Abstract
Humans lacking sclerostin display progressive bone overgrowth due to increased bone formation. Although it is well established that sclerostin is an osteocyte-secreted bone formation inhibitor, the underlying molecular mechanisms are not fully elucidated. We identified in tandem affinity purification proteomics screens LRP4 (low density lipoprotein-related protein 4) as a sclerostin interaction partner. Biochemical assays with recombinant proteins confirmed that sclerostin LRP4 interaction is direct. Interestingly, in vitro overexpression and RNAi-mediated knockdown experiments revealed that LRP4 specifically facilitates the previously described inhibitory action of sclerostin on Wnt1/β-catenin signaling. We found the extracellular β-propeller structured domain of LRP4 to be required for this sclerostin facilitator activity. Immunohistochemistry demonstrated that LRP4 protein is present in human and rodent osteoblasts and osteocytes, both presumed target cells of sclerostin action. Silencing of LRP4 by lentivirus-mediated shRNA delivery blocked sclerostin inhibitory action on in vitro bone mineralization. Notably, we identified two mutations in LRP4 (R1170W and W1186S) in patients suffering from bone overgrowth. We found that these mutations impair LRP4 interaction with sclerostin and its concomitant sclerostin facilitator effect. Together these data indicate that the interaction of sclerostin with LRP4 is required to mediate the inhibitory function of sclerostin on bone formation, thus identifying a novel role for LRP4 in bone.
Keywords: Bone, Human Genetics, Lipoprotein-like Receptor (LRP), Protein-Protein Interactions, Wnt Pathway
Introduction
Bone homeostasis and skeletal integrity require a tightly regulated equilibrium between the activity of bone-resorbing osteoclasts and bone-forming osteoblasts (1, 2). Osteoporosis is a disease where imbalance between both processes leads to low bone mineral density (BMD)5 and deterioration of bone structure. In contrast, some human patients present with abnormal high BMD due to either decreased resorption (osteopetrosis) or increased bone formation (high bone mass). Human genetic studies identified LRP5 as an important regulator of BMD (3). Osteoporosis-pseudoglioma syndrome (OMIM 259770) results from loss-of-function mutations in LRP5, whereas gain-of-function mutations in LRP5 cause high bone mass conditions (4–6). LRP5 is a co-receptor for secreted Wnt ligands and thus mediates Wnt/β-catenin signaling, which is a crucial pathway for embryonic and postnatal bone homeostasis (7). Consequently, bone phenotypes related to LRP5 loss- or gain-of-function mutations are thought to result from disturbed Wnt/β-catenin signaling. Likewise, mutations in Wnt co-receptor LRP6 have been linked to BMD changes in humans (8). In vitro studies demonstrated that LRP5 high bone mass mutations have an impaired interaction with the Wnt/β-catenin signaling inhibitors DKK1 and SOST (sclerostin) (4, 9–12). Sclerostin, encoded by the SOST gene, is a secreted glycoprotein acting as negative regulator of bone formation. Patients afflicted by sclerosteosis (OMIM269500) or Van Buchem disease (OMIM239100) display lifelong massive bone overgrowth with increased BMD and bone strength due to lack of sclerostin protein. Sclerosteosis patients have loss-of-function mutations in SOST, whereas Van Buchem disease patients lack a 52-kb non-coding region 35 kb downstream of the gene that contains an enhancer element implicated in adult SOST expression (13–17). Consistent with these facts and in line with findings in preclinical animal models, anti-sclerostin antibody treatment was found to increase serum markers of bone formation and BMD in healthy postmenopausal women (18, 19). Sclerostin is secreted by osteocytes, which are terminally differentiated cells of the osteoblastic lineage that represent over 90% of all cells in the adult skeleton (20, 21). Osteocytes are embedded within the mineralized bone matrix and are connected through a canalicular network with each other and with cells at the bone surface, including osteoblasts (22). It is thought that sclerostin passes through the osteocytic canalicular network to inhibit osteoblastic canonical Wnt/β-catenin signaling by binding to the Wnt co-receptors LRP5 and LRP6. However, conclusive in vivo proof for this hypothesis is lacking to date. In fact, it has been proposed recently that LRP5 might not impact bone homeostasis locally but rather act indirectly by controlling preosteoblast proliferation via inhibition of serotonin synthesis in the duodenum (23). Moreover, sclerostin was originally identified as a bone morphogenetic protein (BMP) antagonist in bone, based on amino acid sequence similarity to members of the DAN/Cerberus family of cystine knot-containing secreted glycoproteins and its ability to bind BMPs and thereby inhibit BMP signaling (24–26). In summary, although the role of sclerostin as an osteocyte-secreted bone formation inhibitor is well established, the underlying molecular mechanisms are not fully elucidated.
To further study sclerostin's mechanism of action, we performed a proteomics screen aiming at identification of its interaction partners. Here we report that the interaction between sclerostin and LRP4 (low density lipoprotein-related protein 4) is crucial to mediate the inhibitory function of sclerostin on Wnt1/β-catenin signaling and on bone formation. Moreover, we describe the identification of LRP4 mutations, which are associated with bone overgrowth and impaired sclerostin facilitator function.
EXPERIMENTAL PROCEDURES
Tandem Affinity Purification
In order to identify novel partners of sclerostin, we applied a systematic tandem affinity purification (TAP) method combined with mass spectrometry (27, 28). In brief, C-terminally tagged sclerostin amino acids (aa) 21–213 were stably expressed in HEK293T as well as in osteoblastic UMR-106 cells. TAP-tagged sclerostin was found to be secreted into the cell culture supernatant of either cell type. The medium was replaced with fresh medium (DMEM with 10% FCS) 48 h prior harvesting. Cells were harvested by mechanical detachment, washed with excess PBS on ice, and lysed in immunoprecipitation buffer as described (27). A membranous fraction was isolated and used as starting material for the TAP. Purified protein complexes were separated by one-dimensional SDS-PAGE and stained by colloidal Coomassie Blue. Entire gel lanes were systematically cut into slices, and proteins were in-gel digested with trypsin as described by Shevchenko et al. (29). Protein identification by liquid chromatography-tandem mass spectrometry was done using an Eksigent 1D+ HPLC system coupled to an LTQ Orbitrap mass spectrometer (Thermo-Finnigan). Peptide mass and fragmentation data were used to query an in-house curated version of the International Protein Index database using Mascot (Matrix Science). Candidate lists were filtered for secreted factors and membrane proteins by an in-house curated list.
Cell Culture, Plasmids, and Production of Recombinant Proteins
All cell lines used were grown in standard culture conditions and medium (Invitrogen). C28a2 chondrocytic cells were obtained from Dr. M. Goldring (Massachusetts General Hospital). Kusa-A1 (JCRB1119) cells were purchased from the Japan Health Sciences Foundation, Health Science Research Resources Bank. All plasmids were generated using standard molecular biology techniques, and details will be provided upon request. Plasmids for the Wnt signaling assay were described elsewhere (30).
For human LRP4 overexpression experiments in the Wnt signaling assay, the following constructs harboring a CMV promoter were generated. Full-length LRP4 refers to aa 1–1905; LRP4Δcytoplasm refers to aa 1–1748; and LRP4ΔECD refers to aa 1723–1905 (LRP4 numbering refers to NP002325 and corresponds to ENSP00000378623 with 38 exons lacking exon 2). Mutations R1170W and W1186S used in Fig. 5, A and B, were generated by site-directed mutagenesis.
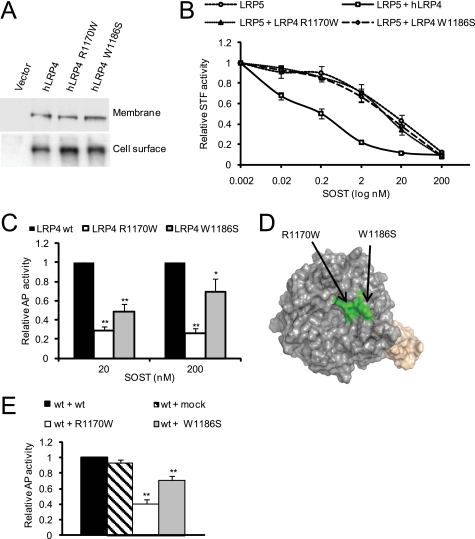
FIGURE 5.
