reported (44, 54), and we (55) and others (56) have established that cAMP-responsive element-binding protein and kruppel-like transcription factor 6 are important transcriptional regulators of this gene.
Despite the prominent roles of sphingolipids in cancer development and the proliferative actions of low doses of genistein, little is known about the relationship between these two factors in cancer progression. Therefore, based on the importance of ASAH1 in sphingolipid metabolism, the role of Cer/S1P balance in carcinogenesis, and the numerous biological effects of genistein in cancer cells, we sought to determine the role of genistein in ASAH1 gene transcription in MCF-7 breast cancer cells. We show that genistein induces ASAH1 gene expression through an ERK1/2-dependent mechanism involving both GPR30 and ERα. Furthermore, we demonstrate that ASAH1 expression is required for genistein-stimulated cyclin B2 expression and MCF-7 cell proliferation.
EXPERIMENTAL PROCEDURES
Reagents
Genistein (5,7-dihydroxy-3-(4-hydroxyphenyl) chromen-4-one), U0126, PP2, pertussis toxin, and LY294002 were purchased from EMD Chemicals Inc. (Gibbstown, NJ). ICI-182780 (Fulvestrant) and 17β-estradiol (E2) were purchased from Sigma. G-1 (1-[4-(6-bromobenzo[1,3]dioxol-5-yl)-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinolin-8-yl]-ethanone) and G-15 ((3a-4–9b)-4-(6-bromo-1,3-benzodioxol-5-yl)-3a,4,5,9b-3H-cyclopenta[c]quinoline) were purchased from Cayman Chemicals (Ann Arbor, MI). All compounds were dissolved in dimethyl sulfoxide, except E2, which was dissolved in ethanol.
Cell Culture
MCF-7 human breast cancer cells were obtained from American Type Culture Collection (ATCC) (Manassas, VA) and cultured in phenol red-free Eagle's minimum essential medium (Mediatech, Inc., Manassas, VA) supplemented with 10% fetal bovine serum (Mediatech, Inc.), 11 mm sodium pyruvate (Sigma), 0.01 mg/ml of bovine insulin (Sigma), antibiotics, and antimycotics. MDA-MB-231 human breast cancer cells were obtained from ATCC and cultured in Dulbecco's modified Eagle's medium (DMEM) (Mediatech, Inc.) supplemented with 10% bovine calf serum (Mediatech, Inc.), antibiotics, and antimycotics.
Real Time RT-PCR
MCF-7 cells were subcultured into 12-well plates and 48 h later treated with 20 nm genistein or 10 nm G-1 for 1–24 h. In some experiments, cells were preincubated with 100–200 nm G-15 for 1 h prior to treatment with genistein or G-1. Total RNA was isolated and quantified by qRT-PCR as previously described (55). Primers used are listed under Table 1. ASAH expression was normalized to β-actin mRNA levels and calculated using the ΔΔ cycle threshold (ΔΔCt) method.
TABLE 1.
Sequences of primers used in quantitative RT-PCR and ChIP assays
| Primer | Forward sequence (5′–3′) | Reverse sequence (5′–3′) |
|---|---|---|
| RT-PCR | ||
| β-Actin | ACGGCTCCGGCATGTGCAAG | TGACGATGCCGTGCTGCATG |
| ASAH1 | GCACAAGTTATGAAGGAAGCCAAG | TCCAATGATTCCTTTCTGTCTCG |
| ASAH2 | GCATCAACACAGGAGAGTC | GGAGGCAGAGGCATAGAG |
| ASAH3 | ATCCGCCTGGTCTTCATC | CTCCTTATTGCTGGTCTTCC |
| ChIP | ||
| ASAH1 (−123/+31) | AGTCCCGCCTCCTCCGAGCGTTCCCCCT | GACTAAGGCGACGCAACTCCGGCCCGGC |
| ASAH1 (−325/−214) | ACGGGTGAAGCTCCCGGCCCCACCTA | GAAAAGGGTGGCGTAGAGAAAGAGAGAG |
| ASAH1 (−500/−278) | GGCCGCTTTTCTCAGAGGGCAAA | GCGTAGAGAAAGAGAGAGAGCC |
RNA Interference (RNAi) and Real Time RT-PCR
MCF-7 cells were subcultured into 12-well plates and 24 h later transfected with 75 nm nonspecific 100 nm GPR30 (L-005563-00-0005, Dharmacon/Thermo Scientific), or 75 nm ERα (L-003401-00-0005, Dharmacon/Thermo Scientific) small interfering RNA (siRNA) oligonucleotides using HiPerfect Transfection Reagent (Qiagen, Valencia, CA). Twenty-four h later, cells were transfected again, incubated for an additional 24 h, and then treated with 20 nm genistein or 10 nm G-1 for 24 h. Total RNA was isolated and quantified by qRT-PCR as described above. Reduction of GPR30 and ERα protein expression at the time of RNA isolation was confirmed by Western blotting.
ERα Transient Transfection
MDA-MB-231 cells were subcultured into 12-well plates and transfected with 1 μg of pCMV-hERα (kindly provided by Dr. Ann Nardulli, University of Illinois, Urbana, IL) using GeneJuice (EMD Biosciences). Twenty-four h after transfection, cells were treated with 20 nm genistein or 5 nm E2 for 24 h. Total RNA was isolated and ASAH1, pS2, and β-actin mRNA levels were quantified by qRT-PCR as described above. Expression of ERα was confirmed by Western blotting 48 h after transfection.
Western Blotting
For quantification of ASAH1 protein expression, MCF-7 cells were treated with 20 nm genistein for 24, 48, or 72 h and harvested into RIPA buffer (1× PBS, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS). Cell lysates were isolated and separated by SDS-PAGE as previously described (55). Western blots were incubated with anti-ASAH1 (HPA005468, Sigma) and anti-GAPDH (sc-25778, Santa Cruz Biotechnology, Santa Cruz, CA) antibodies. For quantification of ERK1/2 activation, MCF-7 cells were serum starved for 40 h, then treated with 20 nm genistein for 0 to 30 min. ERK1/2 phosphorylation was determined by Western blotting using an anti-phospho-ERK1/2 antibody (sc-7383, Santa Cruz) and normalized to total ERK2 expression (sc-154, Santa Cruz). For quantification of phosphorylated ERα, MCF-7 cells were serum starved for 40 h, then treated with 20 nm genistein for 10, 30, or 60 min. ERα phosphorylation at serine 118 was determined by Western blotting using an anti-phospho-Ser118-ERα antibody (clone NL-44, Millipore) and normalized to total ERα expression (H-184, Santa Cruz). For quantification of cyclin B2 protein expression, MCF-7 cells were serum starved for 48 h and then pre-treated with 200 ng/ml of nocodazole (Calbiochem Inc.) for 10 h. Cells were rinsed 3 times with PBS, serum-free minimum essential medium was added to each well, and cells were treated with 20 nm genistein for 6 or 12 h. In some experiments, cells were transfected with 75 nm ASAH1 siRNA oligonucleotides (M-005228-01-0005, Dharmacon/Thermo Scientific) using HiPerfect Transfection Reagent (Qiagen) for 24 h prior to pretreatment with nocodazole. Cell lysates were isolated and separated by SDS-PAGE. Western blots were incubated with anti-cyclin B2 (K0189–3, Cell Signaling), anti-ASAH1 (Sigma), and anti-GAPDH (Santa Cruz) antibodies. Protein expression was detected using an ECF Western blotting kit (GE Healthcare) and visualized using a VersaDoc 4000 Imager (Bio-Rad).
Transient Transfection and Reporter Gene Analysis
Cloning of the human ASAH1 promoter and generation of deletion constructs were previously described (55). MCF-7 cells were subcultured into 24-well plates and transfected with 100 ng of pGL3-ASAH1 or pGL3-ASAH1(EREmutant) and 1.5 ng of pRL-TK (Promega) using GeneJuice (EMD Biosciences). Some cells were co-transfected with 50 ng of pCMV-hERα. Twenty-four h after transfection, cells were treated with 20 nm genistein for 16 h and the transcriptional activity of the ASAH1 reporter gene was determined using a dual luciferase assay kit (Promega). Firefly (pGL3-ASAH1) luciferase activity was normalized to Renilla luciferase activity (pRL-TK) and expressed as fold-change over the mean of the untreated control group.
