Abstract
To enhance understanding of the metabolic indicators of type 2 diabetes mellitus (T2DM) disease pathogenesis and progression, the urinary metabolomes of well characterized rhesus macaques (normal or spontaneously and naturally diabetic) were examined. High-resolution ultra-performance liquid chromatography coupled with the accurate mass determination of time-of-flight mass spectrometry was used to analyze spot urine samples from normal (n = 10) and T2DM (n = 11) male monkeys. The machine-learning algorithm random forests classified urine samples as either from normal or T2DM monkeys. The metabolites important for developing the classifier were further examined for their biological significance. Random forests models had a misclassification error of less than 5%. Metabolites were identified based on accurate masses (<10 ppm) and confirmed by tandem mass spectrometry of authentic compounds. Urinary compounds significantly increased (p < 0.05) in the T2DM when compared with the normal group included glycine betaine (9-fold), citric acid (2.8-fold), kynurenic acid (1.8-fold), glucose (68-fold), and pipecolic acid (6.5-fold). When compared with the conventional definition of T2DM, the metabolites were also useful in defining the T2DM condition, and the urinary elevations in glycine betaine and pipecolic acid (as well as proline) indicated defective re-absorption in the kidney proximal tubules by SLC6A20, a Na+-dependent transporter. The mRNA levels of SLC6A20 were significantly reduced in the kidneys of monkeys with T2DM. These observations were validated in the db/db mouse model of T2DM. This study provides convincing evidence of the power of metabolomics for identifying functional changes at many levels in the omics pipeline.
Keywords: Diabetes, Kidney, Mass Spectrometry (MS), Metabolomics, Urine
Introduction
Type 2 diabetes mellitus (T2DM),4 the most common type of diabetes, is a significant, costly, and rapidly expanding public health concern worldwide. T2DM is associated with microvascular (nephropathy, retinopathy, and neuropathy) and macrovascular (coronary artery disease and peripheral vascular disease) pathologies resulting in a complex, multifactorial metabolic phenotype. Therefore, understanding the molecular pathogenesis and progression of T2DM, its associated and varied complications, and its effects on numerous organ systems is not trivial. The emerging field of small molecule profiling, or metabolomics, has already provided new perspectives on T2DM (1–5). As the end products of all cellular processes, global metabolite profiling may represent the best and largest net with which to capture changes originating from epigenomic, genomic, transcriptomic, and proteomic alterations.
Metabolomics aims to identify and quantify all small molecules as they may be the most accurate indicators of cellular physiology (6). Moreover, metabolomics is an NIH Roadmap Initiative and was listed as a research priority in the American Society of Nephrology Renal Research Report (7). Metabolomic studies of T2DM have utilized a variety of animal models (e.g. db/db mice, streptozotocin-treated mice, Zucker diabetic rats, even hyperinsulinemic horses), as well as T2DM individuals. Several analytical platforms, including high-field 1H nuclear magnetic resonance spectroscopy (NMR), gas chromatography coupled mass spectrometry (GC-MS), and liquid chromatography coupled mass spectrometry (LC-MS) have been employed in metabolomic investigations (8–13). These approaches were useful in demonstrating proof-of-concept for metabolomic analysis as a tool to discriminate normal from T2DM-diseased samples, yet the mechanistic underpinnings and the tissue of origin of the reported metabolites remain uncertain.
The rhesus macaque (Macaca mulatta) represents an ideal animal model for understanding the pathogenesis and complications of T2DM. As primates, rhesus macaques are more similar to humans than rodents, sharing ∼93% DNA sequence identity (14). With respect to T2DM in particular, the rhesus macaque is strikingly similar to humans. Like many groups of T2DM humans, laboratory-maintained ad libitum fed rhesus macaques develop T2DM with a lifetime incidence estimated to be around 30%. Clinically, T2DM rhesus macaques exhibit the same phenotype as T2DM humans: hyperglycemia, glycosuria, polydipsia, polyphagia, excess adiposity, dyslipidemia, insulin resistance, and impaired glucose tolerance (15–17). They also show the same metabolic disturbances (17–23) and develop the same complications, including nephropathy, retinopathy, neuropathy, and other macrovascular changes (24–30). Given its well recognized applicability to many human diseases, including T2DM, and its recently published genome (31, 32), the rhesus macaque is an exceptional model with which to understand T2DM pathogenesis and progression at the metabolic and metabolomic levels.
In the present metabolomic study, urine samples from a cohort of spontaneously T2DM rhesus macaques were compared with samples from normal counterparts. An analytical platform having superior resolution and high mass accuracy, ultra-performance liquid chromatography (UPLC) coupled electrospray ionization quadrupole time-of-flight mass spectrometry (ESI-QTOF-MS), was combined with contemporary machine learning algorithms to identify urinary metabolites capable of discriminating normal from T2DM monkeys. The kidney proximal tubule transporter, SLC6A20 (a Na+-dependent transporter), was implicated in the increased excretion of the urinary metabolites. Similar results were replicated in the db/db mouse model of T2DM. The combination of a unique nonhuman primate colony (rhesus macaque), genetically modified mice, and the latest in metabolomic technology provides a new perspective on the multifactorial T2DM phenotype.
EXPERIMENTAL PROCEDURES
Chemicals
Betaine hydrochloride was purchased from MP Biomedicals, LLC (Aurora, Ohio). N-Methylnipecotic acid was purchased from Oakwood Products Inc. (West Columbia, SC). Chlorpropamide, citrate, creatinine, debrisoquine hemisulfate, glucose, isocitrate, kynurenic acid, α-methyl-l-proline, 4-nitrobenzoic acid, d- and l-pipecolic acid, d- and l-proline, and theophylline were purchased from Sigma. All reagents and solvents were HPLC grade.
Monkey Urine Collection
Rhesus monkeys (M. mulatta) were individually housed and cared for according to the Guide for the Care and Use of Laboratory Animals (National Research Council Institute for Laboratory Animal Resources). All animal protocols were approved by the Institutional Animal Care and Use Committee at the University of South Florida. Each subject was placed on the same diet lot (17% protein, 13% fat, and 70% carbohydrate, 5038, Purina Mills, St. Louis, MO) provided ad libitum together with ad libitum fresh water. Standard 12-h light/dark cycles were used. Spot urines (except for the first urine of the morning) were collected in clean autoclaved steel pans. Urine samples were discarded if found to be contaminated with feces, food, or water. Monkey blood samples used for clinical chemistry were obtained under light anesthesia (ketamine hydrochloride, 10 mg/kg body weight) after a consistent 16-h overnight fast.
Mouse Urine Collection
C57Bl/6J and db/db (B6.BKS(D)-Leprdb/J) male mice 12 weeks of age were obtained from The Jackson Laboratory. Food (NIH31 standard chow) and water was provided ad libitum. Standard 12-h light/dark cycles were used. Mice were placed in metabolic cages to collect 24-h urine samples (Tecniplast USA, Exton, PA) every month for 3 months. Mice were euthanized at 12 (n = 3), 16 (n = 4), 20 (n = 4), and 24 weeks (n = 4) of age and tissue was harvested and immediately flash-frozen in liquid nitrogen. The db/db diabetic phenotype was confirmed in all mice by blood chemistry analysis. All animal studies were approved by the National Cancer Institute Animal Care and Use Committee. Urine samples were diluted with HPLC grade water containing 1 μm chlorpropamide as the internal standard. The samples were centrifuged to remove insoluble debris and the supernatant was transferred to an autosampler vial.
UPLC-ESI-QTOF-MS of Monkey Urine
Monkey spot urines were diluted with an equal volume of solvent comprised of water and acetonitrile (1:1). The samples were centrifuged to remove insoluble debris and the supernatant was transferred to an autosampler vial. Five μl was chromatographed on a 50 × 2.1-mm Acquity 1.7-μm C18 column (Waters Corp, Milford, MA) using an Acquity UPLC system (Waters). To avoid artifacts based on sample injection order, the order was randomized. The gradient mobile phase consisted of 0.1% formic acid (A) and acetonitrile containing 0.1% formic acid (B). A typical 10-min sample run consisted of 0.5 min of 100% solvent A followed by an incremental increase of solvent B up to 100% for the remaining 9.5 min. The flow rate was set to 0.5 ml/min. The eluent was introduced by electrospray ionization into the mass spectrometer (Waters QTOF Premier) operating in positive (ESI+) or negative (ESI−) ionization modes. The capillary and sampling cone voltages were set to 3,000 and 30 V, respectively. Source and desolvation temperatures were set to 120 and 350 °C, respectively, and the cone and desolvation gas flows were set to 50.0 and 650.0 liter/h, respectively. To maintain mass accuracy, sulfadimethoxine at a concentration of 300 pg/μl in 50% acetonitrile was used as a lock mass and injected at a rate of 50 μl/min. For MS scanning, data were acquired in centroid mode from 50 to 850 m/z and for tandem MS the collision energy was ramped from 5 to 35 V.