LRP4 mutations (R1170W and W1186S) impair sclerostin interaction and concomitant sclerostin-enhancer function. A, expression levels of LRP4 mutants (R1170W and W1186S) are similar to wild type LRP4. HEK293 were transfected with LRP4 wild type and mutant expression vectors and subjected to membrane-specific protein extracts (top; quantification compared with WT 100%: R1170W 95% and W1186S 104%) or cell surface proteins isolated by biotinylation (bottom; quantification compared with WT 100%: R1170W 113% and W1186S 114%) prior to anti-LRP4 immunoblotting. B, LRP4 mutants (R1170W and W1186S) have impaired sclerostin enhancer function in Wnt1/β-catenin signaling inhibition. HEK293 cells were transiently transfected with LRP4 wild type and mutants, STF-LUC reporter plasmid, and Wnt signaling-inducing (Wnt1 with or without LRP5) plasmids. Five h after transfection, sclerostin was added in a dose-dependent manner for an additional 19 h. Cells were resuspended in lysis buffer, and luciferase levels were measured. C, mutations (R1170W and W1186S) in LRP4 lead to reduced sclerostin binding properties. Conditioned medium from HEK293 cells transfected for 4 days with wild type and mutant LRP4 ECD were harvested. Levels of secreted proteins were assessed by immunoblotting, and adjusted amounts were used as a source of LRP4 in the sclerostin LRP4 interaction ELISA, as shown in Fig. 1B. *, p < 0.05; **, p < 0.01 versus LRP4 WT. D, co-localization of LRP4 mutations (R1170W and W1186S) at the surface of propeller 3 based on the structural model. E, dominant negative effect of LRP4 mutants (R1170W and W1186S) on the SOST LRP4 wild type interaction. A combination of mutant LRP4 and wild type LRP4 was used as a source of LRP4 in the sclerostin LRP4 ELISA. **, p < 0.01 versus LRP4 WT plus medium. Error bars, S.E.
For production of secreted recombinant human LRP4 ECD, the following construct harboring a CMV promoter was generated: CD33-hsLRP4 aa 21–1763-hIgG1 (LRP4 numbering corresponds to ENSP0000025991 with 39 exons and therefore also takes into account exon 2). The CD33 leader sequence was introduced to allow for efficient secretion and the hIgG1 tag to facilitate protein purification. HKB-11 cells cultivated in suspension culture using a proprietary Novartis medium supplemented with 1% fetal calf serum were transfected at high cell density with 25-kDa linear polyethyleneimine at a ratio of plasmid DNA/polyethyleneimine of 1:3; the total plasmid quantity consisted of equal amounts of huLRP4, huMESD, and huRAP plasmids. Following a regimen of daily feeding of one volume of medium, the cells were transferred to a Wave bioreactor on day 3 post-transfection and expanded to a final volume of 10 liters. The supernatant was harvested 7 days post transfection, concentrated, and purified by affinity chromatography on a Protein A-Sepharose column. To assess the sclerostin binding properties of LRP4 mutants (mutations introduced by site-directed mutagenesis) in the ELISA (Fig. 5, C and D), conditioned medium was harvested 4 days after transient transfection of HEK293 with the different LRP4 ECD (WT, R1170W, and W1186S) variants and equal amounts of huMESD and huRAP plasmids. Levels of LRP4 ECD were assessed by LRP4 immunoblotting (Atlas), and normalized amounts were used as a source of LRP4.
Recombinant human sclerostin was harvested from the supernatant of HKB11 (31) stably expressing sclerostin aa 24–213 with a proprietary 6-aa affinity tag suitable for detection and purification. In addition, sclerostin aa 24–213 was also produced in Escherichia coli bacteria using standard procedures. For both ways of production, quality control was performed by HPLC. Recombinant human DKK1 and LRP6 were purchased from R&D Systems.
ELISA Interaction Assay
Sclerostin was precoated for 24 h prior to the addition for 1 h of LRP4 ECD marked with an hIgG1 tag. Upon blocking in a buffer containing 5% BSA, 10% FCS and Superblock T20 (Thermo) and washing steps (0.1% Tween 20 in PBS), LRP4 was detected with the help of anti-human IgG (Fc-specific) alkaline phosphatase-conjugated antibody (Sigma-Aldrich), alkaline phosphatase solution (Sigma-Aldrich), and standard techniques. In Fig. 1C, isotype control antibody or an antibody against sclerostin (R&D Systems) was added after LRP4 and incubated with the preformed sclerostin LRP4 complex for 1 h.
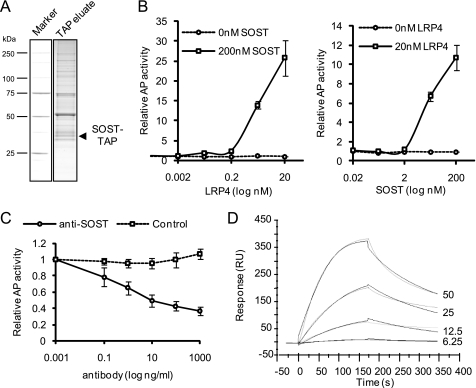
FIGURE 1.
LRP4 is a direct SOST interaction partner. A, LRP4 was identified as sclerostin interaction partner by tandem affinity purification. C-terminal TAP-tagged sclerostin was expressed into HEK293T and affinity-purified from a cellular membrane fraction. The purified material was then loaded on an SDS gel and stained with Coomassie Blue as exemplified for one gel lane. The interaction partners were identified through mass spectrometry. B, sclerostin interacts with LRP4 in an ELISA. Sclerostin was precoated prior to the addition of recombinant LRP4 ECD. After binding and washing, LRP4 was detected by anti-Fc antibody coupled to alkaline phosphatase (AP). The left panel shows an LRP4 dose-response curve with a fixed sclerostin dose of 200 nm. The right panel shows a sclerostin dose-response curve with a fixed LRP4 dose of 20 nm. Interaction is shown as alkaline phosphatase activity (arbitrary units). C, sclerostin antibody disrupts a preformed sclerostin LRP4 complex. Binding of sclerostin (200 nm) to LRP4 (5 nm) was challenged with control antibody or antibody against sclerostin in a dose-response manner. Detection was as described in B. D, sclerostin interacts with LRP4 in surface plasmon resonance. Binding of sclerostin (four concentrations, 6.25, 12.5, 25, and 50 nm, are shown in gray) toward immobilized LRP4 was measured. Sensograms (shown in black) were fitted using a 1:1 binding model. RU, response units. Error bars, S.E.
Surface Plasmon Resonance Interaction Assay
Human sclerostin or human LRP4 ECD was immobilized in 10 mm acetate, pH 4.5 or 4.0, respectively, using a CM5 sensor chip, amine-coupling kit, and Hepes-Buffered-Saline EDTA-Surfactant as running buffer in a BiacoreT100 system. Binding was measured at 20 °C with different concentrations of proteins in running buffer. Sensograms were fitted using a 1:1 binding. Binding of sclerostin to immobilized LRP6 was performed using the same conditions.
Wnt Signaling Reporter Gene Assay
HEK293 and C28a2 cells were transiently transfected with LRP4, STF-LUC reporter plasmid, and a combination of Wnt/β-catenin signaling-inducing plasmids (Wnt1, Wnt1/LRP5, or Wnt1/LRP6). Equal amounts of each plasmid were transfected, and total DNA quantities were adjusted with empty vectors. Five h after transfection, sclerostin or DKK1 was added in a dose-dependent manner for an additional 19 h. Cells were resuspended in lysis buffer, and Dual-Luciferase activities were measured (Promega). For down-regulation experiments, HEK293 cells stably expressing Wnt1 and the STF reporter (30) were transfected with control siRNA or siRNA against LRP4 (all synthesized in house) for 48 h prior to a 24-h incubation with sclerostin or DKK1. Luciferase levels were measured as noted above.
Immunohistochemistry
Human femoral neck bone samples were obtained from hip replacement patients afflicted with coxarthrosis. Bone biopsies were fixed in 4% paraformaldehyde phosphate-buffered solution, pH 7.4, for 3 days at 4 °C, decalcified in 15% EDTA solution, pH 7.4, dehydrated, and embedded in paraffin. Five-μm paraffin sections were processed according to standard protocols and incubated for 3 h at room temperature with a rabbit polyclonal antibody directed against human LRP4 (Orbigen) in 1% BSA, 0.1% Triton X-100 in Tris-buffered saline (TBS; Sigma-Aldrich), followed by a 1-h incubation with biotinylated goat anti-rabbit secondary antibodies and chromogenic detection using the ABC Elite Kit (both from Vector Laboratories) and 3,3′-diaminobenzidine substrate-chromogen (Dako). Prior to mounting, sections were briefly counterstained in Gill's hematoxylin solution (Merck). Images were collected on a Zeiss Axiophot microscope.