Site-directed Mutagenesis
Mutagenesis of the putative ER response element (ERE) at position 475/−457 of the ASAH1 promoter was carried out using a QuikChange site-directed mutagenesis kit (Agilent, Santa Clara, CA) and confirmed by sequencing. ERE was disrupted by mutating 4 consecutive residues to alanine (underlined) using the following primer set: forward 5′-GGG CAA AGA TGG AAA AGG GTG GGA TGT TAC-3′, reverse 5′-ACA TCC CAC CCT TTT CCA TCT TTG CCC TCT-3′.
Chromatin Immunoprecipitation (ChIP)
MCF-7 cells were subcultured into 150-mm dishes and treated for 1 h with 20 nm genistein and ChIP assays were performed as described previously (57). The purified chromatin solutions were precleared and immunoprecipitated using 5 μg of primary antibody (anti-acetylhistone H3, anti-RNA polymerase II, anti-phospho-Ser118-ERα (clone NL-44), anti-SRC-1, or anti-specificity protein-1 (Sp1)) and 30 μl of protein A/G Plus-agarose (Santa Cruz). All antibodies used for ChIP were purchased from Millipore. Quantitative PCR was carried out using 20% output, 5% input (diluted 1:4), the ABsolute qPCR SYBR Green Fluorescein Mix (Thermo Scientific) and the primer sets are listed under Table 1.
Cell Proliferation
For quantitative proliferative assays, MCF-7 cells were seeded in 96-well plates (5 × 103 cells/well) and 24 h later transfected with 75 nm nonspecific siRNA oligonucleotides or siRNA against ASAH1 or siRNA against ASAH2 (M-005229-00-0005, Dharmacon/Thermo Scientific) for 24 h. In some experiments, cells were seeded in 96-well plates (5 × 103 cells/well) and 24 h later transfected with an ASAH1 expression plasmid (0.4 μg/well) for 24 h. Cells were then treated with 20 nm genistein, 10 nm G-1, or vehicle (dimethyl sulfoxide) for 24 h. After treatment, the cultures were incubated for an additional 6 h in the presence of 5-bromo-2-deoxyuridine (BrdU; 10 μm). Cell proliferation was assayed by BrdU incorporation measurements with an ELISA kit (Roche Applied Science). Western blotting was used to confirm reduction of ASAH1 and ASAH2 protein levels 48 h after siRNA transfection. Similarly, ASAH1 overexpression was confirmed by Western blotting 48 h after transfection.
Cell Viability
MCF-7 cells were plated in 96-well plates and 24 h later transfected with 75 nm nonspecific siRNA oligonucleotides or siRNA against ASAH1 (M-005228-01-0005, Dharmacon/Thermo Scientific). Twenty-four h later, cells were treated for 12, 24, or 48 h with 20 nm genistein or dimethyl sulfoxide. Cell viability was assessed using the CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega) following the manufacturer's instructions. Reduction of ASAH1 protein levels was confirmed by Western blotting 48 h after transfection.
Acid Ceramidase Activity
MCF-7 cells were subcultured into 100-mm dishes and treated for 48 or 72 h with 20 nm genistein and in vitro ASAH1 activity assay was performed as previously described (55). TLC plates were visualized by fluorescence scanning on a VersaDoc 4000 imager (Bio-Rad). NBD-dodecanoic acid formation was quantified and normalized to the protein content of each sample.
Statistical Analysis
One-way analysis of variance, Tukey-Kramer multiple comparison, and unpaired Student's t tests were performed using GraphPad InStat software (GraphPad Software Inc., San Diego, CA). Statistically significant differences from a compared value were defined as p < 0.05 denoted by asterisks (*) or carats (∧).
RESULTS
Genistein Induces ASAH1 mRNA Expression
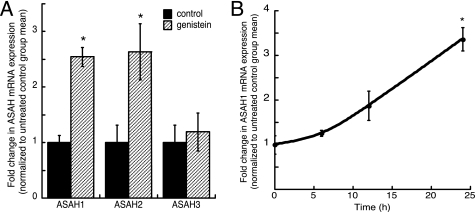
To evaluate the effect of genistein on the transcription of ceramidase genes (ASAH), MCF-7 cells were treated with 20 nm genistein for 24 h. ASAH1 mRNA expression was significantly increased by 2.5-fold in response to low-dose genistein treatment (Fig. 1A). Interestingly, ASAH2 mRNA expression was also induced by 2.6-fold in response to treatment, whereas the expression of ASAH3 was unchanged (Fig. 1A). Because aberrant ASAH1 expression has been reported in cancer cells (46, 49, 50), we focused on the effect of genistein on the transcription of this ceramidase isoform. We carried out experiments to assess the kinetics of the ASAH1 transcriptional response to genistein by treating cells for 6, 12, and 24 h. As shown in Fig. 1B, ASAH1 mRNA levels begin to increase after 6 h of genistein stimulation with maximal induction at the 24-h time point.
FIGURE 1.
Genistein induces ASAH1 transcription. A, MCF-7 cells were treated for 24 h with 20 nm genistein. Total RNA was isolated for analysis of acid (ASAH1), neutral (ASAH2), and alkaline (ASAH3) ceramidase mRNA expression by qRT-PCR. B, MCF-7 cells were treated for 6, 12, or 24 h with 20 nm genistein (gen) and ASAH1 and β-actin mRNA expressions were quantified by qRT-PCR. Data graphed are expressed as fold-change in ASAH mRNA expression normalized to the mRNA expression of β-actin and represent mean ± S.E. of three separate experiments, each done in triplicate. *, statistically different from untreated control group (p < 0.05).
GPR30 and ERα Mediate Genistein-induced ASAH1 Transcription
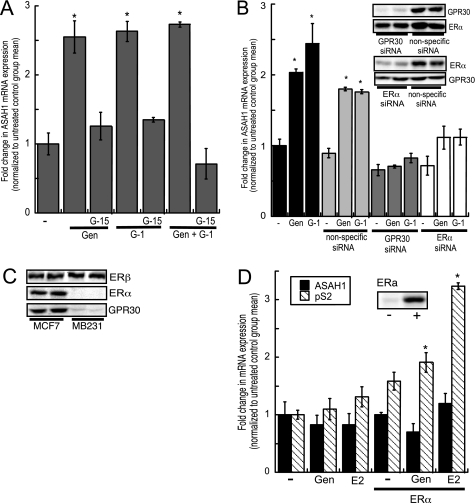
Genistein activates GPR30-mediated non-genomic signaling in thyroid and breast cancer cells (17, 20). Therefore, to investigate the involvement of GPR30 in the up-regulation of ASAH1 expression in response to genistein stimulation, we quantified ASAH1 mRNA levels in MCF-7 cells treated with 10 nm of the high-affinity GPR30 agonist G-1 (58) for 24 h. As shown in Fig. 2A, G-1 significantly increased ASAH1 transcription by 2.6-fold, which parallels the fold-increase elicited by genistein. Furthermore, neither genistein nor G-1 were able to induce ASAH1 expression in the presence of the high-affinity GPR30 antagonist G-15 (59) (Fig. 2A). Significantly, the transcriptional response was not enhanced by G-1 in combination with genistein (Fig. 2A). Because G-1 and G-15 are not ligands for ER-α or -β (58, 59), these data suggest that GPR30 plays a key role in genistein-stimulated ASAH1 transcription. The selectivity of genistein for GPR30 is further supported by the inability of ICI-182780, an ER antagonist (60), to repress genistein-induced ASAH1 mRNA expression (supplemental Fig. S1A). ICI-182780 is also a high-affinity GPR30 agonist (25), which is consistent with an increase in ASAH1 mRNA transcript levels observed in ICI-182780 only treated cells (supplemental Fig. 1A).
FIGURE 2.