Chromatogram Deconvolution
The mass chromatographic data were aligned using MarkerLynx software (Waters) to generate a data matrix consisting of peak areas corresponding to a unique m/z and retention time. The peak area corresponding to protonated creatinine (m/z = 114.0671+, retention time = 0.31 min) was used to normalize the peak areas in a sample. This procedure minimized differences in analyte concentration that were due to variations in renal physiology. Data from ESI+ and ESI− were combined to generate a data matrix suitable for downstream analysis. The conventionally noted clinical parameters of age, weight, fasting plasma glucose, serum triglycerides, HbA1c, and plasma insulin were added to some of the analyses to serve as pre-existing clinically important characterization and biomarkers of T2DM.
Random Forests Analysis
The random forests machine learning algorithm (33) was used to classify monkey urine samples as normal or T2DM. The random forests method uses an ensemble of simple decision trees, each trained on a bootstrapped subset of the samples and a random set of variables. The algorithm provides two particularly useful features: 1) internal cross-validation that assesses model accuracy without a priori specifying independent testing and training sets; and 2) a permutation analysis that estimates the relative “importance score” of each variable in the models. Random forests run-to-run variability can reduce the reproducibility of importance scores, so more robust importance scores were computed using a panel of 25 independent random forests models, each with 10,000 trees, trained on the set of all ions (ESI+ and ESI−). Next, the importance score rank for each variable in each model was computed. The ranks were then averaged over the set of models. Bootstrapping the results from the 25 independent random forests was used to determine the 95% confidence intervals of the variable importance ranks.
The minimal number of variables necessary to build an optimal random forests model for classifying each group was determined with a greedy (i.e. locally optimal but potentially globally suboptimal) approach. The average performance of sets of 25 random forests models were compared, each trained using a subset of the top ranked variables. The set of 25 random forests models were trained using the previously determined top 10, 20, 50, 75, 150, 250, 350, 500, 750, or 1000 most important model variables. The set of models that achieved the best average classification performance was concluded to contain the most meaningful set of variables. Bootstrapping of the 25 forests was used to determine the variation in model performance within each of the sets of 25 models. In the case that more than one set of top performing variables was non-statistically significantly better than another set, the best model was the one that used the fewest variables.
Metabolite Identification and Validation
Elemental compositions of ions were determined using Seven Golden Rules (34) and the Madison Metabolomics Consortium Database (35). Putative ion identities were validated using tandem MS by comparison with authentic compounds.
Metabolite Concentration Measurements
Urine samples were diluted in 10 mm ammonium formate buffer (pH 3.5) and separated on a Phenomenex Synergi Polar-RP column (Torrance, CA). Samples for measurement of glycine betaine, creatinine, and glucose were diluted 1:500 and kynurenic acid samples were diluted 1:10. LC analysis was carried out using a high-performance LC system consisting of a PerkinElmer Series 200 quaternary pump, vacuum degasser, and autosampler (PerkinElmer Life Sciences) with a 100-μl loop interfaced to an API2000 SCIEX triple quadrupole tandem mass spectrometer (Applied Biosystems/MDS Sciex, Foster City, CA). Ions were monitored by either single ion monitoring or multiple reaction monitoring (MRM) modes. Single ion monitoring was used to monitor the negatively charged ion, m/z = 214.9, corresponding to the chloride adduct of glucose with 4-nitrobenzoic acid used as the internal standard (m/z = 165.9). The following MRM transitions were monitored in positive mode: glycine betaine (118.0 → 58.1 m/z), creatinine (114.0 → 86.1 m/z), debrisoquine hemisulfate (176.1 → 134.2 m/z), and kynurenic acid (190.0 → 144.0 m/z). Citrate and its isomer, isocitrate, were separated by UPLC and detected by QTOF-MS by monitoring their sodium adducts in ESI+ mode (m/z = 215.0168). Citrate and isocitrate were quantitated using QuanLynx (Waters).
Mouse serum samples were diluted 20-fold in 66% acetonitrile containing 200 nm chlorpropamide as an internal standard and separated on a 50 × 2.1-mm Acquity 1.7-μm C18 column and introduced via electrospray into a triple quadrupole mass spectrometer (XEVO TQ-MS). MRM transitions for glycine betaine, pipecolic acid, and proline were as described above and below. The MRM transition for chlorpropamide was 275.0 → 189.96 m/z. Results were analyzed and quantitated using TargetLynx (Waters).
Chiral Chromatography
Chiral chromatography for pipecolic acid and proline was performed as described (36) to distinguish the l- from the d-stereoisomer. Briefly, urine samples were diluted 1:1 in 50% aqueous methanol and spiked with internal standards N-methylnipecotic acid (for pipecolic acid measurements) or α-methyl-l-proline (for proline measurements). Samples were chromatographed on an Astec Chirobiotic T (Sigma) HPLC column and introduced via electrospray into an API2000 SCIEX triple quadrupole tandem mass spectrometer. The following MRM transitions were monitored in positive mode: N-methylnipecotic acid (144.1 → 98.2 m/z), pipecolic acid (130.0 → 84.1 m/z), α-methyl-l-proline (130.1 → 84.1 m/z), and proline (116.2 → 70.1 m/z). α-Methyl-l-proline was clearly distinguished from pipecolic acid based on differences in retention time.
RNA Analysis
RNA was extracted from representative portions of flash-frozen kidney using TRIzol reagent (Invitrogen). Quantitative real time reverse transcriptase-PCR (QPCR) was performed with cDNA generated from 1 μg of total RNA with a SuperScript II reverse transcriptase kit (Invitrogen) using random hexamers. Primers were designed using Primer 3 software (37) based on the following NCBI rhesus macaque reference sequences: SLC6A20 (XM_001114465), SLC36A1 XM_001109989), HNF1a (XM_001089567), COLLECTRIN/TMEM27 (NM_001194195), and β-ACTIN (NM_001033084). Mouse primers were designed using qPrimerDepot (mouseprimerdepot.nci.nih.gov). All primers were further validated using NCBI Primer-BLAST. Primer sequences can be found under supplemental Table S1. QPCR was carried out using SYBR Green PCR master mix (Applied Biosystems) using an ABI Prism 7900HT sequence detection system (Applied Biosystems). Values were quantified using the comparative threshold cycle method and samples were normalized to β-actin. Monkey QPCR products were sequenced to confirm proper target amplification.
Isolation of Kidney Membrane Vesicles and Western Blotting Analysis
Flash-frozen necropsied kidney was homogenized in 1 ml of lysis buffer (5 mm Tris, pH 8.0, 2 mm EDTA, and protease inhibitors). The homogenate was centrifuged twice at 500 × g for 15 min at 4 °C. The supernatant was retained each time and then centrifuged at 36,000 × g (Optima Max Ultracentrifuge, Beckman Coulter) for 15 min at 4 °C. The pellet was resuspended in 100 μl of resuspension buffer (75 mm Tris, pH 8.0, 12.5 mm MgCl2, 5 mm EDTA, and protease inhibitors). Protein concentration was determined using the BCA assay (Thermo Scientific, Waltham, MA). 20 μg of total protein was separated on a 4–15% Tris-HCl gel and transferred to PVDF. Membranes were incubated with antibodies against SLC6A20 (Santa Cruz Biotechnology, Inc.) or used as control for loading ferroportin (Alpha Diagnostic International, San Antonio, TX) or actin (Abcam, Cambridge, MA).
Statistical Analysis
Statistical analysis of differences in ion concentration was performed using GraphPad Prism (San Diego, CA). The significance of metabolite concentration differences was determined using an unpaired t test or the Mann-Whitney test (for data of non-normal distribution). Outliers were detected using Grubb's test (QuickCalcs, GraphPad Software, San Diego, CA). p values less than 0.05 were considered significant.