Similar staining procedures were used for mouse femora with the exception that femoral sections were incubated overnight with a rabbit polyclonal anti-LRP4 antibody (Atlas). Negative controls were processed in the same way but without the addition of primary antibodies.
Osteoblastic Differentiation Assays with Lentivirus-mediated shRNA Delivery
Kusa-A1 osteoblastic cells were transduced (multiplicity of infection of 5) with lentiviral particles harboring four different shRNAs against Lrp4 or -2 control shRNA from Sigma. Upon selection with 2 μg/ml puromycin (Invivogen), down-regulation of Lrp4 RNA levels were assessed by real-time quantitative PCR using Taqman gene-specific probes (Applied Biosystems). Two control shRNA and two shLRP4s were subsequently selected for osteoblastic differentiation experiments. The osteogenic mixture contained 5 mm β-glycerophosphate (32). Matrix-associated calcium levels were assessed 4 days after the induction of differentiation using a commercially available kit (AxonLab). This assay is based on the property of 2-cresolphthalein-complexone to react with calcium ion in an alkaline environment and give rise to a purple-red color. The color intensity is directly proportional to the calcium concentration and is measured spectrophotometrically (595 nm).
Material from Patient
Ethical approval for clinical investigations and informed consent by all patients were obtained prior to the study.
Direct Sequencing of PCR-amplified LRP4 DNA
Primer design was done using a template sequence (ENSG00000134569) retrieved from the Ensembl Data base (available on the World Wide Web), taking into account that Ensembl predicts that there are two transcripts in this gene (ENST0000025991 and ENST00000378623). Amplification was performed using GoTaq DNA polymerase-mediated PCR (Promega). Primers for PCR were designed to cover all 38 or 39 (depending on the transcript) coding exons and the respective exon-intron boundaries (primer sequences are available upon request). Amplification of the fragments was verified by agarose gel electrophoresis, simultaneously running a Generuler 100bp Plus DNA Ladder (Fermentas). To remove primers and unincorporated dNTPs, exonuclease I (New England Biolabs) and calf intestine alkaline phosphatase (Roche Applied Science) were used. Sequencing was carried out directly on purified fragments with the ABI 310 Genetic Analyzer (Applied Biosystems), using an ABI Prism BigDye terminator cycle sequencing ready reaction kit, version 1.1 (Applied Biosystems). The primers used here were identical to the primers that were used for amplification. The BigDye XTerminator purification kit was used as purification method for DNA sequencing with the purpose of removing unincorporated BigDye terminators.
Direct Sequencing of Spanish Patient cDNA
Total RNA was isolated from Epstein Barr virus-transformed cell lines with TRIzol according to the manufacturer's instructions (Invitrogen). Concentration and purity of RNA were verified using UV-visible spectrophotometry (NanoDrop ND-1000, Thermo Fischer Scientific) and agarose gel electrophoresis. The Superscript III first strand synthesis system for RT-PCR (Invitrogen) was used to synthesize first strand cDNA from total RNA. More precisely, 2.5 μg of total RNA was reverse transcribed with oligo(dT)20 primers provided with the kit. The cDNA products were directly used as templates for PCR amplification with the following primers: 5′-CCA GTG TCT ACA CCA TGT GC-3′ and 3′-GTT AAA ACC TAG TGG CTG TGC-5′. To improve efficiency of the amplification, we made use of a touchdown approach. This resulted in a 931-bp fragment containing the site of interest. Amplification of the fragment was verified by agarose gel electrophoresis, simultaneously running a Generuler 100bp Plus DNA Ladder. Enzymatic purification and sequencing were carried out as described above, using the same primers as were used for amplification. In addition, one extra primer for sequencing was used to cover the site of interest: 5′-CAC CAC AGG GCT ACA GAC C-3′.
Determination of LRP4 Expression Levels
HEK293 cells were transiently transfected for 24 h with the respective LRP4 expression plasmids. Membrane-enriched extracts performed using Mem-PER eukaryotic membrane protein extraction reagent (Pierce) or hypotonic buffer (10 mm KCl and 10 mm Tris-HCl) were subjected to anti-LRP4 immunoblotting (Atlas). Cell surface protein isolation was performed according to the manufacturer's instructions (Pierce cell surface protein isolation kit) prior to anti-LRP4 immunoblotting. RNA from human femoral neck and rodent calvaria/femor samples was extracted using TRIzol (Invitrogen), cDNA was synthesized, and gene expression was assessed by real-time quantitative PCR using Taqman gene-specific probes (Applied Biosystems).
Structural Modeling
LRP4 structural model was based on Swiss-Prot entry O75096. Sequence alignment identified eight LDL receptor class A repeats, 20 LDL receptor class B repeats, and six EGF-like domains. LRP4 is likely to have four β-propeller domains, each one formed by five LDL receptor class B repeats (referred to as blades) and followed by an EGF-like domain. For the modeling, each blade of the first LRP4 β-propeller was modeled on the structure of the closest blade forming the β-propeller of LDLR (Protein Data Bank code 1IJQ). The Protein Data Bank structure of tachylectin-2 from horseshoe crab (1TL2) was used as template to assemble the whole five-blade propeller domain of LRP4. The model of the EGF-like domain next to LRP4 propeller 1 was built by using as template the structure of the EGF-like 3 domain of LDLR (Protein Data Bank code 1IJQ). The final model of LRP4 β-propeller 1 followed by the EGF-like domain was obtained by superimposing the two built models with the LDLR structure (Protein Data Bank code 1IJQ). The localization of residues mutated in high bone mass present on propeller 3 was mapped on the structural model of propeller 1 by aligning the respective residues.
Statistical Analysis
Data representing mean ± S.E. t test (two-tailed, unpaired) have been used. The following p values were considered as significant: *, p < 0.05; **, p < 0.01.
RESULTS
SOST Interacts with LRP4 in a Direct Manner
To identify novel interaction partners of SOST we performed a series of TAP-MS experiments with a TAP-tagged version of the protein. C-terminal tagged sclerostin was stably expressed in HEK293T cells by retrovirus-mediated gene transfer. Monitoring the expression of tagged sclerostin by Western blotting demonstrated that the protein is robustly secreted into the cell culture supernatant and is present in a cellular membrane fraction (data not shown). Protein complexes were isolated (Fig. 1A) from both fractions (cell culture supernatant and membrane fraction) and were subsequently analyzed by tandem mass spectrometry. Identified candidate interactors were filtered for membrane proteins and secreted factors. We confirmed the binding of the published interaction partners LRP5 and LRP6 while identifying another member of the low density lipoprotein receptor-related protein family, LRP4, as a potential novel candidate interaction partner (Table 1). BMPs were not identified under any tested condition. Association of LRP4 with sclerostin was confirmed in an independent TAP-MS experiment from stably expressing osteoblastic UMR106 cells (data not shown). To test whether the interaction between LRP4 and sclerostin is direct, we produced both proteins as purified recombinant material. For LRP4, we generated constructs allowing for the secretion of the full extracellular domain (LRP4 ECD marked with a hIgG1 tag) consisting of four highly structured β-propellers. We performed ELISA experiments assessing the binding of LRP4 to precoated sclerostin by detection of LRP4 through an alkaline phosphatase-coupled anti-Fc antibody. Recombinant LRP4 dose-dependently bound to recombinant sclerostin (Fig. 1B, left). Varying doses of precoated sclerostin also depicted a dose dependence of the binding (Fig. 1B, right), showing that recombinant LRP4 is correctly folded at least for the part that interacts with sclerostin. The specificity of the sclerostin LRP4 interaction was further studied using a commercial anti-sclerostin antibody. Preincubation of the antibody with sclerostin prevented the binding of LRP4 to sclerostin (Fig. 1C). Moreover, the antibody was able to disrupt a preformed sclerostin LRP4 complex. Surface plasmon resonance experiments demonstrated that recombinant sclerostin bound to immobilized LRP4 with an affinity (KD) of 9 nm (Fig. 1D), whereas the binding of sclerostin to immobilized LRP6 was in the same range as LRP4 (KD of 15 nm). Performing the experiment in the opposite direction yielded similar binding affinities between sclerostin and LRP4 (data not shown). Taken together, these data demonstrate that sclerostin interacts with LRP4 in a direct manner.
TABLE 1.
