Abstract
Many proteins contain a thioredoxin (Trx)-like domain fused with one or more partner domains that diversify protein function by the modular construction of new molecules. The Escherichia coli protein YbbN is a Trx-like protein that contains a C-terminal domain with low homology to tetratricopeptide repeat motifs. YbbN has been proposed to act as a chaperone or co-chaperone that aids in heat stress response and DNA synthesis. We report the crystal structure of YbbN, which is an elongated molecule with a mobile Trx domain and four atypical tetratricopeptide repeat motifs. The Trx domain lacks a canonical CXXC active site architecture and is not a functional oxidoreductase. A variety of proteins in E. coli interact with YbbN, including multiple ribosomal protein subunits and a strong interaction with GroEL. YbbN acts as a mild inhibitor of GroESL chaperonin function and ATPase activity, suggesting that it is a negative regulator of the GroESL system. Combined with previous observations that YbbN enhances the DnaK-DnaJ-GrpE chaperone system, we propose that YbbN coordinately regulates the activities of these two prokaryotic chaperones, thereby helping to direct client protein traffic initially to DnaK. Therefore, YbbN may play a role in integrating the activities of different chaperone pathways in E. coli and related bacteria.
Keywords: Chaperone Chaperonin, Crystal Structure, Heat Shock Protein, Protein Folding, Protein Structure, Thiol, Tetratricopeptide Repeat, Thioredoxin-like
Introduction
Thioredoxin (Trx)2 is a widely distributed and extensively studied thiol-disulfide oxidoreductase that protects organisms from the deleterious effects of reactive oxygen species through the action of two conserved active site cysteine residues (1). In addition, Trx also has a role in cell signaling that is important in higher eukaryotes (2). Although the role of Trx in the oxidative stress response is well established, a large number of proteins from all kingdoms of life contain one or more Trx-like domains fused to various other domains. The diversity of the partner domain(s) in Trx domain-containing proteins is remarkable, with >300 different partners identified from all sequenced genomes by the SUPERFAMILY webserver (3). In combination with the intrinsic functional plasticity of the Trx domain itself (4, 5), these domain fusions generate remarkable functional diversity in the Trx-like proteins. Despite the abundance of proteins containing a Trx domain fused to another domain, the functions and structures of many of these proteins are unknown.
The conserved prokaryotic protein YbbN exemplifies one such family of Trx-domain containing proteins. YbbN is a 31-kDa Escherichia coli protein with an N-terminal Trx domain and a C-terminal domain with poor homology to known conserved domains. YbbN gene expression is significantly induced by heat shock (6) or upon overexpression of the heat shock σ-factor 32 encoded by the rpoH gene (7). Physiologically, ybbN-deficient E. coli have been reported to display an increased sensitivity to heat stress (8) and show defects in DNA synthesis and cell division (9). YbbN physically interacts with multiple components of the DNA polymerase III holoenzyme (9) and with the chaperones DnaK and GroEL (8), suggesting that YbbN may promote the proper folding or assembly of key proteins involved in DNA replication and stress response. Therefore, defects in these processes have been proposed to result in the phenotypes associated with YbbN deficiency.
Although YbbN has been implicated in facilitating proper protein quality control and folding, many of the details of YbbN biochemical activity remain unclear. YbbN was initially reported to possess a weak thiol-disulfide protein oxidoreductase activity in an in vitro RNase A disulfide isomerization assay (10). However, the conserved thioredoxin CXXC active site motif necessary for this activity is mutated to a SXXC sequence in E. coli YbbN. The reported oxidoreductase activity was proposed to result from a non-canonical active site comprising the conserved Cys-38 and a distal but more highly conserved Cys-63 as a second redox active cysteine residue (10). However, a subsequent report from the same group noted that the in vivo relevance of this weak activity is uncertain, as a ybbN-deficient strain of E. coli does not display increased sensitivity to oxidative stress, and no clear evidence was found for YbbN oxidoreductase activity in vivo (8).
More recently, an alternative chaperone activity has been proposed for YbbN. Caldas et al. (10) found that YbbN alone is able to facilitate the refolding of urea-denatured citrate synthase (CS), α-glucosidase or β-clamp DnaN in vitro with an efficiency comparable to chaperones like DnaK and other heat shock proteins. However, a contradictory result was briefly mentioned by Pan and Bardwell (5) using CS as a substrate. Moreover, YbbN alone cannot suppress the heat-induced aggregation of CS at 43 °C, which is an alternative in vitro chaperone assay (10). YbbN physically interacts with GroEL and DnaK (8, 11) as well as other proteins in pulldown experiments, and YbbN deletion strains show reduced levels of these two chaperone proteins (8). Furthermore, YbbN can enhance the rate of citrate synthase refolding by the DnaK-DnaJ-GrpE chaperone complex in vitro (8). Considered together, these results suggest that YbbN may be more active as a co-chaperone for the DnaK system than as a chaperone in isolation. Additionally, YbbN may be a specific chaperone for components of the DNA polymerase holoenzyme, as recently suggested (9).
To clarify the structural basis of YbbN function, we have determined the x-ray crystal structure of YbbN at 1.8 Å resolution. The structure shows that the protein consists of a loosely tethered N-terminal Trx domain and a divergent C-terminal tetratricopeptide repeat (TPR) motif fold that possesses unusual structural features. The protein is negatively charged over most of its solvent-exposed surface and is a highly prolate monomer in solution. The Trx domain of YbbN does not have an active site structure that is conducive to thiol-disulfide exchange chemistry, supporting the conclusion that the weak oxidoreductase activity of the protein is likely not physiologically relevant. Immobilized YbbN interacts with a variety of proteins in pulldown assays, including a strong interaction with GroEL. Unexpectedly, the chaperone and ATPase activities of the GroESL chaperonin complex are mildly inhibited by YbbN in an in vitro refolding assay. In total, these results suggest a hypothesis that YbbN proteins may play a role in coordinating the activities of key bacterial chaperone systems.
EXPERIMENTAL PROCEDURES
YbbN, GroEL, and GroES Cloning, Expression, and Purification
The full-length coding sequences for YbbN, GroEL, and GroES were PCR-amplified from E. coli genomic DNA using primers that introduced NdeI (5′) and XhoI (3′) restriction enzyme sites at the indicated ends of the amplified products. These coding sequences were cloned between the NdeI and XhoI sites of the bacterial expression vector pET15b, and the recombinant proteins were expressed in BL21(DE3) E. coli grown in Luria-Bertani medium supplemented with 100 μg/ml ampicillin at 37 °C with shaking. Once the A600 of the culture reached 0.6–0.8, it was equilibrated at 20 °C for 1 h before induction of protein expression by the addition of 0.2 mm isopropyl β-d-1-thiogalactopyranoside. The induced culture was incubated at 20 °C with shaking overnight and harvested by centrifugation. Cell pellets were stored at −80 °C until needed.