GPR30 and ERα mediate genistein-dependent ASAH1 transcription. A, MCF-7 cells were pre-treated for 1 h with 100 or 200 nm G-15 followed by treatment with 10 nm G-1 or 20 nm genistein (gen) for 24 h. Some cells were treated with both G-1 and genistein in the presence or absence of 200 nm G-15. Total RNA was isolated for analysis of ASAH1 mRNA expression by qRT-PCR. B, MCF-7 cells were transfected twice with 100 nm GPR30, 75 nm ERα, or 75 nm nonspecific siRNA for 48 h (GPR30) or 24 h (ERα) followed by treatment with 20 nm genistein or 10 nm G-1 for 24 h. Total RNA was isolated and ASAH1 mRNA expression was quantified by qRT-PCR and normalized to β-actin. Inset, Western blot of GPR30 and ERα protein expression at the time of RNA isolation. C, ERα, ERβ, and GPR30 protein quantification in MCF-7 and MDA-MB-231 cells. D, ERα was expressed in MDA-MB-231 cells followed by treatment with 20 nm genistein (gen) or 5 nm E2 for 24 h. ASAH1 and pS2 mRNA levels were quantified by qRT-PCR and normalized to β-actin. Inset, ectopic expression of ERα: −, control; +, ERα-transfected cells. Data are graphed as fold-change in ASAH1 mRNA expression normalized to the mRNA expression of β-actin and represent mean ± S.E. of three separate experiments, each done in triplicate.
To determine whether GPR30 is required for ASAH1 transcription, we suppressed GPR30 translation (Fig. 2B, inset) and assessed the effect of reduced GPR30 expression on genistein- and G-1-stimulated ASAH1 mRNA expression. As shown in Fig. 2B, neither genistein nor G-1 were able to induce ASAH1 transcription in cells transfected with GPR30 siRNA oligonucleotides. Because genistein can also signal through ERα (15) and GPR30 can work together with ERα in certain estrogen-mediated activities (29, 61, 62), we also determined the effect of ERα suppression on ASAH1 transcription. Surprisingly, similar to GPR30, ERα suppression by siRNA (Fig. 2B, inset) significantly reduced both genistein- and G-1-stimulated ASAH1 transcription (Fig. 2B). Collectively, these data suggest that both GPR30 and ERα are required for the induction of ASAH1 mRNA expression by genistein.
Consistent with other reports (24, 61), MCF-7 cells express ERα, ERβ, and GPR30 protein, whereas MDA-MB-231 cells only express ERβ (Fig. 2C). Therefore, to further investigate the role of ERα in genistein-mediated ASAH1 transcription, ASAH1 mRNA expression was quantified in genistein-treated MDA-MB-231 cells transfected with an ERα expression plasmid (Fig. 2D, inset). Significantly, although ectopic expression of ERα was capable of inducing pS2 transcription, a well known ERα target gene (also known as TFF1), expression of the receptor was not sufficient to promote ASAH1 mRNA expression in genistein-treated cells (Fig. 2D).
Genistein-induced ASAH1 Transcription Occurs via a Pertussis Toxin-sensitive Pathway That Requires c-Src and ERK1/2 Activation
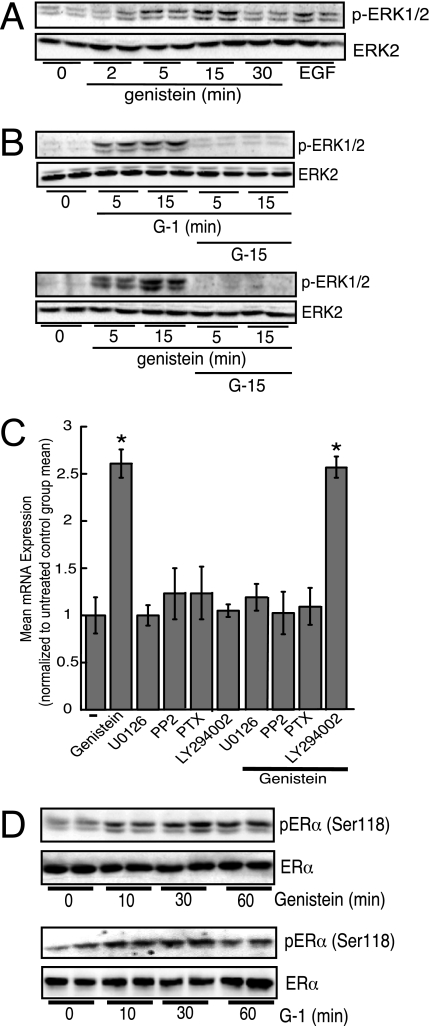
GPR30 signaling activates various downstream signaling cascades including ERK and phosphoinositol-3-kinase (PI3K)/Akt pathways (27). Therefore, to determine which kinases are activated by genistein stimulation, we tested the ability of genistein to increase ERK1/2 phosphorylation. As shown in Fig. 3A, genistein rapidly increased the phosphorylation of ERK1/2 in MCF-7 cells, but genistein did not activate Akt (data not shown). In addition, the effect of genistein on ERK1/2 activation was mimicked by G-1 (Fig. 3B, top panel) and G-15 prevented ERK1/2 phosphorylation induced by both G-1 (top panel) and genistein (bottom panel) (Fig. 3B). Additional experiments geared toward identifying effector kinases in the ERK pathway revealed that genistein also activated Raf-1 and c-Src (supplemental Fig. 1B), and that activation of these kinases was required for ERK phosphorylation (supplemental Fig. 1C). Consistent with these Western blot data, inhibition of ERK, c-Src, and Gαi activation attenuated genistein-stimulated ASAH1 mRNA expression (Fig. 3C). Furthermore, in agreement with the modulation of nuclear ERs by membrane-initiated signaling (reviewed in Ref. 63) and ligand-independent activation of ER by ERK signaling (64), both genistein (top panel) and G-1 (bottom panel) rapidly increased the levels of phospho-Ser118-ERα by 1.6- and 1.5-fold after 30 min, respectively (Fig. 3D).
FIGURE 3.
Genistein-induced ASAH1 mRNA expression requires Gαi, c-Src, and ERK1/2. A, MCF-7 cells were serum-starved for 40 h followed by treatment with 20 nm genistein for 0–30 min or 25 ng/ml of EGF for 10 min. Cell lysates were harvested and separated by SDS-PAGE followed by Western blotting using anti-phospho-ERK1/2 and ERK2 antibodies. B, MCF-7 cells were serum starved for 40 h, pre-treated with G-15 for 1 h, and then treated with 10 nm G-1 (top panel) or 20 nm genistein (lower panel) for 5 or 15 min. Cell lysates were harvested and separated by SDS-PAGE followed by Western blotting using anti-phospho-ERK1/2 and ERK2 antibodies. C, MCF-7 cells pre-treated for 1 h with 1 μm U0126, 10 μm PP2, 1 pg/ml of pertussis toxin, or 10 μm LY294002 were followed by treatment with 20 nm genistein for 24 h. Total RNA was isolated for analysis of ASAH1 mRNA expression by qRT-PCR. Data are graphed as fold-change in ASAH1 mRNA expression normalized to the mRNA expression of β-actin and represent mean ± S.E. of three separate experiments, each done in triplicate. *, statistically different from an untreated control group (p < 0.05). D, MCF-7 cells were serum-starved for 40 h followed by treatment with 20 nm genistein (top panel) or 10 nm G-1 (lower panel) for 0–60 min. Lysates were isolated and separated by SDS-PAGE followed by Western blotting using antibodies against phospho-Ser118-ERα or ERα.