Details of mass spectrometry-based protein identification of sclerostin interaction partners (reverse database-derived false positive rate <1%)
Sclerostin interaction partners were identified through tandem affinity purification screens. The following information is provided for the known interaction partner LRP5 and LRP6 as well as for the newly identified LRP4: representative International Protein Index (IPI) accession number, Entrez gene name, number of total spectral matches to the protein, and distinct identified peptide sequences.
| IPI accession number | Gene name | Mascot score | Total spectrum matches | Peptide sequence |
|---|---|---|---|---|
| IPI00306851.3 | LRP4 | 372 | 11 | ANLDGSER |
| IEAADLNGANR | ||||
| ELVFWSDVTLDR | ||||
| TVLIWENLDRPR | ||||
| VYYTDVFLDVIR | ||||
| ADKGTGSNVILVR | ||||
| AVLINNNLGWPNGLTVDK | ||||
| VLIGSQLPHPFGLTLYGER | ||||
| SEYTLLLNNLENAIALDFHHR | ||||
| IPI00744811.1 | LRP5 | 134 | 5 | NHVTGASSSSSSSTK |
| AIDYDPLDKFIYWVDGR | ||||
| IPI00000203.1 | LRP6 | 936 | 24 | AALDGSDR |
| AIVVNPEK | ||||
| CANGQCIGK | ||||
| IYWTDISLK | ||||
| ANGLTIDYAK | ||||
| CDHNVDCSDK | ||||
| DGATELLLLAR | ||||
| IESSDLSGANR | ||||
| IEVSNLDGSLR | ||||
| ILISEDLEEPR | ||||
| IEVSNLDGSLRK | ||||
| KILISEDLEEPR | ||||
| EASALDFDVTDNR | ||||
| EVIIDQLPDLMGLK | ||||
| AHVTGASSSSSSSTK | ||||
| AIDYDPLDKQLYWIDSR | ||||
| ISLETNNNNVAIPLTGVK | ||||
| ISLDTPDFTDIVLQLEDIR | ||||
| AIVLDPMVGYMYWTDWGEIPK |
Sclerostin-mediated Inhibition of Wnt1/β-Catenin Signaling Is Enhanced by LRP4
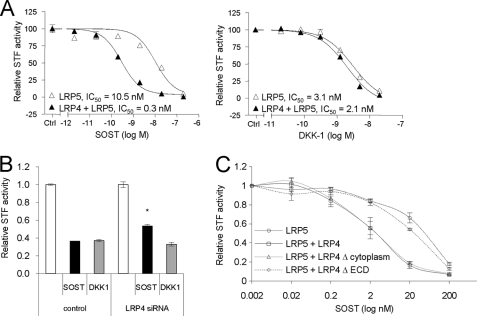
Next, we tested whether modulation of LRP4 levels by in vitro overexpression or RNAi-mediated knockdown experiments affects the ability of sclerostin to inhibit Wnt1/β-catenin signaling. Interestingly, sclerostin-mediated inhibition of Wnt1-induced signaling was increased in the presence of LRP4 in HEK293 (Fig. 2A and supplemental Fig. S1, B and C) and C28a2 cells (supplemental Fig. S1A), whereas the activity of the Wnt inhibitor and LRP4 ligand DKK1 was not modified, suggesting specificity for sclerostin under the tested conditions. These effects were observed in Wnt1/LRP5-induced (Fig. 2A), Wnt1/LRP6-induced (supplemental Fig. S1C), or Wnt1-induced (supplemental Fig. S1B) conditions, and the IC50 of sclerostin was decreased in the presence of LRP4 by 35-, 20-, and 5-fold, respectively. In the C28a2 chondrocytic cell line, where the basal sclerostin activity is lower than in HEK293, we observed not only a shift in potency but also an increased maximal inhibitory effect (supplemental Fig. S1A). Consistent with overexpression studies, partial 75% knockdown of LRP4 (supplemental Fig. S2B) in HEK293 diminished sclerostin- but not DKK1-mediated suppression of Wnt signaling (Fig. 2B). The relief of sclerostin inhibitory function was moderate yet significant and consistent, as documented with four different siRNAs against LRP4 (supplemental Fig. S2A). LRP4 truncation mutation studies proved that the extracellular β-propeller structured domain of LRP4 is required for the sclerostin facilitator activity (Fig. 2C). Indeed, a membrane-anchored LRP4 lacking the whole cytoplasmic tail (LRP4Δcytoplasm) was fully active in terms of sclerostin enhancer effect, whereas a LRP4 variant without the extracellular domain (LRP4ΔECD) was not able to exert the enhancer effect. Overall, the data demonstrate that LRP4 facilitates sclerostin's inhibitory action on the Wnt/β-catenin signaling reporter readout by binding with its extracellular domain to sclerostin.
FIGURE 2.
LRP4 is a specific facilitator of sclerostin-mediated inhibition of Wnt1/β-catenin signaling. A, LRP4 expression specifically facilitates sclerostin inhibitory action on Wnt1/β-catenin signaling. HEK293 cells were transiently transfected with LRP4, STF-LUC reporter plasmid, and Wnt signaling-inducing (Wnt1 + LRP5) plasmids. Five h after transfection, sclerostin or DKK1 was added in a dose-dependent manner for an additional 19 h. Cells were resuspended in lysis buffer, and luciferase levels were measured. B, down-regulation of LRP4 diminishes sclerostin action on Wnt1/β-catenin signaling. Stable HEK293-Wnt1-STF cells were transfected with control siRNA or siRNA against LRP4 for 48 h prior to a 24-h incubation with sclerostin or DKK1. *, p < 0.05 versus SOST in control. C, LRP4 extracellular domain is required to mediate the sclerostin facilitator effect. Membrane-anchored LRP4 truncation mutants (LRP4Δcytoplasm and LRP4ΔECD) were compared with full-length LRP4, as described in A. Error bars, S.E.
Sclerostin-mediated Inhibition of in Vitro Mineralization Is Blunted upon Lrp4 Silencing
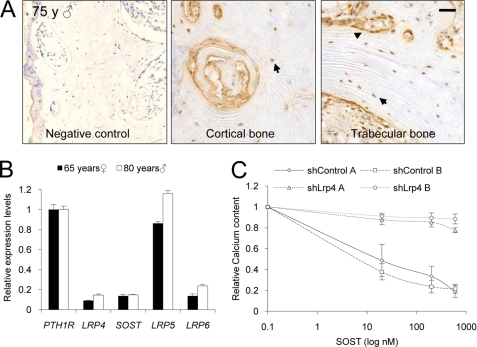
We found Lrp4 RNA to be expressed in calvariae and femora from rodents as well as LRP4 RNA in human femoral neck samples. The LRP4 levels are in the range of SOST and LRP6 and one-sixth of PTH1R (parathyroid hormone receptor 1), which is known for its high bone expression levels (33) and its role in bone homeostasis (Fig. 3B). LRP5 RNA expression levels were comparable with PTH1R levels. Immunohistochemistry of human femoral neck samples and mouse femora confirmed that the protein is present in osteoblasts and osteocytes, both presumed target cells of sclerostin action (Fig. 3A and supplemental Fig. S3). To assess the role of LRP4 in osteogenesis, we performed silencing of Lrp4 by lentivirus-mediated shRNA delivery in osteoblastic cells. To analyze the LRP4 dependence of the sclerostin-mediated inhibition of mineralization, we used of a rapid Kusa-A1 osteoblastic differentiation assay in which mineralization can be inhibited by recombinant sclerostin treatment. The silencing of Lrp4 RNA by >90% (supplemental Fig. S2C) almost completely blocked sclerostin inhibitory action on in vitro bone mineralization as assessed by calcium content measurement (Fig. 3C). These studies identify a role for LRP4 in bone formation. Based on these in vitro findings, we hypothesized that the interaction of sclerostin with LRP4 is required to mediate sclerostin inhibitory function on bone formation and proceeded to investigate whether LRP4 is also implicated in human bone mass regulation.
FIGURE 3.
LRP4 is expressed in human bone, and Lrp4 silencing blunts sclerostin-mediated inhibition of in vitro bone mineralization. A, LRP4 protein is expressed in human osteoblasts (arrowheads) and osteocytes (arrows). Immunohistochemistry of LRP4 in human femoral neck from a male subject aged 75 is shown. Scale bar, 50 μm. B, LRP4 RNA is expressed in samples from human femoral neck. RNA was extracted from a female subject aged 65 and a male subject aged 80. PTH1R, LRP4, SOST, LRP5, and LRP6 RNA levels were assessed by real-time quantitative PCR and normalized with GAPDH, and relative expression compared with PTH1R is shown. C, knockdown of LRP4 in Kusa-A1 blocks the inhibitory effect of sclerostin on in vitro mineralization activity. Kusa-A1 cells were transduced with lentiviral particles harboring shRNA against Lrp4. Reduction of Lrp4 RNA levels was assessed by real-time quantitative PCR, prior to the addition of an osteoblastic differentiation medium. Calcium content was assessed 4 days later. Error bars, S.E.