The three proteins were expressed with thrombin-cleavable N-terminal His6 tags for purification by metal affinity chromatography. For each, the cell pellet was resuspended in extraction buffer (50 mm HEPES, pH 7.5, 300 mm NaCl, 0–10 mm imidazole, 2 mm DTT) supplemented with 1 mg/ml lysozyme and sonicated to complete lysis. Cleared lysate was loaded onto Ni2+ metal affinity His-Select resin (Sigma), washed with imidazole-supplemented extraction buffer (5 mm imidazole for YbbN, 10 mm for GroES, and 5 mm for GroEL), and the recombinant proteins were eluted using 250 mm imidazole in extraction buffer. The N-terminal hexahistidine tag was removed by thrombin cleavage for 4–8 h at 22 °C followed by dialysis against storage buffer (25 mm HEPES, 100 mm KCl, 2 mm DTT) at 4 °C overnight for YbbN and GroEL or incubation at 22 °C for 48 h with thrombin followed by dialysis at 4 °C for GroES. The proteins were passed sequentially over His-Select resin to remove any protein that retained the tag and then benzamidine-Sepharose resin to remove thrombin. For YbbN and GroEL, the proteins were then passed over S-Sepharose resin equilibrated in storage buffer as a subtractive purification step to remove impurities; the purified protein is present in the flow-through. The purified proteins were concentrated using a centrifugal concentrator (Millipore) with a 10-kDa cutoff. YbbN was concentrated to 26 mg/ml as determined by absorbance at 280 nm using a calculated extinction coefficient at 280 nm of 23,000 m−1 cm−1. Concentrations of GroEL and GroES were calculated using ϵ280 values of 10,555 and 1,490 m−1 cm−1, respectively. The purified proteins were stored in storage buffer, snap-frozen in 50–100 μl aliquots on liquid nitrogen, and stored at −80 °C.
Reductive Lysine Methylation of YbbN
Initial sparse matrix crystallization trials using YbbN did not produce diffraction-quality crystals, so the protein was modified by reductive methylation of lysines as described in Rypniewski et al. (12). Briefly, purified YbbN protein was dialyzed against PBS (10 mm Na2HPO4·7H2O, 1.7 mm KH2PO4,137 mm NaCl, 2 mm KCl) buffer, pH 7.4, for 4 h to dilute the DTT present in the protein storage buffer and then was reductively methylated by adding 20 μl/ml of 1 m DMAB solution and 40 μl/ml of 1 m formaldehyde. The reaction was incubated for 2 h at 4 °C followed by two additional cycles of DMAB and formaldehyde addition and incubation and a final addition of 10 μl/ml of 1 m DMAB and incubation at 4 °C for 12 h. Unreacted reagents were removed by dialysis against storage buffer. Methylated YbbN protein was concentrated to 22 mg/ml using a centrifugal concentrator (Millipore) and ran as a single band with an apparent size of ∼32 KDa on overloaded Biosafe (Bio-Rad) Coomassie-stained SDS-PAGE. The methylated protein migrates in SDS-PAGE with a slightly larger apparent size than the unmethylated protein, possibly due to altered interaction of methylated YbbN with the anionic SDS detergent in the gel. This mobility shift was used to semiquantitatively monitor the extent and homogeneity of protein methylation when compared with unmodified protein. The methylated YbbN protein was snap-frozen on liquid nitrogen and stored at −80 °C. The specific lysines that were methylated by this procedure were determined by trypsin digestion of the protein overnight followed by liquid chromatography MS-MS of the resulting peptides at the University of Nebraska Mass Spectrometry Core facility.
Crystallization and Data Collection
For all crystallization experiments, methylated YbbN protein at 22 mg/ml in storage buffer was crystallized using the sitting drop vapor diffusion method with drops containing 2 μl of protein and 2 μl of reservoir solution. Initial conditions were obtained from a commercial sparse matrix screen (Hampton Research) and optimized. Tabular crystals in space group P21 grew within 2 days at room temperature against a reservoir solution of 20% polyethylene glycol 8000, 0.2 m Ca(CH3COO)2, 0.1 m MES, pH 6.0. All crystals were cryoprotected by serial transfer through increasing concentrations of ethylene glycol in the reservoir solution to a final concentration of 25% (v/v), harvested in nylon loops, and cryocooled by immersion into liquid nitrogen.
Diffraction data were collected at the Advanced Photon Source, GM/CA-CAT beamline 23 ID-D from a single crystal of YbbN maintained at 100 K. The crystal was illuminated with incident X-rays of 12.66 KeV (0.98 Å), and a MARmosaic 300 CCD detector was used to collect the data. The crystal was exposed to x-rays for 2 s per 1 degree of oscillation for a total of 360 degrees, and the resulting diffraction data were integrated and scaled using HKL2000 (13). Final data statistics for each data set are provided in Table 1.
TABLE 1.
Data collection and refinement statistics
| Data collection | |
| X-ray source | APS GM/CA 23ID-D |
| X-ray wavelength (Å) | 0.98 |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 29.52, 62.45, 79.98 |
| β (degree) | 96.57 |
| Molecules in asymmetric unit | 1 |
| Wilson B factor (Å2) | 23 |
| Resolution (Å)a | 100-1.8 |
| Rmergeb | 0.05 (0.37) |
| 〈I〉/〈σ(I)〉 | 11.4 (2.3) |
| Completeness (%) | 95.0 (79.2) |
| Redundancy | 3.6 (2.5) |
| Refinement | |
| PDB code | 3QOU |
| Program | REFMAC5 |
| Resolution (Å) | 79-1.8 |
| No. reflections | 25553 |
| Rworkc; Rfreed; Ralle (%) | 18.7; 23.9; 18.9 |
| r.m.s.d. | |
| Bond lengths (Å) | 0.009 |
| Bond angles (deg.) | 1.01 |
| Ramachandran plot: favored; allowed; forbidden (%) | 96; 100; 0 |
a Values in parentheses are for highest resolution shell.
b Rmerge = ΣhklΣi|Ihkli − 〈Ihkl〉|/ΣhklΣiIhkli, where i is the ith observation of a reflection with indices h,k,l, and angle brackets indicate the average over all i observations.
c Rwork = Σhkl|Fhklo − Fhklc|/ΣhklFhklo, where Fhklc is the calculated structure factor amplitude with index h,k,l, and Fhklo is the observed structure factor amplitude with index h,k,l.
d Rfree is calculated as Rwork, where the Fhklo is taken from a test set comprising 5% of the data that were excluded from the refinement.
e Rall is calculated as Rwork, where the Fhklo includes all measured data (including the Rfree test set).
Structure Determination, Refinement, and Validation
The structure of YbbN was determined using maximum likelihood molecular replacement as implemented in PHASER (14), part of the CCP4 suite of programs (15). The search model used was based on 2R5S, a protein of unknown function from Vibrio parahaemolyticus whose structure determined by the Midwest Center for Structural Genomics. Although 2R5S was the closest sequence homologue of known structure to YbbN in a BLAST search (46% sequence identity), the aligned region is limited to the C-terminal TPR domains (residues 111–284) of YbbN. Despite high sequence identity, molecular replacement attempts using the entire 2R5S model, a homology model built using 2R5S, as well as various thioredoxin structures as search models were all unsuccessful. Successful molecular replacement solutions could only be obtained by using two truncated versions (residues 115–193 and 194–284) of the YbbN homology model built using 2R5S in Swissmodel (16) for sequential molecular replacement searches. The initial model was improved by automated model building in ARP/wARP (17) as implemented in CCP4, which produced an excellent model for residues 115–284 of YbbN. The quality of the electron density in the N-terminal thioredoxin domain of YbbN, however, remained poor due to disorder and required manual model building in COOT (18).
The model was refined against an amplitude-based maximum likelihood target function using Refmac5 (19) in the CCP4 suite. The model was refined against all measured data with no intensity or amplitude cutoff, and a bulk solvent correction was used to allow the inclusion of low resolution reflections. Manual adjustments to the model, including construction of the solvent model, were performed in COOT. Dimethyl lysine modifications resulting from the reductive methylation protocol were introduced into the model at sites where the electron density clearly supported the inclusion or where trypsinization/mass spectrometry indicated a modified lysine. The restraint files for dimethyl lysine used in refinement were generated using the PRODRG webserver (20). Upon convergence of the Refmac5 refinement with isotropic atomic displacement parameters, the translation-libration-screw (TLS) model was refined with three TLS rigid body groups (residues 6–31, 32–99, and 100–284) determined using TLSMD (21). The application of TLS reduced Rfree (22) by ∼2.5%. The final model was validated using MolProbity (23) and the validation tools in COOT (18). Asp-180 is the only marginal residue in a Ramachandran plot and acts as a bidentate ligand to a bound calcium ion. This residue is well ordered and in unambiguous electron density. All structural figures were made using POVScript+ (24).