RESULTS
Stratification of T2DM Monkeys by Clinical Indices
Monkeys were classified into two groups according to the definition recommended by the American Diabetes Association for humans, but with slight modifications to account for a lower upper limit-of-normal fasting plasma glucose (FPG) in rhesus monkeys (3.6 mmol/liter) compared with that for humans (4.7 mmol/l) (38). Monkeys that had two consecutive FPG values of 7 mmol/liter or above were considered T2DM. Normal monkeys had FPG values consistently below 4.4 mmol/liter. Normal monkeys used in the metabolomic studies had the following average values: age, 14.3 ± 6.1 years; body weight, 12.1 ± 4.0 kg; FPG, 3.3 ± 0.5 mmol/liter; serum triglycerides, 0.7 ± 0.3 mmol/liter; HbA1c, 4.2 ± 0.3%; and plasma insulin, 239.4 ± 125.5 pmol/liter. In comparison, monkeys with T2DM used in the metabolomic studies had the following average values: age, 21.1 ± 3.9 years (p < 0.01); body weight, 15.8 ± 3.0 kg (p < 0.05); FPG, 10.2 ± 2.3 mmol/liter (p < 0.001); triglyceride, 9.7 ± 7.1 mmol/liter (p < 0.001); HbA1c, 9.7 ± 0.9% (p < 0.001); and plasma insulin, 815.1 ± 865.4 pmol/liter (p < 0.01). Normal monkeys used in the gene expression and biochemistry studies had the following average values: age, 23.5 ± 6.4 years; body weight, 11.2 ± 6.2 kg; FPG, 3.9 ± 0.6 mmol/liter; triglyceride, 1.2 ± 0.8 mmol/liter; HbA1c, 4.3 ± 0.3%; and plasma insulin, 282 ± 92.3 pmol/liter. Monkeys with T2DM used in the gene expression and biochemistry studies had the following average values: age, 22.9 ± 4.0 (p > 0.05); body weight, 13.8 ± 2.4 kg (p > 0.05); FPG, 14.8 ± 3.8 (p < 0.01); triglyceride, 7.5 ± 8.1 mmol/liter (p < 0.01); HbA1c, 9.7 ± 1.4% (p < 0.01); and plasma insulin, 241.8 ± 139.9 pmol/liter (p < 0.05). Individual values are shown in supplemental Table S2 (monkeys used for metabolomics studies) and supplemental Table S3 (deceased monkeys used for gene expression and biochemistry studies).
Random Forests Parameters and Models
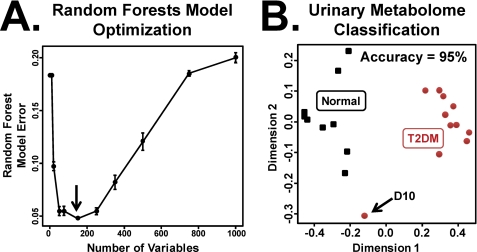
Random forests was used to classify urine as either from normal or T2DM rhesus macaques. A single data matrix consisting of ESI+ and ESI− urine metabolomics data were generated such that each m/z and retention time was represented by a unique variable. Because of the inherent chromatographic noise and spectral artifacts present in LC-MS-based analyses, multiple random forests models were employed to identify the minimal set of variables necessary to build a classifier of normal versus T2DM monkeys. The top 150 ordered variables generated the most accurate random forests model, with an average random forests misclassification error of 5% (Fig. 1A). Non-significant differences in performance were observed between training models based on 150 and 250 variables. The five most important variables (and other notable ones) are described under Table 1.
FIGURE 1.
MDS plots of normal and T2DM monkeys. A, optimization of the number of features used in the random forests model. Models were trained on 10, 20, 50, 75, 150, 250, 350, 500, 750, or 1000 variables. 150 of the top variables were determined to be the most informative (indicated by arrow). Errors bars represent 1 S.D. B, an MDS plot derived from the random forests proximity matrix using the top 150 variables. Normal (n = 10) and T2DM (n = 11) monkeys were classified with an accuracy of 95%. Monkey D10, whereas clinically defined as type 2 diabetic, was misclassified as normal.
TABLE 1.
Summary of biomarker candidates identified by random forests analysis
| Identity | m/z | Retention time | Empirical formula | Mass error | Mean rank (95% CI) |
|---|---|---|---|---|---|
| min | ppm | ||||
| Top biomarker candidates (and adducts) | |||||
| Kynurenic acid | 190.0510+ | 2.78 | C10H7NO3 | 3.2 | 2.0 (1.56–2.52) |
| Pipecolic acid | 130.0873+ | 0.37 | C6H11NO2 | 3.8 | 2.3 (1.96–2.72) |
| Glycine betainea | |||||
| a | 140.0691+ | 0.32 | C5H11NO2Na | 2.9 | 2.4 (2.04–2.64) |
| b | 118.0858+ | 0.36 | C5H11NO2 | 8.5 | 5.6 (5.24–6.08) |
| c | 156.0434+ | 0.31 | C5H11NO2K | 4.5 | 7.2 (6.48–7.84) |
| Glucose | |||||
| d | 215.0351− | 0.34 | C6H12O6Cl | 13.5 | 3.3 (2.88–3.64) |
| Other notable biomarker candidates | |||||
| Citric acid | |||||
| a | 215.0166+ | 0.65 | C6H8O7Na | 0.9 | 42.8 (36.68–48.8) |
a Where a = [Na+] adduct; b = [H+] protonated molecular ion; c = [K+] adduct, and d = [Cl−] adduct.
Random forests-based sample proximity matrices were used to construct multidimensional scaling (MDS) plots for the normal (n = 10) and T2DM (n = 11) monkeys using the top 150 metabolomic variables. When MDS plots were based on the top 150 metabolomic variables alone, the normal and T2DM samples were well separated (Fig. 1B) with a classification accuracy of 95%. Subject D10, although clinically defined as having T2DM, clustered outside of both the normal and T2DM monkeys.
Identification of Biochemical Constituents in the Monkey Urine Metabolome with High Variable Importance Scores
Ions with the highest variable importance scores were analyzed to determine their elemental compositions and identities using Seven Golden Rules (34) and the online Madison Metabolomics Consortium Database (35). The ions 190.0510+ and 130.0873+ ranked as first and second in the variable importance score list, respectively (Table 1). Chemical formula calculations on those ions showed that they corresponded to kynurenic acid (C10H7NO3) and pipecolic acid (C6H11NO2), respectively. Ions 140.0691+ and 215.0351− were found to correspond to the sodium adduct of glycine betaine (C5H11NO2Na+) and the chloride adduct of glucose (C6H12O6Cl−), respectively. Analysis of the other top ranking ions identified 118.0858+ as glycine betaine (C5H11NO2) and 156.0434+ as the potassium adduct of glycine betaine. Another notable ion included the sodium adduct of citrate (215.0166+). The errors in mass measurement ranged from 0.9 to 13.5 ppm (mean = 6.2).
For glycine betaine, kynurenic acid, and pipecolic acid, tandem MS fragmentation patterns and retention times obtained from the urine samples were compared with those obtained from authentic compounds to identify each ion unequivocally. Tandem MS of authentic glycine betaine generated the following m/z fragments (values in parentheses indicate relative percent): 118.0833+ (100), 59.0713+ (6), and 58.0648+ (19). Tandem MS of putative glycine betaine in urine generated the following m/z fragment ions (relative abundance): 118.0827+ (100), 59.0732+ (3), and 58.0642+ (11). Similarly, kynurenic acid was confirmed by comparison of the tandem MS fragmentation patterns of the authentic compound (m/z fragment ions: 190.0531+ (36), 144.0504+ (100), 116.0566+ (14), and 89.0521+ (8)) and putative urinary kynurenic acid (m/z fragment ions: 190.0500+ (33), 144.0486+ (100), 116.0562+ (18), and 89.0428+ (10)). Pipecolic acid was confirmed by comparison of the tandem MS fragmentation patterns of the authentic compound (m/z fragment ions: 130.0835+ (86) and 84.0778+ (100)) and putative urinary pipecolic acid (m/z fragment ions: 130.0848+ (67) and 84.0766+ (100)). For validation by tandem MS, citrate and isocitrate were best observed in ESI− mode because the sodium adduct cannot generate detectable fragment ions in ESI+ mode (data not shown). Isocitrate was found to elute before citrate. Citrate was confirmed by comparison of the tandem MS fragmentation patterns of the authentic compound (m/z fragment ions: 191.0206− (30), 173.0074− (5), 111.0067− (100), 87.0070− (35), and 85.0287− (30)) and putative urinary citric acid (191.0210− (30), 173.0112− (3), 111.0179− (100), 87.0183− (52), and 85.0411− (27)). Isocitrate was eliminated from further analyses because the retention time of citrate closely matched that of the candidate urinary biomarker identified by random forests. Glucose detection was increased in sensitivity by monitoring its chloride adduct (215.0315−) and its identity was validated by comparison with an authentic glucose standard (data not shown).