Clinical Description of Two Isolated Sclerosteosis Cases, Which Are Not Related to Mutations in SOST or LRP5
The patients referred to us with a sclerosing high bone mass bone phenotype were first screened for mutations in both the exons of the SOST gene and all exons of the LRP5 gene. However, in a number of patients, analysis of both SOST and LRP5 did not reveal any mutations, indicating further genetic heterogeneity. Based on our functional findings, these samples were used for the mutation analysis of LRP4, and two patients indeed presented with findings of high relevance for testing of our hypothesis.
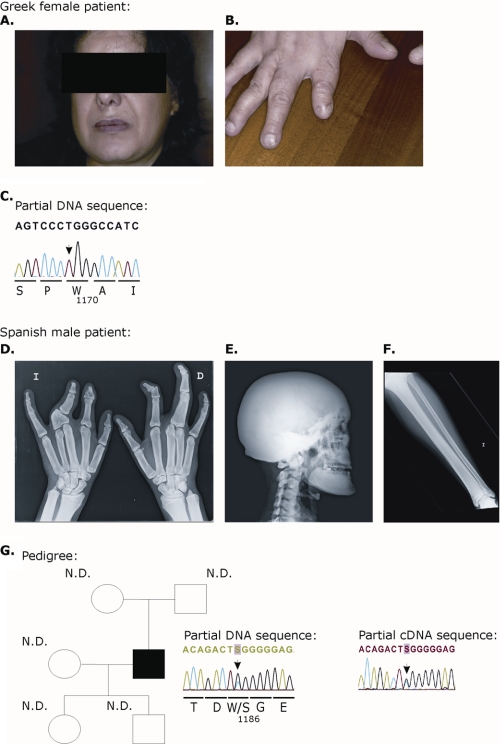
The first patient was a 45-year-old female of Greek origin, whose clinical history was previously reported elsewhere (Fig. 4, A and B) (34). She had syndactyly of the fingers with hypoplasia of the second finger on both hands as well as dysplastic nails, which were separated in two parts, especially on both index fingers (Fig. 4B). She had been hospitalized several times because of multiple complications, such as dizziness, difficulties with gait and coordination, vertigo, and spastic and ataxic gait disturbances. Several neurological complications, such as facial nerve palsy and hearing loss, were present due to bone compression of cranial nerves. X-rays revealed severe thickening of the calvaria and the cortices of the long bones. The parents of the patient were reported to be normal. Overall, the findings were fully consistent with the diagnosis of sclerosteosis. The second patient, a young Spanish male, was also described in an earlier publication (Fig. 4, D–F) (35). This tall male (>97th percentile) was reported with syndactyly and shortening of several phalanges in his hands (Fig. 4D) as well as mild facial nerve palsy. X-rays revealed a sclerotic calvaria and a very dense base of the skull (Fig. 4E). Furthermore, all long bones were showing cortical hyperostosis (Fig. 4F). No abnormalities were noticed in the parents. Again a diagnosis of sclerosteosis was made.
FIGURE 4.
Clinical features and LRP4 mutation analysis of a Greek female (A–C) and a Spanish male sclerosteosis patient (D–G). A, front view of the patient's face showing right facial asymmetry. B, dysplastic finger with shortening and nail malformation with unguis bipartitus. C, partial sequence chromatogram displaying the DNA sequence of the patient. The arrowhead indicates the presence of a homozygous c.3508C→T missense mutation (in exon 26 of the LRP4 gene), resulting in a R1170W substitution. D, plain radiographs of the hands; modeling defect of the metacarpal bones and complex fusion anomaly of the individual bones of the hands. E, plain radiograph of the skull (lateral view). Shown is extensive sclerosis of the calvaria, maxilla, mandible, and cervical spine. F, plain radiograph of the left lower leg. Shown is thickening of the cortical bone of the diaphysis of the tibia with extension into the proximal metaphysis. G, left, family pedigree. All of the proband's relatives are unaffected. Direct sequencing analysis of the proband demonstrated a heterozygous c.3557G→C missense mutation (in exon 27), resulting in a W1186S substitution (arrowhead). The mutation was absent in the DNA of both the parents, suggesting that the mutation is a de novo mutation. The mutation is not segregated to either child. N.D., mutation not detected. Right, partial sequence chromatogram displaying the cDNA (mRNA) sequence of the Spanish male, confirming the heterozygous presence of the c.3557G→C missense mutation. The numbering refers to the transcript with ID ENST00000378623 in the Ensembl data base.
Mutations R1170W and W1186S in LRP4 Identified in the Two Sclerosteosis Patients
In the Greek patient, we identified a homozygous R1170W mutation in exon 26 (Fig. 4C) (the numbering refers to the transcript with ID ENST00000378623), whereas we found in the Spanish patient a heterozygous W1186S mutation in exon 27 (Fig. 4G, left). cDNA obtained from lymphoblastoid cells from the latter patient indicated the presence of both wild type and mutant mRNA (Fig. 4G, right). 110 control individuals, including 56 Spanish subjects and 54 with a mixed European descent, were analyzed for the presence of both variations. However, we did not detect any of the variations in these control subjects. Because contact was lost with the Greek patient, DNA from her parents was not accessible for testing. However, we were able to collect blood from the parents, the daughter, and the son of the Spanish patient. None of these four showed any relevant clinical abnormalities. Mutation analysis revealed that the mutation in the patient is a de novo mutation that did not segregate to his children (Fig. 4G, left). These data are fully consistent with the absence of clinical abnormalities in parents and children.
Human LRP4 protein sequence and those of other species were aligned using the Ensembl data base (available on the World Wide Web) and ClustalW multiple sequence alignment program, version 1.83 (available on the World Wide Web). This alignment revealed that both affected residues and the surrounding residues are highly conserved among all species (supplemental Fig. S4A). Moreover, the Greek mutation reported here implies a change from an arginine, which is a positively charged polar amino acid, into a tryptophan, a large and non-polar amino acid. The Spanish mutation, on the other hand, results in a change of a tryptophan into an uncharged and polar serine. The putative consequences of both substitutions were tested in Polyphen and SIFT, two software-based tools that sort intolerant from tolerant amino acid substitutions on the basis of sequence homology (36, 37). According to SIFT, both substitutions are predicted to be not tolerated, whereas Polyphen scores the R1170W as putatively damaging and the W1186S as benign (absolute difference between the profile scores is 1.839 and 1.440, respectively).
Sclerostin Interaction and Sclerostin Facilitator Function Are Impaired in LRP4 (R1170W and W1186S) Mutants
The discovery of the LRP4 (R1170W and W1186S) mutations in individuals with high bone density and a sclerosteosis phenotype prompted us to assess the functional consequence of these mutations on the LRP4 activities previously described in this paper. The expression levels as well as the homing to the cell surface were not impaired because both R1170W and W1186S LRP4 mutants were detected at similar levels as wild type LRP4 in membrane-specific extracts as well as in surface protein isolation experiments (Fig. 5A). When tested for their sclerostin enhancer function in Wnt signaling, the LRP4 mutants were completely inactive (Fig. 5B), suggesting that these variants are not able to interact normally with sclerostin. Indeed, conditioned medium-containing secreted versions of the LRP4 mutants R1170W and W1186S were impaired in their interaction with sclerostin, as demonstrated in the sclerostin LRP4 interaction ELISA (Fig. 5C), indicating that the mutated residues are involved in sclerostin interaction and concomitant sclerostin function. The results suggest comparable behavior of mutations R1170W and W1186S, which is in accordance with our structural model (supplemental Fig. S4B) predicting the adjacent localization at the surface of propeller 3 of both mutations (Fig. 5D). To assess whether the mutants of LRP4 have a dominant negative effect on the wild type LRP4, we utilized the sclerostin LRP4 interaction assay. Both mutants impair the interaction of wild type LRP4 with sclerostin (Fig. 5E), providing a potential explanation for the phenotype observed especially in the heterozygous patient. Our results demonstrated that the two to date best established functions of sclerostin, namely inhibition of Wnt/β-catenin signaling and inhibition of in vitro bone mineralization, are facilitated through its interaction with LRP4. Notably, our data also suggest that the identified LRP4 mutations might be disease-causing by hindering sclerostin function as a negative bone formation regulator.