Sedimentation Equilibrium Ultracentrifugation
The solution molecular mass of YbbN was determined by sedimentation equilibrium ultracentrifugation using a Beckman Coulter XL-I analytical ultracentrifuge as previously described (25). YbbN samples were prepared at 0.25, 0.50, and 1.0 mg/ml in 25 mm HEPES, pH 7.5, 100 mm KCl, and 1 mm DTT, and the experiment was conducted at 20 °C. The partial specific volume of YbbN was calculated as 0.7405 ml g−1 based on amino acid sequence using SedNTerp (26). All nine datasets (three concentrations of protein at three rotor speeds) were combined and globally fit to obtain molecular mass using Origin 6.
Identification of Proteins Interacting with YbbN
YbbN affinity resin was made by covalently binding 40 mg of purified, unmodified YbbN protein in binding buffer (50 mm HEPES, pH 7.5, 100 mm NaCl) onto 2 ml of cyanogen bromide-activated-Sepharose 4 Fast Flow (Sigma) resin following the manufacturer's instructions. For the binding experiments, a 4-g pellet of packed E. coli K-12 strain MG1655 cells was resuspended in 50 mm HEPES, pH 7.5, 100 mm NaCl, 2 mm DTT containing 200 μl of protease inhibitor mixture P8849 (Sigma) and lysed by sonication. The lysate was clarified by centrifugation at 10,000 rpm for 30 min at 4 °C, and the supernatant was collected and incubated with the YbbN affinity resin overnight at 4 °C. The resin was extensively washed with the binding buffer, and the proteins specifically interacting with YbbN were eluted from the column using binding buffer supplemented with 600 mm NaCl followed by a second elution of tightly bound proteins using binding buffer supplemented with 1 m NaCl. The two elutions were handled separately, and the retained proteins were concentrated by precipitation using a 10% trichloroacetic acid (TCA), 80% cold acetone mixture that was incubated on ice for 2 h. Precipitated proteins were recovered by centrifugation, washed with cold 20% TCA in water to remove any precipitated salts, resuspended in SDS-PAGE running buffer, neutralized with 5 n NaOH, and analyzed using SDS-PAGE. The individual bands were excised from the gel and identified by in-gel trypsin digestion followed by LC-MS/MS at the University of Nebraska Mass Spectrometry Core facility. To determine proteins avidly bound to the YbbN resin, a sample of the resin was taken after the 1.0 m NaCl elution, placed in SDS-PAGE loading buffer, heated to 95 °C for 5 min, and the liberated proteins were analyzed by SDS-PAGE.
Chaperone-assisted Refolding of Citrate Synthase
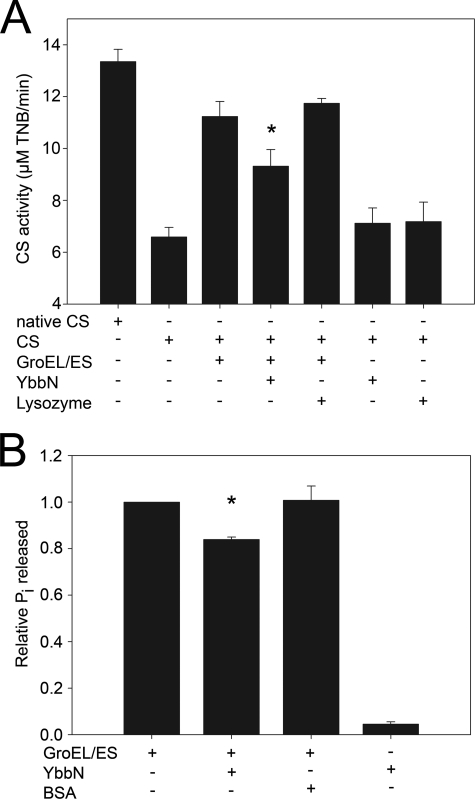
Pig heart citrate synthase (E.C. 2.3.3.1, Sigma) was completely denatured at a monomer concentration of 20 μm in 6 m guanidine-HCl, 50 mm HEPES, pH 7.5, 20 mm dithiothreitol at 22 °C for 60 min. Refolding was initiated by a 100-fold dilution of CS into refolding buffer (100 mm HEPES, pH 7.5, 10 mm KCl, 10 mm MgCl2, and 2 mm ATP) followed by incubation at 22 °C for 100 min. For the chaperone assays, GroEL (2 μm), GroES (4 μm), YbbN (4 μm), or lysozyme (4 μm) was present in the refolding buffer at the indicated concentrations. Upon dilution, the substrate CS was present at 0.2 μm, resulting in a 10–20-fold excess of chaperone to substrate. The enzymatic activity of refolded citrate synthase was measured by adding 950 μl of reaction solution containing 100 mm HEPES, pH 7.5 buffer, 0.047 mm acetyl coenzyme A (Sigma), 0.23 mm oxaloacetic acid (Sigma), and 0.1 mm 5,5′-dithiobis-(2-nitrobenzoic acid) (Invitrogen) to 50 μl of the CS refolding mixture. After mixing with reaction solution, CS activity was measured continuously using the absorption of liberated 5-thio-2-nitrobenzoic acid at 412 nm for 90 s using a Varian Cary50 spectrophotometer. The rate of reaction, which is directly proportional to the amount of correctly folded CS, was determined by measuring the slope of the linear increase in 412-nm absorption between 18 and 90 s. Native CS, treated in the same way as the refolded sample but without denaturation, was used as positive control and lysozyme (E.C. 3.2.1.17, Fisher) was used as negative control. All measurements were repeated at least three times with means and S.D. shown in Fig. 7. Student's t test was used to calculate the probability of the null hypothesis using Microsoft Excel.
FIGURE 7.
GroESL activity is mildly inhibited by YbbN. Panel A shows the amount of CS activity recovered from chemically denatured enzyme that was refolded by dilution into buffer containing the proteins indicated by a + sign in the table at the bottom of the graph. The reaction was followed by monitoring the production of colored 2-nitro-5-thiobenzoate by liberated CoA. Native CS, which had not been denatured before the assay, is the positive control. The GroESL chaperonin leads to a substantial recovery of CS activity that is diminished by the addition of YbbN. The asterisk indicates that the difference between values with and without YbbN is significant to a p value <0.05 by Student's t test for data generated from three independent experiments. YbbN alone is no more effective than the negative control protein lysozyme at refolding CS. Panel B shows the release of inorganic phosphate from ATP by the GroEL ATPase activity as determined using the Malachite Green assay. Samples containing the protein(s) are indicated by a + sign in the table at the bottom of the graph. The addition of YbbN results in a statistically significant (asterisk; p value < 0.05 by Student's t test) decrease in the GroEL ATPase activity, in agreement with the diminution of chaperone activity shown in panel A. Bovine serum albumin (BSA) was used as a negative control. Data are from three independent experiments.
GroEL ATPase Activity Assay
The GroESL ATPase activity assays were performed using the Malachite green phosphate assay kit (BioAssay Systems). GroESL (1 μm) or YbbN (2 μm) were added into the buffer containing 50 mm Tris-HCl, pH 8.0, 10 mm MgCl2, 10 mm KCl, 0.2 mm ATP, and incubated for 10 min at 22 °C. The enzyme reactions were terminated by the addition of working agent from the kit. The absorbance at 620 nm was measured after 30 min of color development at 22 °C. Bovine serum albumin (Invitrogen) was used at 2 μm as negative control. All measurements were repeated at least three times with means and S.D. shown in Fig. 7. Student's t test was used to calculate the probability of the null hypothesis using Microsoft Excel.