Quantitation of Urinary Metabolites in Urine Samples from Normal and T2DM Rhesus Macaques
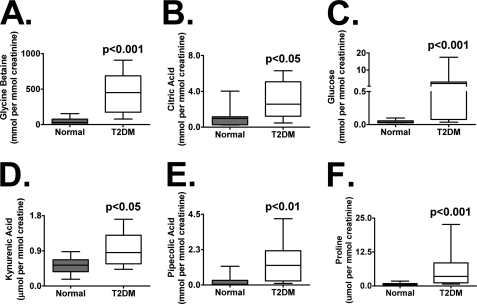
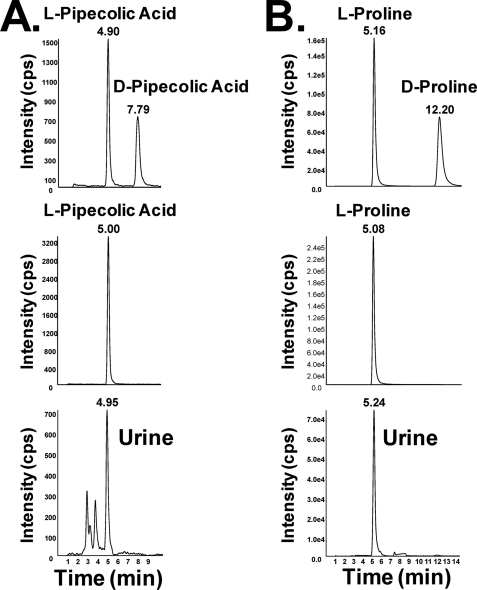
For the ions identified in Table 1, concentration measurements were estimated against calibration curves and expressed as a ratio with creatinine to eliminate variability associated with the collection of spot urines. Glycine betaine (Fig. 2A), kynurenic acid (Fig. 2D), l-pipecolic acid (Fig. 2E), and l-proline (Fig. 2F) concentrations were measured by LC-tandem MS operating in MRM mode. To distinguish the d- and l-isomers of both pipecolic acid and proline, chiral chromatography was used (Fig. 3, A and B). Comparison of mixtures of the dl-stereoisomers, the l-stereoisomer, and urine samples from T2DM monkeys suggested that, in the diabetic condition, the l-stereoisomer predominates. Although not a highly ranked biomarker identified by random forests, l-proline was profiled as it, along with glycine betaine and l-pipecolic acid, is re-absorbed in the kidney proximal tubules by similar transporters. For each biomarker, one normal (C07) and one T2DM (T11) monkey were removed as outliers (as identified by Grubb's test for outliers) having values beyond mean ± 2 S.D. T2DM urines had a 9-fold increase above normal in the glycine betaine concentration (mean ± S.D.; 48 ± 17 to 438 ± 88 μmol/mmol of creatinine; p < 0.001). Kynurenic acid levels were increased in T2DM (0.52 ± 0.07 to 0.95 ± 0.14) monkeys and found to be significantly different (p < 0.05) from normal levels. l-Pipecolic acid concentrations were elevated above normal in T2DMs (0.22 ± 0.13 to 1.42 ± 0.42; p < 0.01). l-Proline was also found to be elevated in T2DMs compared with normal (0.62 ± 0.17 to 4.15 ± 1.04; p < 0.01).
FIGURE 2.
Urine concentration measurements were determined using a standard curve derived from authentic compounds. A, glycine betaine; B, citric acid; C, glucose; D, kynurenic acid; E, l-pipecolic acid; and F, l-proline. l-Pipecolic acid and l-proline were distinguished from their d-isomer by chiral chromatography. Data are plotted after normalizing by creatinine from normal (n = 9) and T2DM (n = 10) urine samples and are expressed as micromoles per millimole of creatinine. Statistical significance when comparing with the normal monkeys was determined by a Mann-Whitney test for non-normal data. Monkeys C07 (normal) and T11 (T2DM) were excluded as outliers by a Grubb test.
FIGURE 3.
Chiral chromatography was used to distinguish the d- and l-stereoisomers of pipecolic acid (A) and proline (B). The top panel in both A and B demonstrates separation and detection of a 50 μm solution containing mixtures of dl-pipecolic acid or dl-proline, respectively. The middle panel in both A and B demonstrates separation and detection of the l-stereoisomer only. The bottom panel in both A and B demonstrates detection of only the l-stereoisomer in a representative diabetic monkey urine sample.
Urinary glucose was measured by LC-MS (in single ion monitoring mode) monitoring the negative ion, 215.0−, corresponding to its chloride adduct (Fig. 2C). Urine glucose in T2DMs was elevated nearly 70-fold compared with normal (45 ± 10 to 3070 ± 165 μmol/mmol of creatinine; p < 0.001). Urinary citric acid was measured using the UPLC to separate it from its isomer, isocitrate (Fig. 2B). Citric acid levels in the T2DMs were elevated 2-fold compared with normal (1060 ± 395 to 2956 ± 634 μmol/mmol of creatinine; p < 0.05).
RNA Analysis and Biochemistry from Kidneys of Normal and T2DM Monkeys
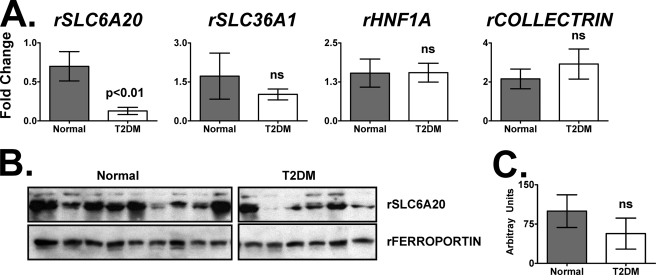
Because it was not practical to biopsy all monkeys in the metabolomic cohort, portions of representative flash-frozen necropsied kidneys (wedges) were taken from deceased monkeys (supplemental Table S3) and used for gene expression and Western blotting analysis. Because the Na+-dependent SLC6A20 transporter in the kidney proximal tubules is known to be important for the re-absorption of imino and N-methylated amino acids such as glycine betaine, l-proline, and l-pipecolic acid (39–41), the mRNA and protein levels of SLC6A20 were measured. QPCR analysis demonstrated a statistically significant (p < 0.05) 4.5-fold decrease in SLC6A20 gene expression (Fig. 4A). There was no significant difference in the expression of mRNAs encoding SLC36A1, HNF1α, or COLLECTRIN/TMEM27 (Fig. 4A). Western blotting of the membrane fraction (Fig. 4B) appeared reduced but there was no significant difference, based on densitometry (Fig. 4C), between SLC6A20 protein levels in normal and T2DM monkey kidneys.
FIGURE 4.
The Na+-dependent transporter SLC6A20 gene expression is reduced in kidneys from normal and T2DM monkeys. A, QPCR analysis of mRNAs encoded by SLC6A20 (4.5-fold reduction), SLC36A1 (no significant change), HNF1A (no significant change), and COLLECTRIN/TMEM27 (no significant change) in normal and T2DM monkeys. B, protein expression of SLC6A20 and FERROPORTIN from kidney membrane fractions isolated from normal and monkeys with T2DM. C, densitometry of the Western blots shown in B. Arbitrary units were calculated by dividing SLC6A20 densitometry values by those for ferroportin. ns, not significant. The r stands for “rhesus.”
Quantitation of Urinary Biomarkers in Urine Samples from Wild-type and db/db Mice
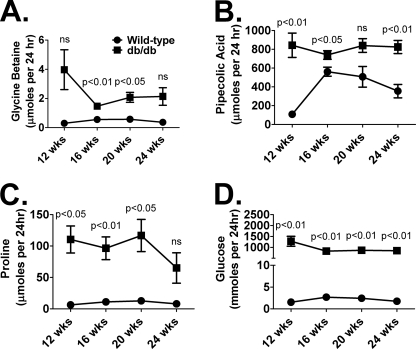
To understand better their association with T2DM and the SLC6A20 transporter, glycine betaine, l-pipecolic acid, and l-proline urinary levels were quantitated in an additional, yet distinct, model of T2DM, the db/db mouse. Urine samples were collected from age-matched, male wild-type and db/db mice at 12-, 16-, 20-, and 24-weeks of age and glycine betaine, l-pipecolic acid, l-proline, and glucose were quantitated (Fig. 5). To account for the large diuresis differences between the wild-type and db/db mice (not shown) and additional factors influencing urinary creatinine excretion in this mouse model (10), results are expressed as micromoles or millimoles per 24 h. Over the three-month sampling period, increased excretion of glycine betaine (Fig. 5A), pipecolic acid (Fig. 5B), proline (Fig. 5C), and glucose (Fig. 5D) was observed. Pipecolic acid (except at 20-weeks) and glucose urinary excretion in db/db mice was significantly different from wild-type at all time points. However, differences in glycine betaine excretion reached statistical significance at 16 (p < 0.01) and 20 weeks (p < 0.05), whereas differences in proline excretion were significant at 12 (p < 0.05), 16 (p < 0.01), and 20 weeks (p < 0.05) of age.