DISCUSSION
The role of sclerostin as an osteocyte-secreted key bone formation inhibitor in humans and in model organisms is well established (16, 25, 38, 39). However, the underlying molecular mechanisms are not fully understood. Although an increasing amount of evidence identifies it as an antagonist of Wnt/β-catenin signaling in bone (7, 10, 11, 25, 40), a direct impact on BMP signaling has been also described (24–26). Our aim was to find, with an unbiased tandem affinity purification proteomics approach, sclerostin interaction partners (27, 28). The identification of the Wnt co-receptors LRP5 and LRP6 as sclerostin interaction partners corroborates previously published in vitro discoveries pinpointing sclerostin as a Wnt/β-catenin signaling antagonist (10, 11, 25, 40). In contrast, no interaction between sclerostin and BMP molecules could be identified under any tested condition, providing thus no further support for the hypothesis that sclerostin exerts its action as a BMP antagonist. Furthermore, LRP4, another member of the lipoprotein-related protein family of membrane receptors, was identified as a candidate sclerostin interaction partner. An involvement of the LRP4 receptor in Wnt/β-catenin signaling has been suggested previously (41), although a link between its inhibitory action on Wnt/β-catenin signaling and the Wnt signaling-related ligands had not been established. Because the proteomics data were supportive of the hypothesis of a predominant role of sclerostin in Wnt/β-catenin signaling, we focused our detailed analysis on the LRP4 sclerostin interaction.
Recently, putative LRP4 ligands, including RAP, Dkk1, Wise, and, interestingly, sclerostin, were described (42, 43). Furthermore, LRP4 has lately been identified as a co-receptor of agrin, a motor neuron-derived ligand that stimulates MuSK phosphorylation and subsequent signaling at the neuromuscular synapses (44, 45). Knowledge about the in vivo function of LRP4 has been obtained to date using genetically designed or naturally occurring mutations in mice and cattle (46–49). Lrp4 null mutations in mice result in limb development defects and in perinatal death due to a lack of neuromuscular junction formation (50). Animals with partial loss-of-function mutations (hypomorphic) are viable and present with different degrees of polysyndactyly (41, 49). A recent study identified a role of Lrp4 in bone metabolism based on a mouse model harboring a stop codon just upstream of the transmembrane domain of LRP4 (42). Ten-week-old growing hypomorph animals presented with increased bone formation and resorption and thus increased bone turnover associated with decreased BMD. However, the dysfunctional receptor, lacking the membrane anchor, could still interact with its extracellular ligands. Consequently, although implicating Lrp4 for the first time in bone metabolism, one cannot conclusively judge from the published data whether the model represented a gain- or loss-of-function model.
We found LRP4 RNA and LRP4 protein to be expressed in human femoral neck samples. Osteoblasts and osteocytes from humans of both sexes stained positive for LRP4, whereas no expression was observed in osteoclasts. The above described paper on the Lrp4 role in bone metabolism describes Lrp4 expression in osteoblasts based on extraction of RNA from mineralized bone (42). Taking into account that osteocytes represent 90% (21) of the cells in mineralized bone and the extraction method used, the data are in agreement with our findings. It is tempting to speculate that LRP4, localized at the osteocyte cell surface, may modulate osteocyte function in concert with interaction partners like sclerostin and potentially agrin, whose bone tissue expression is also established (51). LRP4 expression on osteoblasts may mediate paracrine effects of sclerostin. This hypothesis is supported by our data demonstrating that sclerostin cannot exert its inhibitory effect on osteoblastic in vitro bone mineralization upon Lrp4 silencing. Tightly regulated expression of LRP4 might be required to mediate response to specific signaling pathways at different stages of osteoblastic differentiation involving diverse ligands, including sclerostin. Consistent with the recent identification of several putative LRP4 ligands and thus additional roles of LRP4, we observed that Lrp4 silencing also promotes osteoblast differentiation in the absence of sclerostin as a ligand (data not shown).
The presence of LRP4 on cells of the osteoblastic lineage and the identification of LRP4 function as sclerostin facilitator prompted us to search for the presence of LRP4 mutations in patients with a sclerosing high bone mass phenotype, who do not present with mutations in SOST or LRP5, which are known to give rise to such phenotypes (4–6, 9–13, 15). The subsequent discovery of mutations p.R1170W and p.W1186S in LRP4 in two high bone mass patients of Mediterranean origin represents a novel finding. Both patients share several typical sclerosteosis features, such as syndactyly of the fingers, facial asymmetry, and thickening of the corticalis of long bones and skull. However, the phenotype of the Greek female patient is more severe, with additional neurological complications. Our functional studies clearly indicated that both missense mutations result in a complete loss of sclerostin facilitator function of the LRP4 protein. In the Greek patient, the p.R1170W mutation was present in the homozygous state, implying the absence of any LRP4 sclerostin facilitator function.
The W1186S mutation, on the other hand, was found in the heterozygous state. We confirmed the presence of wild type mRNA along with mutated mRNA at the cDNA level, excluding the possibility of a compound heterozygous state. The genetic analysis of the parents and two children of the patient, all of them unaffected, further confirmed the de novo nature of this mutation, supporting causality of the heterozygous mutation for the observed phenotype. In line with the heterozygous state of the mutation, the patient presented with a relatively mild phenotype because he is, aside from increased bone mass, his hand abnormalities, and a mild facial palsy, asymptomatic. Whether the phenotype can be explained by haploinsufficiency for LRP4 or by a dominant negative effect of the mutation, which is supported by in vitro evidence, is difficult to conclude at this point.
Despite a potential role for LRP4 in neuromuscular junction formation, the two patients were not reported to display any sign of a dysfunctional locomotor system. Interestingly, the mandibular overgrowth, as seen in SOST-associated sclerosteosis and Van Buchem patients (13–15, 17), is less severe or absent in these subjects in analogy to some patients with gain-of-function mutations in LRP5 (4, 6). Both identified mutations reside in the third propeller of the extracellular domain of LRP4. Our structural model predicts that the mutated residues co-localize at the surface of the propeller and could thereby form a region of interaction for sclerostin. We hypothesize that the mutations in LRP4 preclude sclerostin from binding and thus from exerting its negative role on bone formation, thereby mimicking the phenotype observed upon sclerostin loss of function in sclerosteosis and Van Buchem patients. Our bone overgrowth findings in humans suggest a LRP4 sclerostin facilitator loss-of-function phenotype and might thereby explain the discrepancy with the bone phenotype reported in the Lrp4 mutant mice, which express the LRP4 ECD that can still interact with its multiple extracellular ligands.
Recently, Li et al. (52) reported the identification of missense mutations in LRP4 underlying Cenani-Lenz syndrome. These findings are clearly supportive of our data because these patients also show syndactyly. This phenotype is also observed in patients with mutations in SOST and related to the recently elucidated role of sclerostin modulation of Wnt/β-catenin signaling in limb development (53). However, in comparison with our patients, patients with Cenani-Lenz syndrome have additional kidney problems (hypoplasia and agenesis) and synostoses into radius and ulna, whereas no increase in cortical thickening of the long bones was reported. None of the mutations found are localized within the third propeller domain, where both sclerosteosis mutations cluster. Taking into account the highly restricted expression of sclerostin to bone, the data support the hypothesis that the sclerosteosis mutations in LRP4 might indeed selectively affect the binding with sclerostin, whereas in Cenani-Lenz syndrome, most likely the binding to other ligands is disturbed, explaining the divergent clinical features. The binding properties of the LRP4 high bone mass mutants (R1170W and W1186S) toward other ligands will allow us in the future to determine whether these residues are involved in additional interactions with LRP4 sclerostin.
In contrast to previous publications using LRP4 proteins from conditioned medium (44), most of our binding assays were performed with purified recombinant proteins. The correct folding of the produced LRP4 ECD was documented by its interaction with sclerostin and by the fact that we can perform sandwich ELISA by precoating an anti-LRP4 antibody and detecting LRP4 as described previously (data not shown). In addition, we could also detect a direct interaction between recombinant LRP4 and DKK1 (data not shown), consistent with the recent report identifying multiple ligands for LRP4, including DKK1 (42). The lack of impact of LRP4 overexpression on DKK1 inhibition of Wnt1/β-catenin signaling in our experimental setting does not exclude the possibility that LRP4 impacts DKK1 action under other conditions. To date, we could demonstrate the functional consequence of the binding of sclerostin to LRP4 Wnt1/β-catenin signaling. Our findings demonstrate that the cytoplasmic domain of LRP4 is dispensable for sclerostin responsiveness, as reported for agrin responsiveness (44). Thus, the signaling and/or clustering functions of the cytoplasmic domain of LRP4 remain to be elucidated in future studies.