RESULTS
Description of the Structure
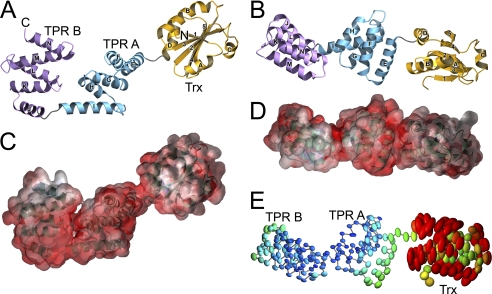
YbbN is a two-domain protein comprising an N-terminal Trx domain (residues 1–111) and a set of four TPR motifs (residues 112–284) with an intervening extended two helix region (180–212) that composes the C-terminal domain. The molecular surface of YbbN is defined by the pendant Trx domain and the curved, saddle-shaped C-terminal TPR domain (Fig. 1). The Trx domain is loosely tethered to the TPR domain, and weak electron density in this region provides clear evidence that the Trx domain is mobile in the crystal. This mobility was modeled using the TLS model (27) with three rigid domains (residues 6–31, 32–99, and 100–284) as determined using the TLSMD webserver (21). The anisotropic atomic displacement parameters calculated from the refined TLS model show dramatically higher and more anisotropic mobility of the Trx domain compared with the C-terminal TPR domain (Fig. 1E). This is supported by the much larger average B-factors for atoms in the Trx domain (87 Å2) compared with those in the remainder of the protein (24 Å2). The Trx domain bridges crystal contacts between neighboring molecules in the crystal but makes no direct packing contacts with the C-terminal domain, suggesting that Trx domain mobility is likely to be more pronounced in solution.
FIGURE 1.
Structure, electrostatics, and disorder in YbbN. In panel A, YbbN is shown as a ribbon diagram, with the N and C termini labeled. The Trx domain is in gold, and the two TPR-containing subdomains are in blue (subdomain A) and purple (subdomain B). Helices are lettered and strands are numbered in both panels A and B. The protein is highly prolate, and the Trx domain makes no direct contacts with the TPR domain. In panel B the ribbon diagram of YbbN is shown rotated by 90 degrees about the horizontal. The electrostatic surface for YbbN is shown in panels C and D in the same orientations as in panels A and B, with red representing negative electrostatic potential and blue representing positive. YbbN presents a predominantly negatively charged surface to solution, particularly in the cleft between the two TPR subdomains. However, there is a basic (positive) patch of residues near the C-terminal region of YbbN. Units of electrostatic potential are kT/e, and the temperature was 300 K. In panel E thermal ellipsoids at 75% probability level for all Cα atoms are shown for the refined TLS model of domain mobility in YbbN, emphasizing the greatly elevated disorder of the Trx domain compared with the better-ordered TPR domains. Ellipsoid color indicates the displacement magnitude, ranging from B values of 10 Å2 (blue) to 80 Å2 (red).
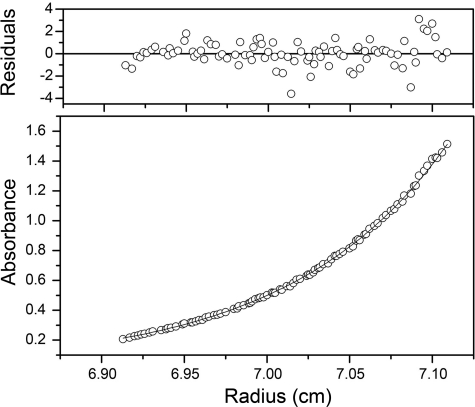
YbbN is an acidic protein (predicted pI 4.5), and the electrostatic surface potential calculated by APBS (28, 29) indicates that both domains of the protein present a predominantly negatively charged surface to solution (Fig. 1, C and D). However, there are two patches of neutral and basic residues in YbbN; one located in the N-terminal Trx domain and the other in the two C-terminal α-helices comprising residues 255–284 (Fig. 1, C and D). The C-terminal cluster is the more basic of the two. The molecule is also highly prolate, measuring ∼90 Å along the long axis and ∼25 Å along the two shorter axes, which would be expected to result in unusual hydrodynamic behavior. This is relevant because a previously reported gel filtration experiment indicated that reduced YbbN was dimeric, with an estimated mass of 65 kDa (10). However, an analysis of lattice contacts in the crystal structure shows no evidence of a significant dimeric interface between neighboring molecules. The solution behavior of YbbN was investigated using sedimentation equilibrium centrifugation, which is insensitive to the shape of the molecule and indicates that the reduced protein is monomeric in solution over the 8–30 μm concentration range. An adequate fit to the data can be obtained assuming a single ideal monomeric species of 31.6 kDa, which is in excellent agreement with the calculated monomeric mass of 32.1 kDa (Fig. 2). Inclusion of a more complex self-association model does not improve the fit as judged by the residuals or the reduced χ2 value. Therefore, we propose that YbbN is a monomer in solution whose prolate shape results in an anomalous hydrodynamic volume that may have confounded previous mass estimates using gel filtration.
FIGURE 2.
YbbN is a monomer in solution. Sedimentation equilibrium ultracentrifugation of YbbN was performed at three rotor speeds and protein concentrations and globally fit to determine molecular mass. A representative run is shown. The best-fit model (shown in the solid line in the lower panel) corresponds to the YbbN monomer and agrees well with the measured absorbance of the protein at 280 nm as a function of radius (open circles). The residuals (top panel) between data and model are randomly distributed and lack a systematic trend, indicating adequate fit.
The Trx Domain Contains a Redox-inert Active Site
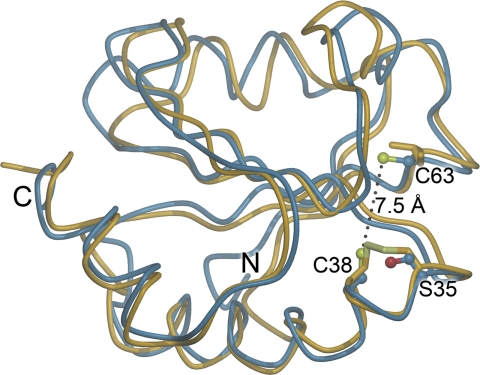
Sequence alignment has shown that the Trx domain of YbbN contains only one of the two catalytic cysteine residues in the Trx active site. In E. coli Trx, Cys-32 and Cys-35 define the active site and participate in thiol-disulfide redox chemistry, with the more reactive Cys-32 having a pKa value of ∼7, whereas Cys-35 has a pKa value of >10 (30). The lower pKa value for Cys-32 is catalytically important, as it promotes thiolate formation at physiological pH and initiates attack at disulfide bonds in substrates (30). In YbbN, Cys-38 corresponds to the high pKa Cys-35 in E. coli Trx, whereas YbbN has redox-inactive Ser-35 in the location equivalent to the more reactive Cys-32 in E. coli Trx, as previously noted based on a homology model of the YbbN Trx domain (10). YbbN possesses a second cysteine at residue 63 that has been proposed to serve as an atypical second member of the two-Cys active site (10) and is well conserved in YbbN-like proteins. This residue is 7.5 Å away from Cys-38 (Fig. 3), which is large compared with the 3.8 Å distance between thiols in reduced E. coli thioredoxin (31). Therefore, disulfide formation between Cys-38 and Cys-63 would require significant conformational changes in the Trx domain that would have to bring these two thiols together across an intervening β-strand, which seems unlikely. Due to the absence of a canonical Trx-like active site containing two cysteines and relatively poor sequence conservation in this region, YbbN is mostly likely not a thiol-disulfide oxidoreductase in vivo, consistent with recent biochemical data (8). We note that although the absence of a two-Cys motif in the YbbN Trx domain makes a classical thiol-disulfide oxidoreductase activity unlikely, it does not completely exclude other potential redox-relevant activities.