FIGURE 5.
Wild-type and db/db urinary measurements of glycine betaine (A), pipecolic acid (B), proline (C), and glucose (D) at 12 (n = 3), 16 (n = 4), 20 (n = 4), and 24 weeks (n = 4) of age. Values are expressed as micromoles or millimoles per 24 h. Significance was determined using an unpaired t test. ns, not significant.
Gene Expression and Western Blotting Analysis from Kidneys of Wild-type and db/db Mice
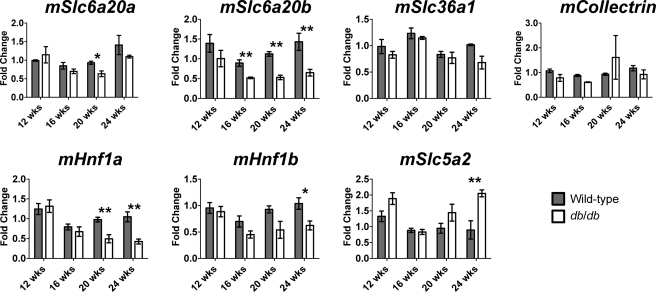
Unlike in monkeys (and humans), the SLC6A20 transporter exists as a gene duplication in mice. Therefore, expression of both the homologues Slc6a20a and Slc6a20b was assessed in the kidneys of wild-type and db/db mice at 12, 16, 20, and 24 weeks of age. Slc6a20a expression was significantly reduced at 20 weeks (p < 0.05) of age in the db/db mouse (Fig. 6). However, Slc6a20b expression was significantly (p < 0.01) reduced at 16, 20, and 24 weeks of age. Expression of mRNAs encoded by the kidney transporter, Slc36a1, and the gene (Collectrin/Tmem27) encoding the docking protein, collectrin, were unchanged. Because Hnf1α/β was shown to be important for regulating the expression of various genes in the kidney (42–48), the expression of mRNA encoding Hnf1α and Hnf1β was assessed. Hnf1α mRNA was significantly reduced at 20 and 24 weeks of age, whereas Hnf1β mRNA was significantly reduced at 24 weeks of age. To determine whether these changes were due to a general decline in kidney gene expression, the gene expression of Slc5a2, a sodium/glucose co-transporter, was examined and found to be significantly up-regulated in the db/db mouse at 24 weeks of age.
FIGURE 6.
QPCR analysis of the kidney proximal kidney transporters and nuclear receptors in kidneys from wild-type and db/db mice at 12 (n = 3), 16 (n = 4), 20 (n = 4), and 24 weeks (n = 4) of age. On average (using all time points), there was a 40% reduction in Slc6a20b gene expression in kidneys obtained from db/db mice. Hnf1a mRNA expression was significantly reduced in the db/db mouse at 20 and 24 weeks of age and Hnf1b mRNA was significantly reduced at 24 weeks. Slc65a2 mRNA was significantly up-regulated at 24 weeks of age in the db/db mouse. Significance was determined using an unpaired t test. *, p < 0.05, **, p < 0.01.
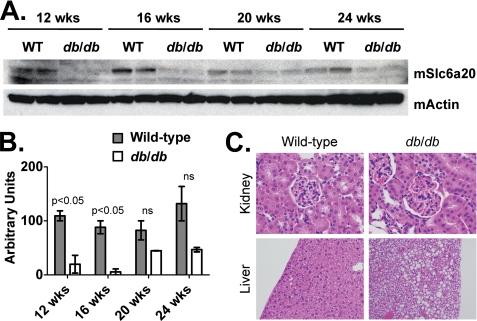
Western blotting analysis revealed significant reduction of Slc6a20 protein levels in kidney membrane fractions of wild-type and db/db mice (Fig. 7A). Although only reaching statistical significance at 16 and 20 weeks, there was a general decline (50% or greater) in Slc6a20 protein levels at all time points (Fig. 7B). The antibody used in this experiment did not differentiate between levels of Slc6a20a and Slc6a20b proteins. Interestingly, even at the 24-week time point, there was no significant change in kidney histology (Fig. 7C, top panels), yet the liver, as expected, was significantly steatotic (Fig. 7C, bottom panels).
FIGURE 7.
Wild-type and db/db protein expression of (A) SLC6A20 and ACTIN in kidney membrane fractions isolated at 12-, 16-, 20-, and 24-weeks of age. B, densitometry of the Westerns blots shown in A. Arbitrary units were calculated by dividing SL6A20 densitometry values by those for ACTIN. ns, not significant. On average (using all time points), there was a 70% reduction in protein levels of SLC6A20 in kidney membrane fractions obtained from db/db mice. The m stands for “mouse.”
Quantitation of Serum Biomarkers in Wild-type and db/db Mice
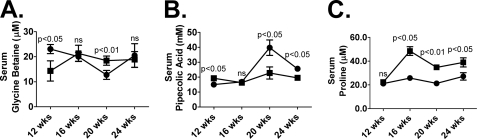
To better understand if the increased urinary excretion of metabolites such as glycine betaine, pipecolic acid, and proline results in decreased circulating levels in the blood, the metabolites were quantitated in the serum. There was no consistent pattern of serum glycine betaine levels over the 4-week study (Fig. 8). Serum glycine betaine levels in the db/db mouse were significantly depleted (p < 0.05) at 12 weeks but significantly elevated (p < 0.01) at 20 weeks of age (Fig. 8A). Serum pipecolic acid levels in the db/db mouse were significantly depleted at 20 (p < 0.05) and 24 weeks (p < 0.05) of age (Fig. 8B). Serum proline levels (Fig. 8C), however, were significantly elevated in the db/db mouse at 16 (p < 0.05), 20 (p < 0.01), and 24 weeks (p < 0.05) of age.
FIGURE 8.
Wild-type (circles) and db/db (squares) serum measurements of glycine betaine (A), pipecolic acid (B), and proline (C) at 12 (n = 3), 16 (n = 4), 20 (n = 4), and 24 weeks (n = 4) of age. Values are expressed as micromolar. Significance was determined using an unpaired t test. ns, not significant.
DISCUSSION
In this study, urine from a well characterized cohort of normal and T2DM rhesus macaques was profiled using a UPLC-ESI-QTOF-MS-based metabolomic approach. A random forests-based algorithm was used to classify normal and T2DM monkeys on the basis of urine metabolomic measurements alone. Four molecules, glycine betaine, glucose, kynurenic acid, and pipecolic acid, were identified and quantified. In the monkey, the kidney proximal renal tubule transporter, SLC6A20, was expressed at significantly lower levels (p < 0.01) in monkeys with T2DM and correlated with reduced SLC6A20 protein. These observations were confirmed in the db/db mouse model (reduced Slc6a20a/Slc6a20b gene expression, reduced Slc6a20 protein, and increased urinary excretion of glycine betaine, proline, pipecolic acid, and glucose) thus suggesting that a decline in SLC6A20 function may be an important and perhaps prognostic indicator of overall kidney health and function.