Finally, there are several examples, including the LRP5 and SOST gene, where the role of certain proteins in bone homeostasis was initially identified by the study of monogenic bone disorders (4, 6, 13, 14), which was subsequently confirmed by genetic association studies illustrating that natural variants within these genes had an influence on BMD in the general population (54). Interestingly, a recent genome-wide association study of BMD in individuals of European descent has showed that non-synonymous SNPs in the LRP4 gene are associated, providing further support for a role of LRP4 in bone metabolism (55, 56).
In summary, our data reveal a novel role for LRP4 in bone and provide further understanding of sclerostin's mode of action as a negative regulator of bone formation. Better understanding of the interaction between LRP4 and sclerostin may have wider implications for the development of therapeutic agents for the treatment of osteoporosis.
Supplementary Material
Acknowledgments
We are grateful to Prof. Dr. A. Kurth and Dr. M. Tonak for providing femoral neck bone samples. We thank Pascale Brebbia, Gaby Guiglia, Tanja Grabenstaetter, Lilian Hartmann, Nelly Laplanche, Renzo Schumpf, Viviane Trendelenburg, and Andrea Venturiere for excellent technical assistance. We are thankful to Rita Schmitz, Mauro Zurini, Lukas Leder, and the protein production team for recombinant proteins. We also thank Carsten Jacobi for the setup of ELISA assays as well as François Natt for the synthesis of siRNA against LRP4.
This work was supported by the European Commission (HEALTH-F2-2008-201099, TALOS) and by “Fonds voor Wetenschappelijk Onderzoek” Grant G.0117.06 and by a grant from the Special Research Funds (Bijzonder Onderzoeksfonds TOP and Nieuwe Onderzoeksinitiatieven) of the University of Antwerp (all to W. V. H.). O. L., C. H., S. H., I. K., F. M., T. B., M. S., B. B., N. C., H. E., S. G., C. X. L., A. B., and M. K. are employees of Novartis.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- BMD
- bone mineral density
- BMP
- bone morphogenetic protein
- TAP
- tandem affinity purification
- aa
- amino acids
- ECD
- extracellular domain.
REFERENCES
- 1. Henriksen K., Neutzsky-Wulff A. V., Bonewald L. F., Karsdal M. A. (2009) Bone 44, 1026–1033 [DOI] [PubMed] [Google Scholar]
- 2. Martin T., Gooi J. H., Sims N. A. (2009) Crit Rev. Eukaryot. Gene Expr. 19, 73–88 [DOI] [PubMed] [Google Scholar]
- 3. Balemans W., Van Hul W. (2007) Endocrinology 148, 2622–2629 [DOI] [PubMed] [Google Scholar]
- 4. Boyden L. M., Mao J., Belsky J., Mitzner L., Farhi A., Mitnick M. A., Wu D., Insogna K., Lifton R. P. (2002) N. Engl. J. Med. 346, 1513–1521 [DOI] [PubMed] [Google Scholar]
- 5. Gong Y., Slee R. B., Fukai N., Rawadi G., Roman-Roman S., Reginato A. M., Wang H., Cundy T., Glorieux F. H., Lev D., Zacharin M., Oexle K., Marcelino J., Suwairi W., Heeger S., Sabatakos G., Apte S., Adkins W. N., Allgrove J., Arslan-Kirchner M., Batch J. A., Beighton P., Black G. C., Boles R. G., Boon L. M., Borrone C., Brunner H. G., Carle G. F., Dallapiccola B., De Paepe A., Floege B., Halfhide M. L., Hall B., Hennekam R. C., Hirose T., Jans A., Jüppner H., Kim C. A., Keppler-Noreuil K., Kohlschuetter A., LaCombe D., Lambert M., Lemyre E., Letteboer T., Peltonen L., Ramesar R. S., Romanengo M., Somer H., Steichen-Gersdorf E., Steinmann B., Sullivan B., Superti-Furga A., Swoboda W., van den Boogaard M. J., Van Hul W., Vikkula M., Votruba M., Zabel B., Garcia T., Baron R., Olsen B. R., Warman M. L. (2001) Cell 107, 513–523 [DOI] [PubMed] [Google Scholar]
- 6. Little R. D., Carulli J. P., Del Mastro R. G., Dupuis J., Osborne M., Folz C., Manning S. P., Swain P. M., Zhao S. C., Eustace B., Lappe M. M., Spitzer L., Zweier S., Braunschweiger K., Benchekroun Y., Hu X., Adair R., Chee L., FitzGerald M. G., Tulig C., Caruso A., Tzellas N., Bawa A., Franklin B., McGuire S., Nogues X., Gong G., Allen K. M., Anisowicz A., Morales A. J., Lomedico P. T., Recker S. M., Van Eerdewegh P., Recker R. R., Johnson M. L. (2002) Am. J. Hum. Genet. 70, 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Johnson M. L., Kamel M. A. (2007) Curr. Opin. Rheumatol. 19, 376–382 [DOI] [PubMed] [Google Scholar]
- 8. Mani A., Radhakrishnan J., Wang H., Mani A., Mani M. A., Nelson-Williams C., Carew K. S., Mane S., Najmabadi H., Wu D., Lifton R. P. (2007) Science 315, 1278–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Balemans W., Devogelaer J. P., Cleiren E., Piters E., Caussin E., Van Hul W. (2007) J. Bone Miner. Res. 22, 708–716 [DOI] [PubMed] [Google Scholar]
- 10. Ellies D. L., Viviano B., McCarthy J., Rey J. P., Itasaki N., Saunders S., Krumlauf R. (2006) J. Bone Miner. Res. 21, 1738–1749 [DOI] [PubMed] [Google Scholar]
- 11. Li X., Zhang Y., Kang H., Liu W., Liu P., Zhang J., Harris S. E., Wu D. (2005) J. Biol. Chem. 280, 19883–19887 [DOI] [PubMed] [Google Scholar]
- 12. Balemans W., Piters E., Cleiren E., Ai M., Van Wesenbeeck L., Warman M. L., Van Hul W. (2008) Calcif. Tissue Int. 82, 445–453 [DOI] [PubMed] [Google Scholar]
- 13. Balemans W., Ebeling M., Patel N., Van Hul E., Olson P., Dioszegi M., Lacza C., Wuyts W., Van Den Ende J., Willems P., Paes-Alves A. F., Hill S., Bueno M., Ramos F. J., Tacconi P., Dikkers F. G., Stratakis C., Lindpaintner K., Vickery B., Foernzler D., Van Hul W. (2001) Hum. Mol. Genet. 10, 537–543 [DOI] [PubMed] [Google Scholar]
- 14. Balemans W., Patel N., Ebeling M., Van Hul E., Wuyts W., Lacza C., Dioszegi M., Dikkers F. G., Hildering P., Willems P. J., Verheij J. B., Lindpaintner K., Vickery B., Foernzler D., Van Hul W. (2002) J. Med. Genet. 39, 91–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brunkow M. E., Gardner J. C., Van Ness J., Paeper B. W., Kovacevich B. R., Proll S., Skonier J. E., Zhao L., Sabo P. J., Fu Y., Alisch R. S., Gillett L., Colbert T., Tacconi P., Galas D., Hamersma H., Beighton P., Mulligan J. (2001) Am. J. Hum. Genet. 68, 577–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Loots G. G., Kneissel M., Keller H., Baptist M., Chang J., Collette N. M., Ovcharenko D., Plajzer-Frick I., Rubin E. M. (2005) Genome Res. 15, 928–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Staehling-Hampton K., Proll S., Paeper B. W., Zhao L., Charmley P., Brown A., Gardner J. C., Galas D., Schatzman R. C., Beighton P., Papapoulos S., Hamersma H., Brunkow M. E. (2002) Am. J. Med. Genet. 110, 144–152 [DOI] [PubMed] [Google Scholar]
- 18. Paszty C., Turner C. H., Robinson M. K. (2010) J. Bone Miner. Res. 25, 1897–1904 [DOI] [PubMed] [Google Scholar]
- 19. Padhi D., Jang G., Stouch B., Fang L., Posvar E. (2011) J. Bone Miner. Res. 26, 19–26 [DOI] [PubMed] [Google Scholar]