Genistein Promotes the Recruitment of ERα and Sp1 to the ASAH1 Promoter
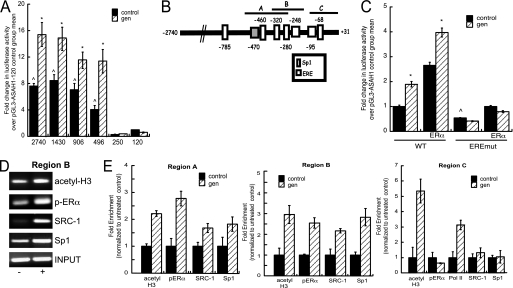
To further define the mechanism by which genistein modulates ASAH1 mRNA expression, MCF-7 cells were transfected with a luciferase reporter construct that contains 2.7 kb of the ASAH1 promoter and treated with 20 nm genistein for 16 h. As shown in Fig. 4A, genistein treatment significantly increased the transcriptional activity of the 2.7-kb reporter gene by 1.8-fold. To identify the genistein-responsive region(s) on the ASAH1 promoter, different lengths of the ASAH1 promoter (55) were tested for their ability to activate a luciferase reporter construct. Although deletion of the region between −2740 to −496 bp had no significant effect on the ability of genistein to stimulate reporter gene activity, removal of 250 bp (from the −496 bp construct) completely abolished the genistein response (Fig. 4A).
FIGURE 4.
Genistein stimulation promotes the recruitment of ERα and Sp1 to the ASAH1 promoter. A, MCF-7 cells were transfected with reporter gene plasmids (pGL3-ASAH1) containing varying regions of the ASAH1 promoter and a Renilla luciferase plasmid (pRL-TK). Twenty-four h after transfection, cells were treated with 20 nm genistein for 16 h and luciferase activity was quantified by luminometry. Data are graphed as fold-change ± S.E. of three separate experiments, each done in triplicate, and normalized to −120-bp construct. Asterisks (*) and carats (∧) denote statistically significant differences from untreated control within each transfection group or from the untreated −120-bp construct, respectively (p < 0.05). B, diagram of putative binding sites for Sp1 and ER within the genistein-responsive region of the ASAH1 promoter. Letters A, B, and C denote regions amplified by each primer set used for ChIP assay. The ERE at position −475/−457 bp that was mutated for further analysis in C is denoted by a gray-shaded square. C, MCF-7 cells were transfected with pGL3-ASAH1–496 (WT) or pGL3-ASAH1–496-EREmutant (EREmut) reporter gene plasmid, pCMV-hERα, and a Renilla luciferase plasmid (pRL-TK). Twenty-four h after transfection, cells were treated with 20 nm genistein for 16 h and luciferase activity was quantified by luminometry. Data are graphed as fold-change ± S.E. of three separate experiments, each done in triplicate and normalized to untreated WT-transfected controls. Asterisks and carats denote statistically significant differences from untreated control within each transfection group or from untreated WT controls, respectively (p < 0.05). D, representative agarose gels of the PCR products from ChIP assays obtained for region B (−325/−214). − and + denote untreated or genistein-treated, respectively. E, MCF-7 cells were treated for 1 h with 20 nm genistein, cross-linked with 1% formaldehyde, and the sheared chromatin immunoprecipitated with antibodies against phospho-Ser118-ERα, Sp1, RNA polymerase II, acetyl-histone H3, or SRC-1 and recruitment to regions A (−500/−278), B (−325/−214), or C (−123/+34) of the ASAH1 promoter assessed by qPCR. DNA purified was quantified by real time PCR and normalized to the ΔCt values of input DNA. Data are graphed as fold-enrichment over untreated control ± S.D.
Next we performed in silico promoter analysis and identified putative binding sites for Sp1 and ER (ER response element, ERE) within the first 500 bp upstream of the transcription initiation site of the ASAH1 gene (Fig. 4B). To define key ER transcriptional elements within the genistein-responsive region, a putative ERE at position −475/−457 bp of the ASAH1 promoter (Fig. 4B) was mutated and genistein-dependent ASAH1 reporter gene activity was determined in luciferase assays. As shown in Fig. 4C, ERα significantly increased the transcriptional activity of the wild type ASAH1 promoter, but failed to increase the reporter gene activity in cells transfected with an ASAH1 reporter plasmid harboring a mutation in the ERE. Interestingly, mutation of this ERE not only abolished the ability of genistein to stimulate promoter activity but also significantly decreased reporter gene activity in untreated cells, suggesting that this ERE may play a role in maintaining basal transcription (Fig. 4C). We next performed ChIP analysis on chromatin isolated from MCF-7 cells treated for 1 h with 20 nm genistein. Phospho-Ser118-ERα is enriched by 2.8-fold at region A (−500/−278) and 2.5-fold at region B (−325/−214) of the ASAH1 promoter in response to genistein (Fig. 4, D and E). Sp1 and the coactivator SRC-1 were also recruited to regions A and B in response to genistein stimulation (Fig. 4E). Furthermore, genistein increased acetylation of histone H3 in all regions, indicating that genistein-stimulated phospho-Ser118-ERα and Sp1 binding occurs concomitant with histone H3 modification (Fig. 4E).
Genistein Increases ASAH1 Protein Expression and Enzymatic Activity
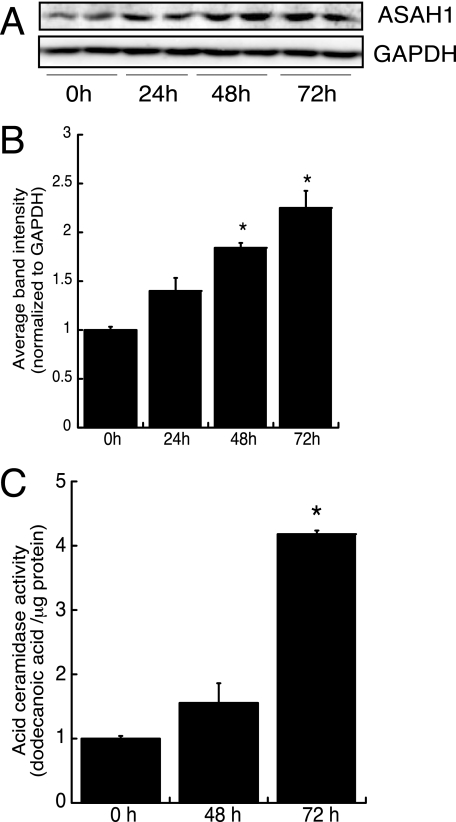
Next, we determined if genistein also increased protein expression by carrying out Western blot analysis of lysates isolated from MCF-7 cells that were treated for 24, 48, or 72 h with 20 nm genistein. As shown in Fig. 5, A and B, genistein significantly increased ASAH1 protein levels by 1.8- and 2.3-fold after 48 or 72 h treatment, respectively. This increase in ASAH1 protein content resulted in increased ceramidase activity (Fig. 5C).
FIGURE 5.
Genistein increases ASAH1 protein expression and enzymatic activity. A, MCF-7 cells were treated for 24, 48, or 72 h with 20 nm genistein. Total cell lysates were harvested and separated by SDS-PAGE followed by Western blotting analysis using anti-ASAH1 and anti-GAPDH antibodies. B, graphical analysis of data obtained from densitometric analysis of Western blots. Data represent mean ± S.D. of two separate experiments, each done in duplicate. *, statistically different from untreated control group (p < 0.05). C, MCF-7 cells were treated for 48 or 72 h with 20 nm genistein and cell lysates were isolated for in vitro ASAH1 activity assays as described under “Experimental Procedures.” TLC plates were visualized by fluorescence scanning. NBD-dodecanoic acid formation was quantified and normalized to the protein content of each sample. Data represent mean ± S.D. of three separate experiments, each done in duplicate. *, Statistically different from untreated control group (p < 0.05).