FIGURE 3.
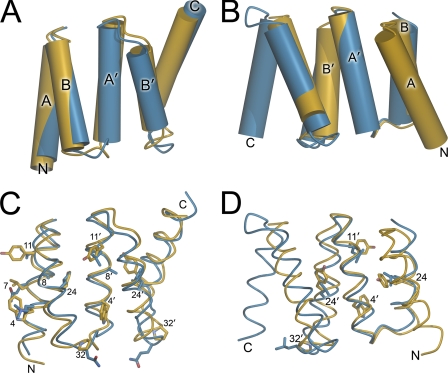
The Trx domain of YbbN lacks a conventional CXXC active site. A superposition of the Trx domain of YbbN (blue) and oxidized E. coli Trx (PDB 2TRX (49); yellow) is shown with the key cysteine residues of both proteins shown in stick representation. Residue number corresponds to YbbN, and the N and C termini of the domains are indicated. YbbN lacks the classical CXXC motif because residue 35, which is the more reactive cysteine in Trx, is a serine in YbbN. Although another cysteine residue (Cys-63) is present in YbbN, it is too distant from Cys-38 to participate in Trx-like thiol-disulfide exchange chemistry. This position is occupied by an isoleucine in Trx.
YbbN Contains a Cryptic TPR Domain
The TPR domain of YbbN contains two similar subdomains of five helices each that are separated by a kink at residue 198. The structure of this domain reveals that these subdomains each contain two tandem TPRs; however, several sequence-based bioinformatics tools failed to identify this region as containing TPR motifs before structure determination. TPRpred (32) is the best-performing of these programs and correctly predicted regions 114–148, 148–181, and 216–249 as TPRs with p values of ∼2 × 10−4 and a whole protein p value of 5 × 10−8, corresponding to a 53% chance of YbbN being a TPR protein according to the program. This prediction is accurate; however, the YbbN TPRs diverge from the canonical TPR motif and have, on average, only 11% identity with a consensus TPR sequence (33).
Despite sharing only modest sequence similarity to other TPR proteins, the peptide backbones of two C-terminal subdomains of YbbN individually superimpose well with the crystal structure of a designed consensus TPR motif (33). Subdomain A of YbbN agrees better with the canonical TPR motif (Cα r.m.s.d. of 1.4 Å from residues 112–198) than does subdomain B, which superimposes more poorly (Cα r.m.s.d. of 1.6 Å) and only over a more restricted core region (Fig. 4). In subdomain B, the best alignment was one in which YbbN subdomain B lacks the first helix of the first consensus TPR motif and where there is one “extra” capping helix at the C terminus (Fig. 4, B and D). This was a surprising result that violates the standard topology of TPR motifs (34) but was the optimal structural alignment using brute force least squares alignment optimization in LSQMAN (35). We note that it is possible to align subdomain B and the idealized TPR crystal structure in a way that preserves the standard TPR topology and places all helices into correspondence, but the quality of this alignment is markedly poorer. Additionally, many core TPR “signature residues” (33, 34) are substituted with other amino acids in YbbN, with a notable preference for replacement of Tyr in the consensus sequence with Leu (Fig. 4, C and D). Although the Leu and Tyr are both hydrophobic residues, they differ significantly in side-chain volume, and YbbN favors the smaller Leu at most consensus Tyr positions in the TPR sequence.
FIGURE 4.
The C-terminal domain of YbbN contains atypical TPR motifs. The YbbN TPR motifs (blue) are superimposed with the crystal structure of an idealized TPR motif (PDB code 1NA3; yellow). In panels A and B, the helices are represented as cylinders and are labeled A or B to indicate the corresponding helix of the idealized TPR motif. N and C termini of the domains are labeled. Panel A shows the close agreement between TPR subdomain A of YbbN (blue) and the idealized TPR motif (yellow). The capping helices are located on the C terminus of the domains. In panel B, the best superposition of YbbN TPR subdomain B (blue) and the idealized TPR motif (yellow) involves an unusual staggered alignment where the YbbN TPR subdomain B lacks the N-terminal A helix and has two C-terminal capping helices. Panels C and D provide a more detailed view of the superimposed TPR domains in the same orientations as panels A and B. Highly conserved TPR consensus residues that differ between YbbN and the idealized TPR motif are shown in stick representation, with standard TPR numbering as described in Main et al. (33).
TPR motifs often assemble into superhelical structures whose large surface area provides a platform for protein interaction (34, 36). This type of superhelical TPR assembly in YbbN would require oligomerization and is prevented by the capping helices on the C terminus of TPR subdomain B and the Trx domain at the protein N terminus. It is possible that these two structural features on either end of the molecule function to prevent multimerization of the protein and thereby help maintain soluble, monomeric YbbN. In addition, the C terminus of the protein coordinates a calcium ion in this structure, and residues in this region of YbbN are highly conserved (supplemental Figs. 1 and 2). The Ca2+ binding site is composed of residues from two molecules in the lattice and thus is likely an artifact of the crystallization condition (which contains 200 mm Ca(CH3COO)2) and lattice packing. However, it is noteworthy that the C-terminal regions of the related crystal structures 2R5S and 2QDN (see below) also interact with neighboring molecules and buffer components and, furthermore, that this region of YbbN is electrostatically distinct from the rest of the protein (Fig. 1, C and D), hinting at a possible functional significance of the C-terminal region.
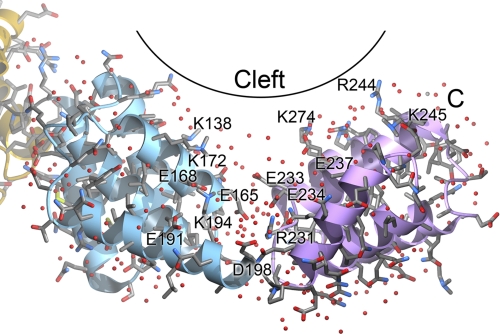
The two TPR motif subdomains in YbbN define a solvent-filled groove that is rich in charged residues and may provide a binding site for other proteins (Fig. 5). This cleft defines the underside of the TPR domain “saddle” and is ∼26 Å long and 12–14 Å across the solvent-rich portion. A joint located at a break between the C-terminal α-helix in TPR subdomain A and the N-terminal helix of subdomain B may allow for segmental flexibility of the two TPR subdomains that could modulate the cleft dimensions. The base of the cleft is rich in negatively charged residues; however, several lysine and arginine residues are present farther up on the cleft walls, creating a shallow and electrostatically varied surface that is reminiscent of the interaction surface of the TPR-containing Hop protein with the C-terminal peptide of Hsp90 (37). Several of these basic residues are well conserved in YbbN homologues (supplemental Fig. 1), including Arg-231, Arg-244, Lys-245, Arg-274, Arg-276, and Arg-277.
FIGURE 5.
A potential binding cleft in the TPR domain. The two TPR subdomains form a solvent-rich cleft in YbbN that may form an interaction surface with other proteins. The YbbN peptide backbone is shown as a ribbon diagram and is colored as in Fig. 1. A variety of charged amino acids are located in this cleft, creating an electrostatically varied surface that may bind to other proteins. Ordered water molecules are depicted as red spheres.