- 20. Poole K. E., van Bezooijen R. L., Loveridge N., Hamersma H., Papapoulos S. E., Löwik C. W., Reeve J. (2005) FASEB J. 19, 1842–1844 [DOI] [PubMed] [Google Scholar]
- 21. Bonewald L. F. (2006) Bonekey. Osteovision 3, 7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bonewald L. F. (2007) Ann. N.Y. Acad. Sci. 1116, 281–290 [DOI] [PubMed] [Google Scholar]
- 23. Yadav V. K., Ryu J. H., Suda N., Tanaka K. F., Gingrich J. A., Schütz G., Glorieux F. H., Chiang C. Y., Zajac J. D., Insogna K. L., Mann J. J., Hen R., Ducy P., Karsenty G. (2008) Cell 135, 825–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. ten Dijke P., Krause C., de Gorter D. J., Lowik C. W., van Bezooijen R. L. (2008) J. Bone Joint Surg. Am. 90, Suppl. 1, 31–35 [DOI] [PubMed] [Google Scholar]
- 25. van Bezooijen R. L., Roelen B. A., Visser A., van der Wee-Pals L., de Wilt E., Karperien M., Hamersma H., Papapoulos S. E., ten Dijke P., Löwik C. W. (2004) J. Exp. Med. 199, 805–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Winkler D. G., Sutherland M. K., Geoghegan J. C., Yu C., Hayes T., Skonier J. E., Shpektor D., Jonas M., Kovacevich B. R., Staehling-Hampton K., Appleby M., Brunkow M. E., Latham J. A. (2003) EMBO J. 22, 6267–6276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bürckstümmer T., Bennett K. L., Preradovic A., Schütze G., Hantschel O., Superti-Furga G., Bauch A. (2006) Nat. Methods 3, 1013–1019 [DOI] [PubMed] [Google Scholar]
- 28. Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Séraphin B. (1999) Nat. Biotechnol. 17, 1030–1032 [DOI] [PubMed] [Google Scholar]
- 29. Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 30. Huang S. M., Mishina Y. M., Liu S., Cheung A., Stegmeier F., Michaud G. A., Charlat O., Wiellette E., Zhang Y., Wiessner S., Hild M., Shi X., Wilson C. J., Mickanin C., Myer V., Fazal A., Tomlinson R., Serluca F., Shao W., Cheng H., Shultz M., Rau C., Schirle M., Schlegl J., Ghidelli S., Fawell S., Lu C., Curtis D., Kirschner M. W., Lengauer C., Finan P. M., Tallarico J. A., Bouwmeester T., Porter J. A., Bauer A., Cong F. (2009) Nature 461, 614–620 [DOI] [PubMed] [Google Scholar]
- 31. Cho M. S., Yee H., Chan S. (2002) J. Biomed. Sci. 9, 631–638 [DOI] [PubMed] [Google Scholar]
- 32. Kawashima N., Shindo K., Sakamoto K., Kondo H., Umezawa A., Kasugai S., Perbal B., Suda H., Takagi M., Katsube K. (2005) J. Bone Miner. Metab. 23, 123–133 [DOI] [PubMed] [Google Scholar]
- 33. Mannstadt M., Jüppner H., Gardella T. J. (1999) Am. J. Physiol. 277, F665–F675 [DOI] [PubMed] [Google Scholar]
- 34. Itin P. H., Keserü B., Hauser V. (2001) Dermatology 202, 259–260 [DOI] [PubMed] [Google Scholar]
- 35. Bueno M., Oliván G., Jiménez A., Garagorri J. M., Sarría A., Bueno A. L., Bueno M., Jr., Ramos F. J. (1994) J. Med. Genet. 31, 976–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ng P. C., Henikoff S. (2003) Nucleic Acids Res. 31, 3812–3814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ramensky V., Bork P., Sunyaev S. (2002) Nucleic Acids Res. 30, 3894–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li X., Ominsky M. S., Niu Q. T., Sun N., Daugherty B., D'Agostin D., Kurahara C., Gao Y., Cao J., Gong J., Asuncion F., Barrero M., Warmington K., Dwyer D., Stolina M., Morony S., Sarosi I., Kostenuik P. J., Lacey D. L., Simonet W. S., Ke H. Z., Paszty C. (2008) J. Bone Miner. Res. 23, 860–869 [DOI] [PubMed] [Google Scholar]
- 39. Lin C., Jiang X., Dai Z., Guo X., Weng T., Wang J., Li Y., Feng G., Gao X., He L. (2009) J. Bone Miner. Res. 24, 1651–1661 [DOI] [PubMed] [Google Scholar]
- 40. Semënov M., Tamai K., He X. (2005) J. Biol. Chem. 280, 26770–26775 [DOI] [PubMed] [Google Scholar]
- 41. Johnson E. B., Hammer R. E., Herz J. (2005) Hum. Mol. Genet. 14, 3523–3538 [DOI] [PubMed] [Google Scholar]
- 42. Choi H. Y., Dieckmann M., Herz J., Niemeier A. (2009) PLoS. One. 4, e7930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ohazama A., Johnson E. B., Ota M. S., Choi H. Y., Porntaveetus T., Oommen S., Itoh N., Eto K., Gritli-Linde A., Herz J., Sharpe P. T. (2008) PLoS One 3, e4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim N., Stiegler A. L., Cameron T. O., Hallock P. T., Gomez A. M., Huang J. H., Hubbard S. R., Dustin M. L., Burden S. J. (2008) Cell 135, 334–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang B., Luo S., Wang Q., Suzuki T., Xiong W. C., Mei L. (2008) Neuron 60, 285–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Drögemüller C., Leeb T., Harlizius B., Tammen I., Distl O., Höltershinken M., Gentile A., Duchesne A., Eggen A. (2007) BMC. Genet. 8, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Duchesne A., Gautier M., Chadi S., Grohs C., Floriot S., Gallard Y., Caste G., Ducos A., Eggen A. (2006) Genomics 88, 610–621 [DOI] [PubMed] [Google Scholar]
- 48. Johnson E. B., Steffen D. J., Lynch K. W., Herz J. (2006) Genomics 88, 600–609 [DOI] [PubMed] [Google Scholar]
- 49. Simon-Chazottes D., Tutois S., Kuehn M., Evans M., Bourgade F., Cook S., Davisson M. T., Guénet J. L. (2006) Genomics 87, 673–677 [DOI] [PubMed] [Google Scholar]
- 50. Weatherbee S. D., Anderson K. V., Niswander L. A. (2006) Development 133, 4993–5000 [DOI] [PubMed] [Google Scholar]
- 51. Hausser H. J., Ruegg M. A., Brenner R. E., Ksiazek I. (2007) Histochem. Cell Biol. 127, 363–374 [DOI] [PubMed] [Google Scholar]
- 52. Li Y., Pawlik B., Elcioglu N., Aglan M., Kayserili H., Yigit G., Percin F., Goodman F., Nürnberg G., Cenani A., Urquhart J., Chung B. D., Ismail S., Amr K., Aslanger A. D., Becker C., Netzer C., Scambler P., Eyaid W., Hamamy H., Clayton-Smith J., Hennekam R., Nürnberg P., Herz J., Temtamy S. A., Wollnik B. (2010) Am. J. Hum. Genet. 86, 696–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Collette N. M., Genetos D. C., Murugesh D., Harland R. M., Loots G. G. (2010) Dev. Biol. 342, 169–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li W. F., Hou S. X., Yu B., Li M. M., Férec C., Chen J. M. (2010) Hum. Genet. 127, 249–285 [DOI] [PubMed] [Google Scholar]
- 55. Styrkarsdottir U., Halldorsson B. V., Gretarsdottir S., Gudbjartsson D. F., Walters G. B., Ingvarsson T., Jonsdottir T., Saemundsdottir J., Center J. R., Nguyen T. V., Bagger Y., Gulcher J. R., Eisman J. A., Christiansen C., Sigurdsson G., Kong A., Thorsteinsdottir U., Stefansson K. (2008) N. Engl. J. Med. 358, 2355–2365 [DOI] [PubMed] [Google Scholar]
- 56. Styrkarsdottir U., Halldorsson B. V., Gretarsdottir S., Gudbjartsson D. F., Walters G. B., Ingvarsson T., Jonsdottir T., Saemundsdottir J., Snorradóttir S., Center J. R., Nguyen T. V., Alexandersen P., Gulcher J. R., Eisman J. A., Christiansen C., Sigurdsson G., Kong A., Thorsteinsdottir U., Stefansson K. (2009) Nat. Genet. 41, 15–17 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.