A likely scenario by which to explain the increased excretion of pipecolic acid and glycine betaine is defective or reduced activity of transporters located in the kidney proximal tubules. The solute carrier family 6 (SLC6) is important for the reabsorption of small hydrophilic compounds, coupled with Na+ and Cl− ions, such as glycine betaine and l-pipecolic acid (41). SLC6A20 (XT3, SIT1, Xtrp3) was demonstrated to transport these molecules in addition to l-proline (39–41). Therefore, it was hypothesized that the increased excretion of these urinary constituents was due to reduced levels of SLC6A20 in the kidney of monkeys with T2DM. Indeed, both mRNA and Western blotting analysis results from T2DM monkeys and the db/db mouse support this hypothesis but raise several important questions. First, is this observation nonspecific due to kidney damage associated with diabetic nephropathy? Because no change was observed in the mRNA encoding COLLECTRIN/TMEM27 (a protein necessary for docking SLC6A20 at the luminal membrane) or SLC36A1 (an amino acid transporter) in the T2DM monkeys and mice, it is unlikely that the observed SLC6A20 attenuation is due to a general decline in the kidney proximal tubule architecture secondary to complications associated with T2DM. Furthermore, the Na(+)/glucose co-transporter, SLC5A2/SGLT, was found to be induced in the db/db mouse kidney as previously reported (49). Second, is the reduced expression of SLC6A20 contributing to the pathogenesis of T2DM and its associated complications, or is its reduced expression an indication of changes in upstream signaling pathways responsible for regulation of the SLC6A20 gene? The present results in the db/db mouse model indicate that Hnf1α and/or Hnf1β are important for regulating Slc6a20a/b gene expression. Indeed, recent studies have shown that Hnf1 is an important regulator of Slc6a20 in the mouse (50, 51). However, what remains is to determine which homologue contributes to the observed phenotype, particularly because others have shown that Slc6a20b is non-functional when expressed in Xenopus laevis oocytes (52). Future studies using genetically modified mice are essential to determine which homologue is most critical for reabsorption of metabolites in the kidney proximal tubules in vivo. Additionally, whereas not directly addressed in the study by Mishra et al. (54), a search of GEO Profiles (53) revealed that Slc6a20b gene expression was reduced nearly 7-fold in the kidneys of 16-week-old diabetic db/db mice compared with age-matched controls. That observation, along with data presented herein, strongly suggests that the decline in SLC6A20 gene expression is tightly associated with T2DM.
Other published studies looking at urinary glycine betaine levels in T2DM have associated elevated excretion of this metabolite with proximal tubular dysfunction (55). Interestingly, elevated excretion was not simply due to hyperglycemia (56) or renal disease (57). Furthermore, there was no apparent correlation with blood levels of glycine betaine (55, 57, 58). In the present study, serum glycine betaine levels also showed no correlation with urinary levels in the db/db mouse. However, pipecolic acid levels in the db/db mouse were depleted compared with wild-type mice suggesting that increases in this urinary metabolite might more accurately reflect the status of SLC6A20. Although the potential of these metabolites as early indicators of renal disease and/or dysfunction is speculative, it warrants further studies.
Alternatively, increased excretion of pipecolic acid may reflect other physiologic changes. Pipecolic acid has not previously been identified as a urinary biomarker of T2DM but has been associated with a high-fat diet and liver stress in rats (59). Elevated serum pipecolic acid, or hyperpipecolic acidemia, is most commonly associated with peroxisomal defect syndromes (Zellweger and its infantile form, Refsum) and has been reported in some non-peroxisomal diseases (60–62). The peroxisomal disorders are associated with, among many other symptoms, hepatomegaly, neuropathy, renal complications, and both eye and craniofacial abnormalities (63). Genetically, Zellweger and Refsum syndromes, along with many other peroxisomal biogenesis disorders, are thought to result from defects in the PEX gene family (63, 64). In relation to T2DM, the observed hyperpipecolic acidemia may simply reflect peroxisomal stress associated with excessive serum triglycerides and may be an early sign of liver damage. A recent metabolomic study observed several nicotinamide metabolites to be elevated in the urine of both T2DM rodents and T2DM humans. Interestingly, these metabolites were also reported to be biomarkers for peroxisome proliferator-activated receptor α activation (10, 65, 66). Given the clear role of peroxisome proliferator-activated receptor α and its family members in fatty acid metabolism, pipecolic acid and nicotinamide metabolites as diagnostic biomarkers for T2DM in human patients should be evaluated.
Increased excretion of glycine betaine in T2DM was reported for both rodents and humans (13, 55, 56). Glycine betaine is primarily diet-derived but can be synthesized from choline in the liver and kidney as well as by gut microbiota (10, 55, 56, 67, 68). Although the precise reason for its elevation in concentration is uncertain, it might also be an indicator of the compensatory increase of organic osmolytes of the body, particularly in the proximal tubules of the kidney, in response to high blood glucose concentrations (55, 56, 69–71). Therefore, glycine betaine may serve as an early indicator of diabetic nephropathy and reduced SLC6A20 function, for which only a few, relatively poor markers currently exist (7).
Kynurenic acid, a tryptophan metabolite, has been associated with the development of dense nuclear cataracts as well as nephropathy, both complications of T2DM (72). Interestingly, elevated kynurenic acid in the urine is thought to be a consequence of reduced plasma vitamin B6 (73). It was recently reported that a mouse model (KK-Ay/Ta) with spontaneous T2DM showed significant improvements in several biochemical parameters as well as improved kidney function when fed a diet supplemented with pyridoxamine (a catalytically active amine form of vitamin B6) (74). The proposed mechanism is based on inhibition of advanced glycation end products by pyridoxamine (75).
Increased urinary concentrations of glucose in T2DM animals was not unexpected given the high fasting plasma glucose measurements. Surprisingly, however, urine glucose was not the most important variable that distinguished between T2DM and normal individuals in the random forests analysis. This observation, along with the other urinary metabolites identified in this study, exemplify the multifactorial phenotype of T2DM and despite decades of research, additional comprehensive comparative analyses of normal and T2DM urinary metabolomes is indicated.
CONCLUSION
A novel perspective of the T2DM metabolic phenotype was developed by profiling the urinary metabolomes of rhesus macaques from a unique colony consisting of normal and T2DM cohorts. A combination of UPLC-ESI-QTOF-MS and random forests biomarker-mining approaches revealed several urinary constituents whose concentrations were elevated to a statistically significant extent. One compound in particular, l-pipecolic acid, has not previously been reported in the diabetes literature, but, intriguingly, it has been linked to peroxisome proliferator-activated receptor α, peroxisomal function, and peroxisomal disease. Furthermore, this approach has identified a unique target, the proximal tubule transporter SLC6A20, thus providing potential biomarkers associated with diabetic nephropathy. The complex metabolomic phenotype of T2DM described here provides a non-glycocentric view of this disease that may furnish an information base for future studies that monitor the pathogenesis and progression of T2DM in human populations.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants NO1-AG-3-1012 (to B. C. H.), AG-42100 (to B. C. H.), and HHSN26300800022C and the National Cancer Institute, Intramural Research Program.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3.
- T2DM
- type 2 diabetes mellitus
- UPLC
- ultra-performance liquid chromatography
- QPCR
- quantitative PCR
- FPG
- fasting plasma glucose
- MDS
- multidimensional scaling.