Genistein Induces MCF-7 Cell Proliferation and Viability in an ASAH1-dependent Manner
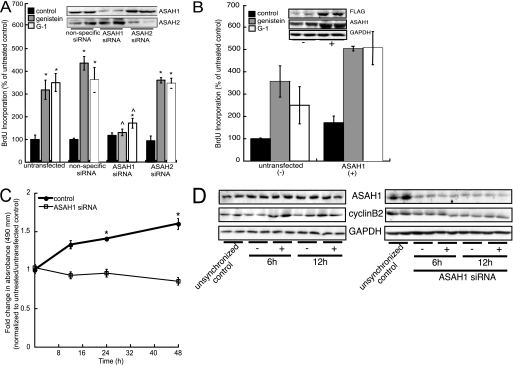
Given that genistein-induced cell growth is mediated by GPR30 signaling (17), we examined the role of ASAH1 in cell proliferation in cells transfected with nonspecific, ASAH1, or ASAH2 siRNAs (Fig. 6A, inset). The increase in BrdU incorporation elicited by both genistein and G-1 was dependent on ASAH1 expression (Fig. 6A). Although G-1 was able to induce cell proliferation by 1.3-fold in ASAH1-depleted cells, this increase was significantly lower than the 3.4-fold increase observed in untransfected cells (Fig. 6A). Importantly, suppression of ASAH2 had no effect on genistein- or G-1-induced cell proliferation (Fig. 6A). Moreover, ASAH1 overexpression potentiated cell proliferation in response to genistein and G-1 stimulation by 1.4- and 2.1-fold, respectively (Fig. 6B). The role of ASAH1 in genistein-dependent cell proliferation was further supported by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assays (Fig. 6C). Finally, to define the functional significance of ASAH1 on the cell cycle, we determined the effect of ASAH1 silencing on the phosphorylation state of cyclin-dependent kinase (CDK) 7 and CDK2, and cyclin B2 protein levels in genistein-treated MCF-7 cells. As shown in Fig. 6D, whereas cyclin B2 expression was increased by 2.3- and 2.5-fold in response to 6- or 12-h genistein treatment, respectively, this increase was attenuated in cells transfected with ASAH1 siRNA oligonucleotides. Although genistein induced the phosphorylation of both CDK7 and CDK2, these events were independent of ASAH1 expression (data not shown).
FIGURE 6.
Genistein increases cell proliferation, cell viability, and cyclin B2 expression in an ASAH1-dependent manner. A, MCF-7 cells were seeded in 96-well plates at 5 × 103 cells/well and 24 h later transfected with 75 nm ASAH1, ASAH2, or nonspecific siRNA for 24 h. Cells were then treated with 20 nm genistein or 10 nm G-1 and 24 h later, cell proliferation was measured with a BrdU incorporation ELISA kit. Data are expressed as mean % of control ± S.E. of four separate experiments, each done in quadruplicate. Inset, Western blot of ASAH1 and ASAH2 protein expression in siRNA-transfected cells 48 h after transfection. B, MCF-7 cells were seeded in 96-well plates at 5 × 103 cells/well and 24 h later transfected with 0.4 μg of an ASAH1 expression plasmid for 24 h. Cells were then treated with 20 nm genistein or 10 nm G-1 and 24 h later, cell proliferation was measured with a BrdU incorporation ELISA kit. Data are expressed as mean % of control ± S.E. of three separate experiments, each done in quadruplicate. Inset, Western blot of ASAH1 protein expression in control (−) and transfected (+) cells 48 h after transfection. C, MCF-7 cells were seeded in 96-well plates and transfected with 75 nm ASAH1 or nonspecific siRNA for 24 h. Cells were then treated with 20 nm genistein for 12, 24, or 48 h and cell viability was measured with an 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay kit. Data represent mean ± S.E. of three separate experiments, each performed in triplicate. Asterisks (*) indicate statistically significant differences from untreated/untransfected or untreated/siRNA-transfected control groups, respectively (p < 0.05). D, MCF-7 cells were transfected with 75 nm ASAH1 siRNA for 24 h, pre-treated with 200 ng/ml of nocodazole for 10 h, and then treated with 20 nm genistein for 6 or 12 h. Total lysates were harvested and separated by SDS-PAGE followed by Western blotting analysis using anti-ASAH1, anti-cyclin B2, or anti-GAPDH antibodies. − and + denote untreated or genistein-treated groups, respectively. Unsynchronized controls denote cells grown in regular minimum essential medium neither treated with nocodazole nor genistein.
DISCUSSION
Genistein mediates many estrogen-dependent pathways by binding to ERs (11, 18) and it has been reported that this phytoestrogen can promote breast cancer cell growth in a dose-dependent manner (10, 15, 16, 65, 66). Although estrogen-activated non-genomic signaling is well documented (reviewed in Ref. 63), the mechanisms by which phytoestrogens evoke changes in cell growth are less defined. Therefore, we investigated the role of genistein in ASAH1 expression in MCF-7 cells.
GPR30 is increasingly recognized as an important mediator of rapid non-genomic estrogenic action (27, 28, 31, 35). We show that 20 nm genistein and the high-affinity GPR30 agonist G-1 induce ASAH1 transcription (Fig. 1) through a GPR30-dependent (Fig. 2, A and B) ERK1/2 signaling pathway (Fig. 3). Consistent with our findings, genistein has been shown to activate ERK1/2 through GPR30 in MCF-7 and thyroid cancer cells (17, 20). In addition, Maggiolini et al. (20) has shown that genistein induces c-FOS gene expression through a GPR30-dependent ERK1/2 cascade that requires c-Src kinase activity.
Importantly, we show that the transcriptional response elicited by genistein requires both GPR30 and ERα (Fig. 2B). Studies have demonstrated that these two receptors act cooperatively to mediate cell proliferation in ER-positive cancer cells (29, 62). Although it is possible that GPR30 and ERα are acting in mutually exclusive signaling pathways, three lines of evidence suggest that these two receptors are part of the same signaling cascade: (a) genistein and G-1 do not have a synergistic effect on ASAH1 transcription (Fig. 2A), (b) abrogation of either GPR30 or ERα expression by siRNA prevents genistein-dependent ASAH1 gene expression (Fig. 2B), and (c) ERα suppression inhibits G-1-stimulated ASAH1 gene expression (Fig. 2B). Reconstitution of this pathway in ERα/GPR30-deficient MDA-MB-231 cells (Fig. 2C) revealed that ERα alone is not sufficient to promote ASAH1 transcription in response to genistein, although E2-induced pS2 expression is restored in the presence of the receptor (Fig. 2D).
ER-mediated transcription can be stimulated by ligand-independent mechanisms involving second messenger signaling systems (64). We show that stimulation with genistein and G-1 leads to phosphorylation of ERα at Ser118, a major target of MAPK signaling (64, 67). Furthermore, promoter analysis revealed that an ERE at position −475 bp of the ASAH1 promoter is required for ERα-dependent ASAH1 reporter gene activity (Fig. 4C), suggesting that ERα is recruited to this site following genistein stimulation. Indeed, ChIP studies demonstrated that phospho-Ser118-ERα is recruited to the ASAH1 promoter in response to genistein stimulation (Fig. 4D). Sp1, which has been shown to interact with ERα to transactivate target genes by binding to Sp1xERE motifs (68), is also recruited to the same promoter region (Fig. 4D). In agreement with our findings, ASAH1 expression was shown to positively correlate with ER status in breast cancer tumors (50) and ER-positive tumors display higher ASAH1 expression (69), supporting a role for ER in the transcriptional regulation of the ASAH1 gene.