The TPR domain of YbbN is similar to a protein of unknown function from Vibrio parahaemolyticus (PDB code 2R5S), which was the most closely related protein of known structure when the crystal structure of YbbN was being solved and was used to determine the structure of YbbN by molecular replacement. The reported 2R5S crystal structure is only the C-terminal TPR domain of the full protein sequence, which includes an N-terminal Trx domain and is a clear homologue of YbbN. In addition, during the preparation of this manuscript, a crystal structure for a YbbN homologue from Salmonella typhimurium (PDB code 3QDN) was also deposited in the Protein Data Bank. This protein and E. coli YbbN are 87% identical and share all of the same structural features, although slight differences in the relative orientations of the domains give rise to an overall Cα r.m.s.d. of 2.5 Å despite having per-domain Cα r.m.s.d. values of ∼0.6 Å.
YbbN Binds to GroEL
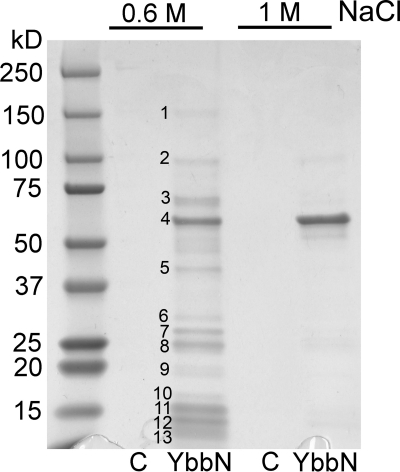
TPR motifs typically bind to other proteins (34), suggesting that protein interaction may be an important component of YbbN function. Supporting this contention, prior studies have identified a variety of YbbN-interacting proteins using the resin-immobilized protein as bait in pulldown studies (8). We sought to determine whether the previously identified interactors could be corroborated in an independent experiment. YbbN was immobilized on cyanogen bromide-derivatized resin and used to pull down interacting soluble proteins from cleared E. coli K-12 lysate, similar to previous work (see “Experimental Procedures”). A number of bound proteins could be eluted using 0.6 m NaCl (Fig. 6), whose identities were determined by trypsinization/mass spectrometry and are shown in Table 2. Some of these proteins were previously identified in other interaction experiments, such as GroEL and DnaK (8). One protein, DNA-directed RNA polymerase β (rpoB), is functionally related to previous identified DNA polymerase III interactors (9). Several of the other interacting proteins are ribosomal proteins as well as the E1 subunit of pyruvate dehydrogenase and phosphoribosylpyrophosphate synthetase. In contrast to previous reports (8), YbbN was not identified as a self-interactor in this experiment, consistent with structural results indicating a monomeric protein. In the more stringent 1 m NaCl wash, highly purified GroEL is the dominant protein visible on a Coomassie-stained SDS-PAGE gel (Fig. 6), indicating avid and direct binding of these two proteins. To identify very strongly associated proteins, a sample of protein-bound resin was heated in denaturing SDS-PAGE loading buffer after the 1.0 m NaCl elution and analyzed by SDS-PAGE. The gel shows two strong bands; one at ∼57 kDa corresponding to bound GroEL and one at ∼32 kDa corresponding to YbbN that had been removed from the resin by harsh denaturing elution (supplemental Fig. 3), supporting the conclusion that GroEL binds robustly to YbbN.
FIGURE 6.
YbbN interacts with a variety of proteins in E. coli lysate. Proteins retained from sonicated E. coli lysate on a YbbN affinity resin were eluted with either a 0.6 m or a 1.0 m NaCl wash. Negative control lanes (C) were those fractions that eluted from unmodified resin that had been incubated with lysate. The gel was stained with Coomassie Blue, and numbers indicate protein bands that were excised and identified using mass spectrometry (see Table 2 for identities). Band 4 is GroEL, which is the dominant protein present in the more stringent 1 m NaCl wash and interacts robustly with YbbN.
TABLE 2.
E. coli proteins that interact with YbbN
| Protein band number | Protein ID by mass spectrometry | MASCOT protein score |
|---|---|---|
| 1 | DNA-directed RNA polymerase β subunit | 1754 |
| 2 | Pyruvate dehydrogenase E1 | 1741 |
| 3a | 30 S ribosomal protein S1; Hsp90, DnaK | 2323; 1436; 1063 |
| 4 | GroEL | 9424 |
| 5 | Elongation factor Tu | 2039 |
| 6 | Phosphoribosyl pyrophosphate synthetase | 1920 |
| 7 | 30 S ribosomal subunit S2 | 700 |
| 8 | 50 S ribosomal subunit L1 | 753 |
| 9 | 50 S ribosomal subunit L6 | 1271 |
| 10b | 30 S ribosomal subunit S2 | 544 |
| 11a | 50 S ribosomal subunit L9; GroES | 2131; 1144 |
| 12 | 50 S ribosomal subunit L24 | 1223 |
| 13 | 50 S ribosomal subunit L27 | 1017 |
a Protein bands for which there were multiple high confidence hits with comparable protein molecular weights in mass spectrometric identification indicate that a mixture of proteins was present in the band. These proteins are named and separated by semicolons in the right-hand column.
b A high scoring hit that shows a disparity in the predicted protein mass and observed band migration.
YbbN Mildly Inhibits GroESL Chaperone and ATPase Activities
Due to the apparently strong interaction between YbbN and GroEL, the ability of YbbN to facilitate refolding of CS either alone or in combination with the GroESL chaperonin system was investigated. Guanidinium HCl-denatured CS was diluted into refolding buffer containing the test protein and allowed to refold for 100 min at room temperature followed by a 5,5′-dithiobis-(2-nitrobenzoic acid)-based activity assay to determine the amount of recovered CS activity (see “Experimental Procedures”). In contrast to a previous report (8), we found that YbbN was no more effective than the negative control protein lysozyme at facilitating refolding of CS, indicating that YbbN lacks foldase chaperone activity in this assay (Fig. 7A). We note, however, that this assay does not directly measure holdase chaperone activities, and thus it remains possible that YbbN could possess such an activity that would not be detected in this experiment. Additionally, although we do not detect any chaperone activity for YbbN against the generic substrate CS, a prior study indicates that YbbN may have a more specific chaperone activity against substrates such as DnaN (9), which we did not test. GroESL significantly enhanced the recovery of CS enzymatic activity compared with the negative control, as expected. Surprisingly, we found that YbbN mildly inhibited GroESL chaperone activity when these proteins were combined at a 2:1 stoichiometry of YbbN to GroEL monomer. The kinetics of CS activity recovery was also monitored, and YbbN inhibited GroESL activity over the entire 120-min duration of the kinetics experiment (supplemental Fig. 4). At early time points (up to 40 min), YbbN almost completely inhibited GroESL-assisted recovery of CS activity. As an independent measure of GroESL activity, we measured the ATPase activity that plays an important role in regulating conformational changes in the GroESL complex, which facilitates folding and release of substrate protein. Consistent with mild inhibition of GroEL chaperone activity, YbbN also decreases the release of inorganic phosphate release from GroEL-bound ATP as monitored using Malachite Green (Fig. 7B). We note that YbbN alone shows a measureable ATPase activity in this assay, although the relevance of this weak activity is unclear.
YbbN may interact with GroEL either as a substrate or, alternatively, as a negative regulator. Although the details of YbbN-GroEL binding are still unclear, the data suggest that YbbN is a negative regulator of GroEL activity in vitro. If YbbN were a GroESL substrate, it could lead to the observed decrease in CS activity recovery by competing with CS for GroESL binding. However, if this model were correct, the GroEL ATPase activity that is coupled to client protein folding and release would either be unaffected or enhanced by YbbN binding as a substrate, which is not observed. In addition, YbbN does not display the characteristics of a poorly folded protein during purification, forms well diffracting crystals, and does not have a significant amount of solvent-exposed hydrophobic surface area in this structure. All of these observations inveigh against the hypothesis that YbbN is a transiently unfolded client of GroESL and suggest that it interacts with GroEL in a regulatory fashion.