REFERENCES
- 1. Lanza I. R., Zhang S., Ward L. E., Karakelides H., Raftery D., Nair K. S. (2010) PLoS One 5, e10538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bain J. R., Stevens R. D., Wenner B. R., Ilkayeva O., Muoio D. M., Newgard C. B. (2009) Diabetes 58, 2429–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang J., Yan L., Chen W., Lin L., Song X., Yan X., Hang W., Huang B. (2009) Anal. Chim. Acta 650, 16–22 [DOI] [PubMed] [Google Scholar]
- 4. Zhang X., Wang Y., Hao F., Zhou X., Han X., Tang H., Ji L. (2009) J. Proteome Res. 8, 5188–5195 [DOI] [PubMed] [Google Scholar]
- 5. Griffin J. L., Nicholls A. W. (2006) Pharmacogenomics 7, 1095–1107 [DOI] [PubMed] [Google Scholar]
- 6. Idle J. R., Gonzalez F. J. (2007) Cell Metab. 6, 348–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. (2005) J. Am. Soc. Nephrol. 16, 1886–1903 [DOI] [PubMed] [Google Scholar]
- 8. Hodavance M. S., Ralston S. L., Pelczer I. (2007) Anal. Bioanal. Chem. 387, 533–537 [DOI] [PubMed] [Google Scholar]
- 9. Major H. J., Williams R., Wilson A. J., Wilson I. D. (2006) Rapid Commun. Mass Spectrom. 20, 3295–3302 [DOI] [PubMed] [Google Scholar]
- 10. Salek R. M., Maguire M. L., Bentley E., Rubtsov D. V., Hough T., Cheeseman M., Nunez D., Sweatman B. C., Haselden J. N., Cox R. D., Connor S. C., Griffin J. L. (2007) Physiol. Genomics 29, 99–108 [DOI] [PubMed] [Google Scholar]
- 11. Wang C., Kong H., Guan Y., Yang J., Gu J., Yang S., Xu G. (2005) Anal. Chem. 77, 4108–4116 [DOI] [PubMed] [Google Scholar]
- 12. Williams R., Lenz E. M., Wilson A. J., Granger J., Wilson I. D., Major H., Stumpf C., Plumb R. (2006) Mol. Biosyst. 2, 174–183 [DOI] [PubMed] [Google Scholar]
- 13. Williams R. E., Lenz E. M., Evans J. A., Wilson I. D., Granger J. H., Plumb R. S., Stumpf C. L. (2005) J. Pharm. Biomed. Anal. 38, 465–471 [DOI] [PubMed] [Google Scholar]
- 14. Gibbs R. A., Rogers J., Katze M. G., Bumgarner R., Weinstock G. M., Mardis E. R., Remington K. A., Strausberg R. L., Venter J. C., Wilson R. K., Batzer M. A., Bustamante C. D., Eichler E. E., Hahn M. W., Hardison R. C., Makova K. D., Miller W., Milosavljevic A., Palermo R. E., Siepel A., Sikela J. M., Attaway T., Bell S., Bernard K. E., Buhay C. J., Chandrabose M. N., Dao M., Davis C., Delehaunty K. D., Ding Y., Dinh H. H., Dugan-Rocha S., Fulton L. A., Gabisi R. A., Garner T. T., Godfrey J., Hawes A. C., Hernandez J., Hines S., Holder M., Hume J., Jhangiani S. N., Joshi V., Khan Z. M., Kirkness E. F., Cree A., Fowler R. G., Lee S., Lewis L. R., Li Z., Liu Y. S., Moore S. M., Muzny D., Nazareth L. V., Ngo D. N., Okwuonu G. O., Pai G., Parker D., Paul H. A., Pfannkoch C., Pohl C. S., Rogers Y. H., Ruiz S. J., Sabo A., Santibanez J., Schneider B. W., Smith S. M., Sodergren E., Svatek A. F., Utterback T. R., Vattathil S., Warren W., White C. S., Chinwalla A. T., Feng Y., Halpern A. L., Hillier L. W., Huang X., Minx P., Nelson J. O., Pepin K. H., Qin X., Sutton G. G., Venter E., Walenz B. P., Wallis J. W., Worley K. C., Yang S. P., Jones S. M., Marra M. A., Rocchi M., Schein J. E., Baertsch R., Clarke L., Csürös M., Glasscock J., Harris R. A., Havlak P., Jackson A. R., Jiang H., Liu Y., Messina D. N., Shen Y., Song H. X., Wylie T., Zhang L., Birney E., Han K., Konkel M. K., Lee J., Smit A. F., Ullmer B., Wang H., Xing J., Burhans R., Cheng Z., Karro J. E., Ma J., Raney B., She X., Cox M. J., Demuth J. P., Dumas L. J., Han S. G., Hopkins J., Karimpour-Fard A., Kim Y. H., Pollack J. R., Vinar T., Addo-Quaye C., Degenhardt J., Denby A., Hubisz M. J., Indap A., Kosiol C., Lahn B. T., Lawson H. A., Marklein A., Nielsen R., Vallender E. J., Clark A. G., Ferguson B., Hernandez R. D., Hirani K., Kehrer-Sawatzki H., Kolb J., Patil S., Pu L. L., Ren Y., Smith D. G., Wheeler D. A., Schenck I., Ball E. V., Chen R., Cooper D. N., Giardine B., Hsu F., Kent W. J., Lesk A., Nelson D. L., O'Brien W. E., Prüfer K., Stenson P. D., Wallace J. C., Ke H., Liu X. M., Wang P., Xiang A. P., Yang F., Barber G. P., Haussler D., Karolchik D., Kern A. D., Kuhn R. M., Smith K. E., Zwieg A. S. (2007) Science 316, 222–23417431167 [Google Scholar]
- 15. Hansen B. C. (2004) in Diabetes Mellitus: A Fundamental and Clinical Text (Leroith D., Olefsky J. M., Taylor S. eds) pp. 1059–1074, Lippincott Williams and Williams, Philadelphia, PA [Google Scholar]
- 16. Tigno X. T., Erwin J. M., Hansen B. C. (2007) in The Laboratory Primate, The Handbook of Experimental Animals (Wolfe-Coote S. ed) pp. 451–468, Academic Press, Elsevier Science, The Netherlands [Google Scholar]
- 17. Ding S. Y., Tigno X. T., Hansen B. C. (2007) Metab. Clin. Exp. 56, 838–846 [DOI] [PubMed] [Google Scholar]
- 18. Ortmeyer H. K., Adall Y., Marciani K. R., Katsiaras A., Ryan A. S., Bodkin N. L., Hansen B. C. (2005) Am. J. Physiol. Regul. Integr. Comp. Physiol. 288, R1509–1517 [DOI] [PubMed] [Google Scholar]
- 19. Tigno X. T., Ding S. Y., Hansen B. C. (2006) Clin. Hemorheology Microcirc. 34, 273–282 [PubMed] [Google Scholar]
- 20. Nadeau K. J., Ehlers L. B., Aguirre L. E., Moore R. L., Jew K. N., Ortmeyer H. K., Hansen B. C., Reusch J. E., Draznin B. (2006) Am. J. Physiol. Endocrinol Metab. 291, E90–98 [DOI] [PubMed] [Google Scholar]
- 21. Tigno X. T., Gerzanich G., Hansen B. C. (2004) J. Gerontol. 59, 1081–1088 [DOI] [PubMed] [Google Scholar]
- 22. Bodkin N. L., Alexander T. M., Ortmeyer H. K., Johnson E., Hansen B. C. (2003) J. Gerontol. 58, 212–219 [DOI] [PubMed] [Google Scholar]
- 23. Standaert M. L., Ortmeyer H. K., Sajan M. P., Kanoh Y., Bandyopadhyay G., Hansen B. C., Farese R. V. (2002) Diabetes 51, 2936–2943 [DOI] [PubMed] [Google Scholar]
- 24. Wagner J. D., Cline J. M., Shadoan M. K., Bullock B. C., Rankin S. E., Cefalu W. T. (2001) Toxicol. Pathol. 29, 142–148 [DOI] [PubMed] [Google Scholar]
- 25. Paré M., Albrecht P. J., Noto C. J., Bodkin N. L., Pittenger G. L., Schreyer D. J., Tigno X. T., Hansen B. C., Rice F. L. (2007) J. Comp. Neurol. 501, 543–567 [DOI] [PubMed] [Google Scholar]
- 26. Otsuji T., McLeod D. S., Hansen B., Lutty G. (2002) Exp. Eye Res. 75, 201–208 [DOI] [PubMed] [Google Scholar]
- 27. Kim S. Y., Johnson M. A., McLeod D. S., Alexander T., Otsuji T., Steidl S. M., Hansen B. C., Lutty G. A. (2004) Invest. Ophthalmol. Vis. Sci. 45, 4543–4553 [DOI] [PubMed] [Google Scholar]
- 28. Kim S. Y., Johnson M. A., McLeod D. S., Alexander T., Hansen B. C., Lutty G. A. (2005) Diabetes 54, 1534–1542 [DOI] [PubMed] [Google Scholar]
- 29. Johnson M. A., Lutty G. A., McLeod D. S., Otsuji T., Flower R. W., Sandagar G., Alexander T., Steidl S. M., Hansen B. C. (2005) Exp. Eye Res. 80, 37–42 [DOI] [PubMed] [Google Scholar]