Significantly, we show that ASAH1 expression is required for the increase in cell proliferation by genistein (Fig. 6A) and ASAH1 overexpression potentiates this proliferative response (Fig. 6B). These data suggest that ASAH1 mediates genistein-stimulated cell proliferation. Furthermore, we show that genistein increases cyclin B2 protein expression in an ASAH1-dependent manner (Fig. 6D). Cyclin B2 expression during the G2/M phase of the cell cycle facilitates the activation of CDK1, regulates mitotic progression (70), and prevents DNA re-replication (71). Thus, our data suggests that endogenous ASAH1 mediates progression through the cell cycle in response to low doses of genistein, at least in part, by triggering cyclin B2 expression. Of note, alkaline ceramidase 3 and neutral ceramidase (ASAH2) were both reported to modulate cell proliferation either by up-regulating cyclin-dependent kinase inhibitor p21CIP1/WAF1 expression (72) or inducing cell cycle arrest at G0/G1 and Rb protein dephosphorylation (73), respectively. Genistein has been shown to promote cell proliferation of pancreatic β-cells (74), and breast (2, 18, 20), thyroid (17), and prostate (75) cancer cells through multiple signaling pathways. Because genistein has a dose-dependent effect on the growth of breast cancer cells (10, 15, 16), it is noteworthy that the effect of genistein on ASAH1 transcription was only observed at nanomolar concentrations (micromolar concentrations had no effect on ASAH1 transcription, data not shown), the same dosage at which it promotes tumor growth in vivo and in vitro (2, 10, 11, 22, 65, 66). Because genistein also induced the transcription of ASAH2 (Fig. 1A) and SPHK1 (data not shown), it is likely that this phytoestrogen may promote sphingolipid metabolic changes by regulating the expression of multiple sphingolipid genes. Indeed, mass spectrometric analysis revealed that genistein evoked a 1.3-fold increase in the cellular concentration of S1P.3 Total Cer levels were not significantly altered by genistein stimulation, whereas sphingomyelin levels were modestly increased by 1.2-fold in genistein-treated cells.3 Given the complexity of the sphingolipid metabolic pathway and its many metabolites, temporal genomic transcriptional and lipidomic analysis is required to pinpoint the mechanisms by which genistein alters sphingolipid concentrations. Gupta et al. (76) recently reported a quantitative model of the sphingolipid pathway that illustrates the complexity of sphingolipid flux alterations in response to pharmacological perturbations. Nonetheless, because S1P activates cell growth-related pathways in a paracrine/autocrine manner by binding to S1P receptors (77, 78), it is possible that genistein also promotes S1P secretion. Given that SK1 is overexpressed in breast cancer cells (79), it is likely that an increase in ASAH1 activity could lead to increased S1P production. Of note, E2 was shown to promote breast cancer cell proliferation by activating SK1 (80) and Takabe et al. (81) recently demonstrated that E2 induces ERK1/2 activation in MCF-7 cells by stimulating SK1 activity and promoting a rapid secretion of S1P in an ERα-dependent manner. Further studies are required to establish that genistein evokes the export of S1P via a similar mechanism. Nonetheless, because E2 treatment mirrored the stimulatory effects of genistein on ASAH1 transcription and protein expression (data not shown), we think that regulation of ASAH1 expression by this phytoestrogen adds to our current understanding of the molecular mechanisms underlying estrogen-dependent mammary cell proliferation. Because ceramide degradation is the only source of cellular SPH (82, 83), ASAH1 directly regulates SK1 substrate availability. Thus, the effect of E2 on SK1 activity would benefit from increased ASAH1 expression. Furthermore, long-term exposure to genistein was recently shown to enhance E2 responsiveness in the rat uterus (84) and reverse the inhibitory effects of tamoxifen (85, 86). Therefore, we hypothesize that low doses of genistein may have a detrimental additive effect to E2 in vivo by promoting sphingolipid-mediated breast cell growth.
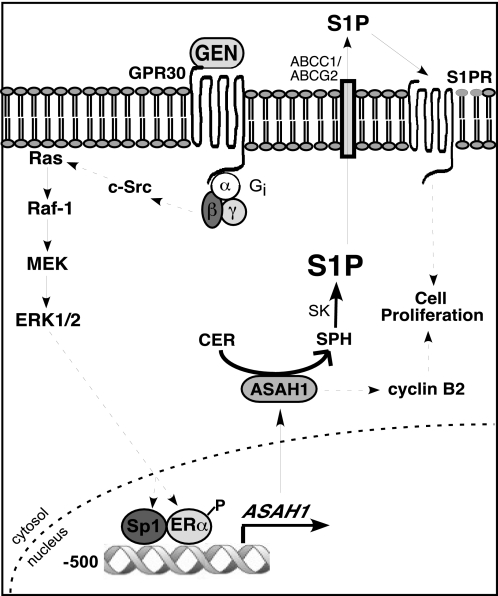
In conclusion, we propose a model whereby genistein induces ASAH1 gene expression by activating a GPR30-dependent pathway that culminates in ERK1/2 phosphorylation (Fig. 7). ERK1/2 activation induces the phosphorylation of ERα and the binding of a complex containing the receptor, Sp1, and SRC1 to the ASAH1 promoter. ASAH1 transcription is followed by increased protein expression and enzymatic activity, which results in increased S1P production. S1P can then be exported (77) and activate proliferative pathways by binding to cell surface S1P receptors (78) (Fig. 7). Concomitantly, ASAH1 mediates cyclin B2 expression, which drives mitotic progression and, consequently, cell growth. Further studies are needed to determine whether S1P is involved in genistein-dependent cyclin B2 expression. In light of the growing interest in the role of phytoestrogens in cancer progression and treatment and the development of chemotherapeutic strategies to modulate sphingolipid metabolism, our results provide evidence for the integral role of ASAH1 in maintaining the cellular proliferative capacity of ER-positive breast cancer cells.
FIGURE 7.
Model pathway for genistein-induced ASAH1 transcription in MCF-7 cells. Genistein (GEN) activates GPR30, which signals through Gαi to activate c-Src and subsequently the MAPK pathway. ERK1/2 activation promotes the phosphorylation of ERα and its recruitment to the ASAH1 promoter. Induction of ASAH1 gene transcription leads to an increase in protein expression and subsequent up-regulation of enzymatic activity, which results in the degradation of ceramide (CER) into sphingosine (SPH) and its phosphorylation into S1P. S1P is exported from cells via ABC transporters ABCC1 and ABCG2 and acts in a paracrine/autocrine matter to activate proliferative signaling cascades by binding to cell-surface S1P receptors (S1PR). Concomitantly, ASAH1 mediates cyclin B2 expression.
Supplementary Material
Acknowledgment
We greatly appreciate the gift of pCMV-ERα expression plasmid from Dr. Ann Nardulli.
This work was supported, in whole or in part, by National Institutes of Health Grant DK08417 (to M. B. S.) and an American Heart Association Predoctoral Fellowship (to N. C. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
N. C. Lucki and M. B. Sewer, unpublished observations.
- ER
- estrogen receptor
- Sp1
- specificity protein-1
- CDK
- cyclin-dependent kinase
- SPH
- sphingosine
- SK
- sphingosine kinase
- ERE
- ER response element
- G-1
- 1-[4-(6-bromobenzo[1,3]dioxol-5-yl)-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinolin-8-yl]-ethanone
- G-15
- (3a-4–9b)-4-(6-bromo-1,3-benzodioxol-5-yl)-3a,4,5,9b-3H-cyclopenta[c]quinoline
- qRT
- qualitative RT
- Cer
- ceramide
- GPR
- G-protein receptor.