DISCUSSION
Multidomain proteins containing a Trx-like domain are widespread; however, only a few have been structurally characterized in their full-length forms (38). YbbN is the first crystal structure of a Trx-TPR domain fusion protein to be reported in the literature and reveals that the Trx domain is likely catalytically inactive and makes few direct contacts with the rest of the protein. Although the classical Trx active site is not preserved in YbbN, the Trx domain does contain two cysteine residues, and early reports suggested that YbbN possesses a weak oxidoreductase activity (10). Therefore, it is possible that this domain may retain some redox role, possibly similar to other 1-Cys variants of the Trx-fold that have glutaredoxin activities (39). However, we note that the CXXC motif in YbbN homologues is poorly conserved at both cysteine positions (supplemental Fig. 1), and several close YbbN homologues from genera Actinobacillus, Haemophilus, and Vibrio with ∼50% sequence identity to YbbN lack both active site cysteine residues. This observation requires either that some of YbbN homologues are redox active, whereas others are not, or that the entire group of YbbN homologues are not functional oxidoreductases. The later hypothesis is more consistent with recent biochemical data (8).
Because it lacks a classical oxidoreductase activity, the function of the Trx domain in YbbN is unclear. We and others (8) failed to identify any common Trx-interacting proteins in pulldown experiments with YbbN, suggesting that the Trx domain does not function as a specific “bait” domain to facilitate interactions between YbbN and classical Trx binding proteins. Some Trx-fold proteins, such as the peroxiredoxins, can form large annular oligomers in stress conditions and activate a latent chaperone activity (40), and it has been proposed that the Trx-fold may have been the template for an ancient proto-chaperonin (41). Although this would provide an appealing explanation for the role of the Trx domain in YbbN and is consistent with some previous work (10), we found using sedimentation equilibrium centrifugation that YbbN is a monomer is solution, and we could detect no chaperone activity for YbbN using a standard foldase assay. Instead of favoring oligomerization, the crystal structure of YbbN suggests that the N-terminal Trx domain might prevent the association of the C-terminal TPR motifs into larger assemblies, which is a common mode of self-interaction for TPR motifs (33, 36). We therefore speculate that the Trx domain in YbbN may serve a purpose that is similar to that of synthetic Trx domain fusions used in recombinant protein expression; it facilitates the formation of soluble, well folded, monomeric YbbN protein and prevents unwanted aggregation. This hypothesis is amenable to direct testing both in vitro and in vivo.
Protein interaction studies by multiple groups (8, 11) have shown that YbbN interacts with GroEL, which we corroborate in this work. In this study YbbN bound GroEL more strongly than any other detected protein in E. coli lysate, as judged by its elution behavior from a YbbN affinity resin. Poorly folded proteins often bind to GroEL tenaciously during overexpression and purification (42), and we cannot rule out that YbbN samples a partially unstructured conformation that binds to GroEL in this experiment. However, His6-tagged YbbN did not co-purify with large amounts of GroEL during purification, contrary to the expected copurification of the two proteins if YbbN were partially unstructured and strongly binding to GroEL as a substrate, suggesting that YbbN does not act as a substrate for GroEL under these conditions. It would be interesting to know how these two proteins interact and how this interaction modulates GroEL activity. Although the binding interface between YbbN and GroEL is not yet known, TPR domains are protein interaction motifs that are known to bind to chaperones (37, 43, 44), and thus we propose that the C-terminal TPR domain of YbbN is likely involved in this interaction.
We found that YbbN mildly inhibits GroESL chaperone function in refolding chemically denatured CS and also inhibits GroESL ATPase activity. The inhibitory effect of YbbN on GroESL was surprising and suggests a potential role for YbbN in regulating GroESL function. Previous work has shown that YbbN enhances the chaperone activity of the DnaK-DnaJ-GrpE complex (8), an alternative prokaryotic chaperone that is homologous to eukaryotic Hsp70. DnaK-DnaJ-GrpE is not functionally redundant with GroESL, and these chaperones have distinct substrate preferences (45). As YbbN appears to be a positive regulator of DnaK-DnaJ-GrpE (8) and a negative regulator of GroESL, we propose the hypothesis that YbbN may function to manage client protein traffic to prokaryotic chaperones by actively favoring the DnaK-DnaJ-GrpE system. This would serve to direct badly misfolded proteins to the DnaK system first, which will then either directly refold these proteins or pass a partially folded intermediate to the GroESL system for completion of renaturation. This is advantageous because initial binding of extensively unfolded proteins to GroESL may result in many futile cycles of substrate binding, abortive incomplete folding, and rebinding of the still-damaged protein to GroESL. The initial action of the DnaK-DnaJ-GrpE system in directing partially folded intermediates to downstream chaperonins is consistent with current models of the integration of chaperone networks in E. coli (46), making such an activity for YbbN plausible. To the best of our knowledge YbbN would be the first protein identified as a coordinate regulator of the GroESL and DnaK chaperone systems, although this hypothesis is still speculative and requires additional testing.
There are other proteins in addition to YbbN that contain both a Trx domain and TPR motifs. The best studied of these proteins is Arabidopsis thaliana TDX, which contains an N-terminal set of three TPR motifs and a C-terminal Trx domain (47), a reversed domain order compared with YbbN. A. thaliana TDX is both a chaperone and an oxidoreductase and can protect Arabidopsis against heat stress when overexpressed. Unlike YbbN, A. thaliana TDX contains a classic CXXC Trx active site (48) and forms higher oligomers that convert the protein from a foldase to a holdase chaperone (47). Although the three-dimensional structure of A. thaliana TDX is not known, it has been proposed that the TPR domain binds to and masks the active site of the Trx domain, implying a direct interaction between these domains (47). This is in sharp contrast to YbbN, which lacks robust oxidoreductase activity, has no contacts between the TPR and Trx domains in the crystal structure, and does not possess a foldase chaperone activity. Therefore, even within the single class of Trx-TPR domain fusions, there is substantial functional diversity among the constituent members.
Supplementary Material
Acknowledgments
We thank Dr. Ronald Cerny and the Nebraska Mass Spectrometry Core Facility for mass spectrometry protein identification, Dr. Nandakumar Madayiputhiya and the Redox Biology Center Mass Spectrometry Core facility for assistance with lysine modification studies, Dr. James Bardwell (University of Michigan) for discussion of YbbN chaperone assays, and Drs. Todd Holyoak and Mark Fisher (University of Kansas Medical Center) for discussion of GroESL chaperone activity. GM/CA-CAT has been funded in whole or in part with federal funds from the National Institutes of Health (NCI, Y1-CO-1020; NIGMS, Y1-GM-1104). Use of the Advanced Photon Source was supported by the United States Department of Energy, Basic Energy Sciences, Office of Science, under Contract DE-AC02–06CH11357.
This work was supported, in whole or in part, by National Institutes of Health Grant R01GM092999-01 (to M. A. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
The atomic coordinates and structure factors (code 3QOU) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- Trx
- thioredoxin
- BLAST
- basic local alignment search tool
- CCD
- charge coupled device
- CS
- citrate synthase
- DMAB
- dimethylaminoborane
- r.m.s.d.
- root mean square deviation
- TLS
- translation-libration-screw
- TPR
- tetratricopeptide repeat.