- 30. Cusumano A. M., Bodkin N. L., Hansen B. C., Iotti R., Owens J., Klotman P. E., Kopp J. B. (2002) Am. J. Kidney Dis. 40, 1075–1085 [DOI] [PubMed] [Google Scholar]
- 31. Pennisi E. (2007) Science 316, 218–221 [DOI] [PubMed] [Google Scholar]
- 32. Pennisi E. (2007) Science 316, 216–218 [DOI] [PubMed] [Google Scholar]
- 33. Breiman L. (2001) Mach. Learning 45, 5–32 [Google Scholar]
- 34. Kind T., Fiehn O. (2007) BMC Bioinformatics 8, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Markley J. L., Anderson M. E., Cui Q., Eghbalnia H. R., Lewis I. A., Hegeman A. D., Li J., Schulte C. F., Sussman M. R., Westler W. M., Ulrich E. L., Zolnai Z. (2007) Pac. Symp. Biocomput. 12, 157–168 [PubMed] [Google Scholar]
- 36. Rashed M. S., Al-Ahaidib L. Y., Aboul-Enein H. Y., Al-Amoudi M., Jacob M. (2001) Clin. Chem. 47, 2124–2130 [PubMed] [Google Scholar]
- 37. Misener S., Krawetz S. A. (2000) Bioinformatics Methods and Protocols, Humana Press, Totowa, NJ [Google Scholar]
- 38. Expert Committee on the Diagnosis and Clasification of Diabetes Mellitus (2003) Diabetes Care 26, Suppl. 1, S5–20 [DOI] [PubMed] [Google Scholar]
- 39. Romeo E., Dave M. H., Bacic D., Ristic Z., Camargo S. M., Loffing J., Wagner C. A., Verrey F. (2006) Am. J. Physiol. Renal Physiol. 290, F376–383 [DOI] [PubMed] [Google Scholar]
- 40. Takanaga H., Mackenzie B., Suzuki Y., Hediger M. A. (2005) J. Biol. Chem. 280, 8974–8984 [DOI] [PubMed] [Google Scholar]
- 41. Verrey F., Singer D., Ramadan T., Vuille-dit-Bille R. N., Mariotta L., Camargo S. M. (2009) Pflugers Arch. 458, 53–60 [DOI] [PubMed] [Google Scholar]
- 42. Cassuto H., Olswang Y., Heinemann S., Sabbagh K., Hanson R. W., Reshef L. (2003) Gene 318, 177–184 [DOI] [PubMed] [Google Scholar]
- 43. Cassuto H., Olswang Y., Livoff A. F., Nechushtan H., Hanson R. W., Reshef L. (1997) FEBS Lett. 412, 597–602 [DOI] [PubMed] [Google Scholar]
- 44. D'Amato E., d'Annunzio G., Calcaterra V., Morsellino V., Larizza D., Lorini R. (2008) Pediatr. Nephrol. 23, 137–140 [DOI] [PubMed] [Google Scholar]
- 45. Frank S., Zoll B. (1998) DNA Cell Biol. 17, 679–688 [DOI] [PubMed] [Google Scholar]
- 46. Freitas H. S., Schaan B. D., David-Silva A., Sabino-Silva R., Okamoto M. M., Alves-Wagner A. B., Mori R. C., Machado U. F. (2009) Mol. Cell. Endocrinol. 305, 63–70 [DOI] [PubMed] [Google Scholar]
- 47. Igarashi P., Shao X., McNally B. T., Hiesberger T. (2005) Kidney Int. 68, 1944–1947 [DOI] [PubMed] [Google Scholar]
- 48. Mache C. J., Preisegger K. H., Kopp S., Ratschek M., Ring E. (2002) Pediatr. Nephrol. 17, 1021–1026 [DOI] [PubMed] [Google Scholar]
- 49. Arakawa K., Ishihara T., Oku A., Nawano M., Ueta K., Kitamura K., Matsumoto M., Saito A. (2001) Br. J. Pharmacol. 132, 578–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bonzo J. A., Patterson A. D., Krausz K. W., Gonzalez F. J. (2010) Mol. Endocrinol. 24, 2343–2355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kikuchi R., Yagi S., Kusuhara H., Imai S., Sugiyama Y., Shiota K. (2010) Kidney Int. 78, 569–577 [DOI] [PubMed] [Google Scholar]
- 52. Bröer A., Cavanaugh J. A., Rasko J. E., Bröer S. (2006) Pflugers Arch. 451, 511–517 [DOI] [PubMed] [Google Scholar]
- 53. Barrett T., Troup D. B., Wilhite S. E., Ledoux P., Rudnev D., Evangelista C., Kim I. F., Soboleva A., Tomashevsky M., Marshall K. A., Phillippy K. H., Sherman P. M., Muertter R. N., Edgar R. (2009) Nucleic Acids Res. 37, D885–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mishra R., Emancipator S. N., Miller C., Kern T., Simonson M. S. (2004) Am. J. Physiol. Renal Physiol. 286, F913–921 [DOI] [PubMed] [Google Scholar]
- 55. Dellow W. J., Chambers S. T., Lever M., Lunt H., Robson R. A. (1999) Diabetes Res. Clin. Pract. 43, 91–99 [DOI] [PubMed] [Google Scholar]
- 56. Dellow W. J., Chambers S. T., Barrell G. K., Lever M., Robson R. A. (2001) Diabetes Res. Clin. Pract. 52, 165–169 [DOI] [PubMed] [Google Scholar]
- 57. Lever M., Sizeland P. C., Bason L. M., Hayman C. M., Robson R. A., Chambers S. T. (1994) Clin. Chim. Acta 230, 69–79 [DOI] [PubMed] [Google Scholar]
- 58. Lever M., Sizeland P. C. B., Bason L. M., Hayman C. M., Chambers S. T. (1994) Bba-Gen. Subjects 1200, 259–264 [DOI] [PubMed] [Google Scholar]
- 59. Fardet A., Llorach R., Martin J. F., Besson C., Lyan B., Pujos-Guillot E., Scalbert A. (2008) J. Proteome Res. 7, 2388–2398 [DOI] [PubMed] [Google Scholar]
- 60. Lam S., Hutzler J., Dancis J. (1986) Biochim. Biophys. Acta 882, 254–257 [DOI] [PubMed] [Google Scholar]
- 61. Peduto A., Baumgartner M. R., Verhoeven N. M., Rabier D., Spada M., Nassogne M. C., Poll-The B. T., Bonetti G., Jakobs C., Saudubray J. M. (2004) Mol. Genet. Metab. 82, 224–230 [DOI] [PubMed] [Google Scholar]
- 62. Baas J. C., van de Laar R., Dorland L., Duran M., Berger R., Poll-The B. T., de Koning T. J. (2002) J. Inherit. Metab. Dis. 25, 699–701 [DOI] [PubMed] [Google Scholar]
- 63. Steinberg S. J., Dodt G., Raymond G. V., Braverman N. E., Moser A. B., Moser H. W. (2006) Biochim. Biophys. Acta 1763, 1733–1748 [DOI] [PubMed] [Google Scholar]
- 64. Gould S. J., Valle D. (2000) Trends Genet. 16, 340–345 [DOI] [PubMed] [Google Scholar]
- 65. Ringeissen S., Connor S. C., Brown H. R., Sweatman B. C., Hodson M. P., Kenny S. P., Haworth R. I., McGill P., Price M. A., Aylott M. C., Nunez D. J., Haselden J. N., Waterfield C. J. (2003) Biomarkers 8, 240–271 [DOI] [PubMed] [Google Scholar]
- 66. Zhen Y., Krausz K. W., Chen C., Idle J. R., Gonzalez F. J. (2007) Mol. Endocrinol. 21, 2136–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Grossman E. B., Hebert S. C. (1989) Am. J. Physiol. Renal Physiol. 256, F107–112 [DOI] [PubMed] [Google Scholar]
- 68. Lohr J., Acara M. (1990) J. Pharmacol. Exp. Ther. 252, 154–158 [PubMed] [Google Scholar]
- 69. Bagnasco S., Balaban R., Fales H. M., Yang Y. M., Burg M. (1986) J. Biol. Chem. 261, 5872–5877 [PubMed] [Google Scholar]
- 70. Garcia-Perez A., Burg M. B. (1990) Hypertension 16, 595–602 [DOI] [PubMed] [Google Scholar]
- 71. Körner A., Eklöf A. C., Celsi G., Aperia A. (1994) Diabetes 43, 629–633 [DOI] [PubMed] [Google Scholar]
- 72. Numa S., Hashimoto T., Nakanishi S., Okazaki T. (1972) Biochem. Soc. Symp. 35, 27–39 [PubMed] [Google Scholar]
- 73. Takeuchi F., Tsubouchi R., Izuta S., Shibata Y. (1989) J. Nutr. Sci. Vitaminol. 35, 111–122 [DOI] [PubMed] [Google Scholar]
- 74. Tanimoto M., Gohda T., Kaneko S., Hagiwara S., Murakoshi M., Aoki T., Yamada K., Ito T., Matsumoto M., Horikoshi S., Tomino Y. (2007) Metab. Clin. Exp. 56, 160–167 [DOI] [PubMed] [Google Scholar]
- 75. Khalifah R. G., Todd P., Booth A. A., Yang S. X., Mott J. D., Hudson B. G. (1996) Biochemistry 35, 4645–4654 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.