REFERENCES
- 1. Yuan-Jing F., Nan-Shan H., Lian X. (2009) Cancer Lett. 284, 189–197 [DOI] [PubMed] [Google Scholar]
- 2. Liu H., Du J., Hu C., Qi H., Wang X., Wang S., Liu Q., Li Z. (2010) J. Nutr. Biochem. 21, 390–396 [DOI] [PubMed] [Google Scholar]
- 3. Brown N. M., Lamartiniere C. A. (2000) Cell Growth & Differ. 11, 255–260 [PubMed] [Google Scholar]
- 4. Cotroneo M. S., Wang J., Fritz W. A., Eltoum I. E., Lamartiniere C. A. (2002) Carcinogenesis 23, 1467–1474 [DOI] [PubMed] [Google Scholar]
- 5. Lamartiniere C. A. (2000) Am. J. Clin. Nutr. 71, 1705S–1707S [DOI] [PubMed] [Google Scholar]
- 6. Messina M. J., Persky V., Setchell K. D., Barnes S. (1994) Nutr. Cancer 21, 113–131 [DOI] [PubMed] [Google Scholar]
- 7. Yang X., Edgerton S. M., Kosanke S. D., Mason T. L., Alvarez K. M., Liu N., Chatterton R. T., Liu B., Wang Q., Kim A., Murthy S., Thor A. D. (2003) Cancer Res. 63, 2425–2433 [PubMed] [Google Scholar]
- 8. Kuiper G. G., Lemmen J. G., Carlsson B., Corton J. C., Safe S. H., van der Saag P. T., van der Burg B., Gustafsson J. A. (1998) Endocrinology 139, 4252–4263 [DOI] [PubMed] [Google Scholar]
- 9. Bowers J. L., Tyulmenkov V. V., Jernigan S. C., Klinge C. M. (2000) Endocrinology 141, 3657–3667 [DOI] [PubMed] [Google Scholar]
- 10. Hsieh C. Y., Santell R. C., Haslam S. Z., Helferich W. G. (1998) Cancer Res. 58, 3833–3838 [PubMed] [Google Scholar]
- 11. Allred C. D., Allred K. F., Ju Y. H., Virant S. M., Helferich W. G. (2001) Cancer Res. 61, 5045–5050 [PubMed] [Google Scholar]
- 12. Gikas P. D., Mokbel K. (2005) Int. J. Fertil. Womens Med. 50, 250–258 [PubMed] [Google Scholar]
- 13. Messina M., McCaskill-Stevens W., Lampe J. W. (2006) J. Natl. Cancer Inst. 98, 1275–1284 [DOI] [PubMed] [Google Scholar]
- 14. Peeters P. H., Keinan-Boker L., van der Schouw Y. T., Grobbee D. E. (2003) Breast Cancer Res. Treat. 77, 171–183 [DOI] [PubMed] [Google Scholar]
- 15. Wang T. T., Sathyamoorthy N., Phang J. M. (1996) Carcinogenesis 17, 271–275 [DOI] [PubMed] [Google Scholar]
- 16. Miodini P., Fioravanti L., Di Fronzo G., Cappelletti V. (1999) Br. J. Cancer 80, 1150–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vivacqua A., Bonofiglio D., Albanito L., Madeo A., Rago V., Carpino A., Musti A. M., Picard D., Andò S., Maggiolini M. (2006) Mol. Pharmacol. 70, 1414–1423 [DOI] [PubMed] [Google Scholar]
- 18. Maggiolini M., Bonofiglio D., Marsico S., Panno M. L., Cenni B., Picard D., Andò S. (2001) Mol. Pharmacol. 60, 595–602 [PubMed] [Google Scholar]
- 19. Buteau-Lozano H., Velasco G., Cristofari M., Balaguer P., Perrot-Applanat M. (2008) J. Endocrinol. 196, 399–412 [DOI] [PubMed] [Google Scholar]
- 20. Maggiolini M., Vivacqua A., Fasanella G., Recchia A. G., Sisci D., Pezzi V., Montanaro D., Musti A. M., Picard D., Andò S. (2004) J. Biol. Chem. 279, 27008–27016 [DOI] [PubMed] [Google Scholar]
- 21. Dang Z. C., Audinot V., Papapoulos S. E., Boutin J. A., Löwik C. W. (2003) J. Biol. Chem. 278, 962–967 [DOI] [PubMed] [Google Scholar]
- 22. Jiang X., Patterson N. M., Ling Y., Xie J., Helferich W. G., Shapiro D. J. (2008) Endocrinology 149, 5366–5373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thomas P., Dong J. (2006) J. Steroid Biochem. Mol. Biol. 102, 175–179 [DOI] [PubMed] [Google Scholar]
- 24. Revankar C. M., Cimino D. F., Sklar L. A., Arterburn J. B., Prossnitz E. R. (2005) Science 307, 1625–1630 [DOI] [PubMed] [Google Scholar]
- 25. Thomas P., Pang Y., Filardo E. J., Dong J. (2005) Endocrinology 146, 624–632 [DOI] [PubMed] [Google Scholar]
- 26. Filardo E. J., Quinn J. A., Frackelton A. R., Jr., Bland K. I. (2002) Mol. Endocrinol. 16, 70–84 [DOI] [PubMed] [Google Scholar]
- 27. Filardo E. J., Quinn J. A., Bland K. I., Frackelton A. R., Jr. (2000) Mol. Endocrinol. 14, 1649–1660 [DOI] [PubMed] [Google Scholar]
- 28. Filardo E. J. (2002) J. Steroid Biochem. Mol. Biol. 80, 231–238 [DOI] [PubMed] [Google Scholar]
- 29. Albanito L., Lappano R., Madeo A., Chimento A., Prossnitz E. R., Cappello A. R., Dolce V., Abonante S., Pezzi V., Maggiolini M. (2008) Environ. Health Perspect. 116, 1648–1655 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30. Vivacqua A., Bonofiglio D., Recchia A. G., Musti A. M., Picard D., Andò S., Maggiolini M. (2006) Mol. Endocrinol. 20, 631–646 [DOI] [PubMed] [Google Scholar]
- 31. Pandey D. P., Lappano R., Albanito L., Madeo A., Maggiolini M., Picard D. (2009) EMBO J. 28, 523–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hsieh Y. C., Yu H. P., Frink M., Suzuki T., Choudhry M. A., Schwacha M. G., Chaudry I. H. (2007) Am. J. Pathol. 170, 1210–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kanda N., Watanabe S. (2003) J. Invest. Dermatol. 121, 1500–1509 [DOI] [PubMed] [Google Scholar]
- 34. Kanda N., Watanabe S. (2004) J. Invest. Dermatol. 123, 319–328 [DOI] [PubMed] [Google Scholar]
- 35. Li Y., Birnbaumer L., Teng C. T. (2010) Mol. Endocrinol. 24, 969–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hannun Y. A., Obeid L. M. (2008) Nat. Rev. Mol. Cell Biol. 9, 139–150 [DOI] [PubMed] [Google Scholar]
- 37. Lavieu G., Scarlatti F., Sala G., Carpentier S., Levade T., Ghidoni R., Botti J., Codogno P. (2008) Methods Mol. Biol. 445, 159–173 [DOI] [PubMed] [Google Scholar]
- 38. Lucki N. C., Sewer M. B. (2008) Subcell. Biochem. 49, 387–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huwiler A., Pfeilschifter J. (2006) Curr. Pharm. Des. 12, 4625–4635 [DOI] [PubMed] [Google Scholar]
- 40. Gómez-Muñoz A. (2006) Biochim. Biophys. Acta 1758, 2049–2056 [DOI] [PubMed] [Google Scholar]
- 41. Maceyka M., Payne S. G., Milstien S., Spiegel S. (2002) Biochim. Biophys. Acta 1585, 193–201 [DOI] [PubMed] [Google Scholar]
- 42. Tani M., Ito M., Igarashi Y. (2007) Cell. Signal. 19, 229–237 [DOI] [PubMed] [Google Scholar]
- 43. Saddoughi S. A., Song P., Ogretmen B. (2008) Subcell. Biochem. 49, 413–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li C. M., Park J. H., He X., Levy B., Chen F., Arai K., Adler D. A., Disteche C. M., Koch J., Sandhoff K., Schuchman E. H. (1999) Genomics 62, 223–231 [DOI] [PubMed] [Google Scholar]
- 45. Spiegel S., Merrill A. H., Jr. (1996) FASEB J. 10, 1388–1397 [DOI] [PubMed] [Google Scholar]
- 46. Saad A. F., Meacham W. D., Bai A., Anelli V., Elojeimy S., Mahdy A. E., Turner L. S., Cheng J., Bielawska A., Bielawski J., Keane T. E., Obeid L. M., Hannun Y. A., Norris J. S., Liu X. (2007) Cancer Biol. Ther. 6, 1455–1460 [DOI] [PubMed] [Google Scholar]