REFERENCES
- 1. Holmgren A. (1985) Annu. Rev. Biochem. 54, 237–271 [DOI] [PubMed] [Google Scholar]
- 2. Saitoh M., Nishitoh H., Fujii M., Takeda K., Tobiume K., Sawada Y., Kawabata M., Miyazono K., Ichijo H. (1998) EMBO J. 17, 2596–2606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gough J., Chothia C. (2002) Nucleic Acids Res. 30, 268–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Atkinson H. J., Babbitt P. C. (2009) PLoS Comput. Biol. 5, e1000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pan J. L., Bardwell J. C. (2006) Protein Sci. 15, 2217–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Richmond C. S., Glasner J. D., Mau R., Jin H., Blattner F. R. (1999) Nucleic Acids Res. 27, 3821–3835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nonaka G., Blankschien M., Herman C., Gross C. A., Rhodius V. A. (2006) Genes Dev. 20, 1776–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kthiri F., Le H. T., Tagourti J., Kern R., Malki A., Caldas T., Abdallah J., Landoulsi A., Richarme G. (2008) Biochem. Biophys. Res. Commun. 374, 668–672 [DOI] [PubMed] [Google Scholar]
- 9. Le H. T., Gautier V., Kthiri F., Kohiyama M., Katayama T., Richarme G. (2011) Biochem. Biophys. Res. Commun. 405, 52–57 [DOI] [PubMed] [Google Scholar]
- 10. Caldas T., Malki A., Kern R., Abdallah J., Richarme G. (2006) Biochem. Biophys. Res. Commun. 343, 780–786 [DOI] [PubMed] [Google Scholar]
- 11. Butland G., Peregrín-Alvarez J. M., Li J., Yang W., Yang X., Canadien V., Starostine A., Richards D., Beattie B., Krogan N., Davey M., Parkinson J., Greenblatt J., Emili A. (2005) Nature 433, 531–537 [DOI] [PubMed] [Google Scholar]
- 12. Rypniewski W. R., Holden H. M., Rayment I. (1993) Biochemistry 32, 9851–9858 [DOI] [PubMed] [Google Scholar]
- 13. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 14. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Collaborative Computational Project Number 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 16. Arnold K., Bordoli L., Kopp J., Schwede T. (2006) Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 17. Perrakis A., Sixma T. K., Wilson K. S., Lamzin V. S. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 448–455 [DOI] [PubMed] [Google Scholar]
- 18. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 19. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 20. Schüttelkopf A. W., van Aalten D. M. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 [DOI] [PubMed] [Google Scholar]
- 21. Painter J., Merritt E. A. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 439–450 [DOI] [PubMed] [Google Scholar]
- 22. Brünger A. T. (1992) Nature 355, 472–475 [DOI] [PubMed] [Google Scholar]
- 23. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fenn T. D., Ringe D., Petsko G. A. (2003) J. Appl. Crystallogr. 36, 944–947 [Google Scholar]
- 25. Lakshminarasimhan M., Maldonado M. T., Zhou W., Fink A. L., Wilson M. A. (2008) Biochemistry 47, 1381–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Laue T. M., Shah B. D., Ridgeway T. M., Pelletier S. L. (1992) in Analytical Ultracentrifugation in Biochemistry and Polymer Science (Harding S. E., Rowe A. J., Horton J. C. eds) pp 90–125, The Royal Society of Chemistry, Cambridge, UK [Google Scholar]
- 27. Winn M. D., Isupov M. N., Murshudov G. N. (2001) Acta Crystallogr. D Biol. Crystallogr. 57, 122–133 [DOI] [PubMed] [Google Scholar]
- 28. Dolinsky T. J., Czodrowski P., Li H., Nielsen J. E., Jensen J. H., Klebe G., Baker N. A. (2007) Nucleic Acids Res. 35, W522–W525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dyson H. J., Jeng M. F., Tennant L. L., Slaby I., Lindell M., Cui D. S., Kuprin S., Holmgren A. (1997) Biochemistry 36, 2622–2636 [DOI] [PubMed] [Google Scholar]
- 31. Jeng M. F., Campbell A. P., Begley T., Holmgren A., Case D. A., Wright P. E., Dyson H. J. (1994) Structure 2, 853–868 [DOI] [PubMed] [Google Scholar]
- 32. Karpenahalli M. R., Lupas A. N., Söding J. (2007) BMC Bioinformatics 8, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Main E. R., Xiong Y., Cocco M. J., D'Andrea L., Regan L. (2003) Structure 11, 497–508 [DOI] [PubMed] [Google Scholar]
- 34. D'Andrea L. D., Regan L. (2003) Trends Biochem. Sci. 28, 655–662 [DOI] [PubMed] [Google Scholar]
- 35. Novotny M., Madsen D., Kleywegt G. J. (2004) Proteins 54, 260–270 [DOI] [PubMed] [Google Scholar]
- 36. Krachler A. M., Sharma A., Kleanthous C. (2010) Proteins 78, 2131–2143 [DOI] [PubMed] [Google Scholar]
- 37. Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., Bartunik H., Hartl F. U., Moarefi I. (2000) Cell 101, 199–210 [DOI] [PubMed] [Google Scholar]
- 38. Gaudet R., Bohm A., Sigler P. B. (1996) Cell 87, 577–588 [DOI] [PubMed] [Google Scholar]
- 39. Fomenko D. E., Marino S. M., Gladyshev V. N. (2008) Mol. Cells 26, 228–235 [PMC free article] [PubMed] [Google Scholar]
- 40. Jang H. H., Lee K. O., Chi Y. H., Jung B. G., Park S. K., Park J. H., Lee J. R., Lee S. S., Moon J. C., Yun J. W., Choi Y. O., Kim W. Y., Kang J. S., Cheong G. W., Yun D. J., Rhee S. G., Cho M. J., Lee S. Y. (2004) Cell 117, 625–635 [DOI] [PubMed] [Google Scholar]
- 41. Dekker C., Willison K. R., Taylor W. R. (2011) Proteins 79, 1172–1192 [DOI] [PubMed] [Google Scholar]
- 42. Katayama H., McGill M., Kearns A., Brzozowski M., Degner N., Harnett B., Kornilayev B., Matković-Calogović D., Holyoak T., Calvet J. P., Gogol E. P., Seed J., Fisher M. T. (2009) J. Struct. Funct. Genomics 10, 57–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cortajarena A. L., Wang J., Regan L. (2010) FEBS J. 277, 1058–1066 [DOI] [PubMed] [Google Scholar]
- 44. Prasad B. D., Goel S., Krishna P. (2010) PLoS One 5, e12761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schröder H., Langer T., Hartl F. U., Bukau B. (1993) EMBO J. 12, 4137–4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hartl F. U., Hayer-Hartl M. (2009) Nat. Struct. Mol. Biol. 16, 574–581 [DOI] [PubMed] [Google Scholar]
- 47. Lee J. R., Lee S. S., Jang H. H., Lee Y. M., Park J. H., Park S. C., Moon J. C., Park S. K., Kim S. Y., Lee S. Y., Chae H. B., Jung Y. J., Kim W. Y., Shin M. R., Cheong G. W., Kim M. G., Kang K. R., Lee K. O., Yun D. J., Lee S. Y. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 5978–5983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim S. G., Chi Y. H., Lee J. S., Schlesinger S. R., Zabet-Moghaddam M., Chung J. S., Knaff D. B., Kim S. T., Lee S. Y., Kim S. K. (2010) Biochim. Biophys. Acta 1804, 2213–2221 [DOI] [PubMed] [Google Scholar]
- 49. Katti S. K., LeMaster D. M., Eklund H. (1990) J. Mol. Biol. 212, 167–184 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.