- 47. Mahdy A. E., Cheng J. C., Li J., Elojeimy S., Meacham W. D., Turner L. S., Bai A., Gault C. R., McPherson A. S., Garcia N., Beckham T. H., Saad A., Bielawska A., Bielawski J., Hannun Y. A., Keane T. E., Taha M. I., Hammouda H. M., Norris J. S., Liu X. (2009) Mol. Ther. 17, 430–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Elojeimy S., Liu X., McKillop J. C., El-Zawahry A. M., Holman D. H., Cheng J. Y., Meacham W. D., Mahdy A. E., Saad A. F., Turner L. S., Cheng J., Day T., Dong J. Y., Bielawska A., Hannun Y. A., Norris J. S. (2007) Mol. Ther. 15, 1259–1263 [DOI] [PubMed] [Google Scholar]
- 49. Seelan R. S., Qian C., Yokomizo A., Bostwick D. G., Smith D. I., Liu W. (2000) Genes Chromosomes Cancer 29, 137–146 [DOI] [PubMed] [Google Scholar]
- 50. Ruckhaberle E., Holtrich U., Engels K., Hanker L., Gatje R., Metzler D., Karn T., Kaufmann M., Rody A. (2009) Climacteric 12, 1–12 [DOI] [PubMed] [Google Scholar]
- 51. Selzner M., Bielawska A., Morse M. A., Rüdiger H. A., Sindram D., Hannun Y. A., Clavien P. A. (2001) Cancer Res. 61, 1233–1240 [PubMed] [Google Scholar]
- 52. Samsel L., Zaidel G., Drumgoole H. M., Jelovac D., Drachenberg C., Rhee J. G., Brodie A. M., Bielawska A., Smyth M. J. (2004) Prostate 58, 382–393 [DOI] [PubMed] [Google Scholar]
- 53. Bai A., Szulc Z. M., Bielawski J., Mayroo N., Liu X., Norris J., Hannun Y. A., Bielawska A. (2009) Bioorg. Med. Chem. 17, 1840–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li C. M., Hong S. B., Kopal G., He X., Linke T., Hou W. S., Koch J., Gatt S., Sandhoff K., Schuchman E. H. (1998) Genomics 50, 267–274 [DOI] [PubMed] [Google Scholar]
- 55. Lucki N., Sewer M. B. (2009) Biochim. Biophys. Acta 1791, 706–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Park J. H., Eliyahu E., Narla G., DiFeo A., Martignetti J. A., Schuchman E. H. (2005) Biochim. Biophys. Acta 1732, 82–87 [DOI] [PubMed] [Google Scholar]
- 57. Dammer E. B., Leon A., Sewer M. B. (2007) Mol. Endocrinol. 21, 415–438 [DOI] [PubMed] [Google Scholar]
- 58. Bologa C. G., Revankar C. M., Young S. M., Edwards B. S., Arterburn J. B., Kiselyov A. S., Parker M. A., Tkachenko S. E., Savchuck N. P., Sklar L. A., Oprea T. I., Prossnitz E. R. (2006) Nat. Chem. Biol. 2, 207–212 [DOI] [PubMed] [Google Scholar]
- 59. Dennis M. K., Burai R., Ramesh C., Petrie W. K., Alcon S. N., Nayak T. K., Bologa C. G., Leitao A., Brailoiu E., Deliu E., Dun N. J., Sklar L. A., Hathaway H. J., Arterburn J. B., Oprea T. I., Prossnitz E. R. (2009) Nat. Chem. Biol. 5, 421–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Howell A., Osborne C. K., Morris C., Wakeling A. E. (2000) Cancer 89, 817–825 [DOI] [PubMed] [Google Scholar]
- 61. Wang C., Dehghani B., Magrisso I. J., Rick E. A., Bonhomme E., Cody D. B., Elenich L. A., Subramanian S., Murphy S. J., Kelly M. J., Rosenbaum J. S., Vandenbark A. A., Offner H. (2008) Mol. Endocrinol. 22, 636–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Albanito L., Madeo A., Lappano R., Vivacqua A., Rago V., Carpino A., Oprea T. I., Prossnitz E. R., Musti A. M., Andò S., Maggiolini M. (2007) Cancer Res. 67, 1859–1866 [DOI] [PubMed] [Google Scholar]
- 63. Levin E. R. (2005) Mol. Endocrinol. 19, 1951–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lannigan D. A. (2003) Steroids 68, 1–9 [DOI] [PubMed] [Google Scholar]
- 65. Allred C. D., Ju Y. H., Allred K. F., Chang J., Helferich W. G. (2001) Carcinogenesis 22, 1667–1673 [DOI] [PubMed] [Google Scholar]
- 66. Ju Y. H., Allred C. D., Allred K. F., Karko K. L., Doerge D. R., Helferich W. G. (2001) J. Nutr. 131, 2957–2962 [DOI] [PubMed] [Google Scholar]
- 67. Kato S., Endoh H., Masuhiro Y., Kitamoto T., Uchiyama S., Sasaki H., Masushige S., Gotoh Y., Nishida E., Kawashima H., Metzger D., Chambon P. (1995) Science 270, 1491–1494 [DOI] [PubMed] [Google Scholar]
- 68. Safe S., Kim K., Kim K. (2008) J. Mol. Endocrinol. 41, 263–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ruckhäberle E., Rody A., Engels K., Gaetje R., von Minckwitz G., Schiffmann S., Grösch S., Geisslinger G., Holtrich U., Karn T., Kaufmann M. (2008) Breast Cancer Res. Treat. 112, 41–52 [DOI] [PubMed] [Google Scholar]
- 70. Gong D., Pomerening J. R., Myers J. W., Gustavsson C., Jones J. T., Hahn A. T., Meyer T., Ferrell J. E., Jr. (2007) Curr. Biol. 17, 85–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bellanger S., de Gramont A., Sobczak-Thépot J. (2007) Oncogene 26, 7175–7184 [DOI] [PubMed] [Google Scholar]
- 72. Hu W., Xu R., Sun W., Szulc Z. M., Bielawski J., Obeid L. M., Mao C. (2010) J. Biol. Chem. 285, 7964–7976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wu B. X., Zeidan Y. H., Hannun Y. A. (2009) Biochim. Biophys. Acta 1791, 730–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fu Z., Zhang W., Zhen W., Lum H., Nadler J., Bassaganya-Riera J., Jia Z., Wang Y., Misra H., Liu D. (2010) Endocrinology 151, 3026–3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. El Touny L. H., Banerjee P. P. (2009) Cancer Res. 69, 3695–3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Gupta S., Maurya M. R., Merrill A. H., Jr., Glass C. K., Subramaniam S. (2011) BMC Syst. Biol. 5, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kim R. H., Takabe K., Milstien S., Spiegel S. (2009) Biochim. Biophys. Acta 1791, 692–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Alvarez S. E., Milstien S., Spiegel S. (2007) Trends Endocrinol. Metab. 18, 300–307 [DOI] [PubMed] [Google Scholar]
- 79. French K. J., Schrecengost R. S., Lee B. D., Zhuang Y., Smith S. N., Eberly J. L., Yun J. K., Smith C. D. (2003) Cancer Res. 63, 5962–5969 [PubMed] [Google Scholar]
- 80. Sukocheva O. A., Wang L., Albanese N., Pitson S. M., Vadas M. A., Xia P. (2003) Mol. Endocrinol. 17, 2002–2012 [DOI] [PubMed] [Google Scholar]
- 81. Takabe K., Kim R. H., Allegood J. C., Mitra P., Ramachandran S., Nagahashi M., Harikumar K. B., Hait N. C., Milstien S., Spiegel S. (2010) J. Biol. Chem. 285, 10477–10486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wang E., Norred W. P., Bacon C. W., Riley R. T., Merrill A. H., Jr. (1991) J. Biol. Chem. 266, 14486–14490 [PubMed] [Google Scholar]
- 83. Spiegel S., Milstien S. (2002) J. Biol. Chem. 277, 25851–25854 [DOI] [PubMed] [Google Scholar]
- 84. Möller F. J., Diel P., Zierau O., Hertrampf T., Maass J., Vollmer G. (2010) Toxicol. Lett. 196, 142–153 [DOI] [PubMed] [Google Scholar]
- 85. Liu B., Edgerton S., Yang X., Kim A., Ordonez-Ercan D., Mason T., Alvarez K., McKimmey C., Liu N., Thor A. (2005) Cancer Res. 65, 879–886 [PubMed] [Google Scholar]
- 86. Ju Y. H., Doerge D. R., Allred K. F., Allred C. D., Helferich W. G. (2002) Cancer Res. 62, 2474–2477 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.