Abstract
Aberrant expression of the human hyaluronan synthase 2 (HAS2) gene has been implicated in the pathology of malignancy, pulmonary arterial hypertension, osteoarthritis, asthma, thyroid dysfunction, and large organ fibrosis. Renal fibrosis is associated with increased cortical synthesis of hyaluronan (HA), an extracellular matrix glycosaminoglycan, and we have shown that HA is a correlate of interstitial fibrosis in vivo. Our previous in vitro data have suggested that both HAS2 transcriptional induction and subsequent HAS2-driven HA synthesis may contribute to kidney fibrosis via phenotypic modulation of the renal proximal tubular epithelial cell (PTC). Post-transcriptional regulation of HAS2 mRNA synthesis by the natural antisense RNA HAS2-AS1 has recently been described in osteosarcoma cells, but the antisense transcript was not detected in kidney. In this study, PTC stimulation with IL-1β or TGF-β1 induced coordinated temporal profiles of HAS2-AS1 and HAS2 transcription. Constitutive activity of the putative HAS2-AS1 promoter was demonstrated, and transcription factor-binding sequence motifs were identified. Knockdown of Sp1/Sp3 expression by siRNA blunted IL-1β induction of both HAS2-AS1 and HAS2, and Smad2/Smad3 knockdown similarly attenuated TGF-β1 stimulation. Inhibition of IL-1β-stimulated HAS2-AS1 RNA induction using HAS2-AS1-specific siRNAs also suppressed up-regulation of HAS2 mRNA transcription. The thermodynamic feasibility of HAS2-AS1/HAS2 heterodimer formation was demonstrated in silico, and locus-specific cytoplasmic double-stranded RNA was detected in vitro. In summary, our data show that transcriptional induction of HAS2-AS1 and HAS2 occurs simultaneously in PTCs and suggest that transcription of the antisense RNA stabilizes or augments HAS2 mRNA expression in these cells via RNA/mRNA heteroduplex formation.
Keywords: Antisense RNA, Extracellular Matrix, Gene Transcription, Hyaluronate, Kidney, Hyaluronan Synthase 2, Natural Antisense HAS2-AS1, Noncoding RNA, Post-transcriptional Regulation, Renal Fibrosis
Introduction
The linear non-sulfated glycosaminoglycan hyaluronan (HA)5 is a ubiquitous component of the vertebrate extracellular matrix with a multiplicity of cellular functions in both physiological and pathophysiological contexts (1–8). HA is synthesized at the cell membrane by the HA synthase (HAS) enzymes, encoded in humans by the corresponding multigene family HAS1–3 (9–11).
Frequently associated with the fibrotic response in major organs, HA is a highly effective biomarker for liver fibrosis (12, 13). Similarly, in lung fibrosis, accumulation of HA has been observed (14, 15), and in some cases, up-regulated HAS2 transcription has also been reported (14). For all progressive renal diseases, the rate of progression correlates with the degree of corticointerstitial fibrosis. Increased HA synthesis and extracellular matrix expansion in the renal corticointerstitium are common features of kidney fibrosis (16), and our recent studies on renal biopsy samples from diabetic nephropathy patients demonstrate that HA is a correlate of interstitial fibrosis in vivo (17). A number of our in vitro studies have suggested that HA may contribute to renal fibrosis via regulation of the renal proximal tubular epithelial cell (PTC) phenotype and differentiation of fibroblasts to myofibroblasts (18, 19), processes in which up-regulated HAS2 transcription may also play a role (20, 21).
The HAS2 gene, with the current reference mRNA sequence NM_005328.2, is located at 8q24.13 on the minus strand of human chromosome 8 and spans positions 122625271 to 122653630 according to the February 2009 freeze at the UCSC Genome Bioinformatics site. Our work has extended this HAS2 reference mRNA sequence by 130 upstream nucleotides (22) and has shown that the ubiquitously expressed zinc finger transcription factors Sp1 and Sp3 (specificity proteins 1 and 3) bind to multiple recognition sites upstream of this repositioned transcription start site to mediate constitutive HAS2 transcription (23). In collaboration, we have also described activity at the HAS2 promoter of retinoic acid response elements in response to all-trans-retinoic acid and of NF-κB following tumor necrosis factor-α stimulation (24); all-trans-retinoic acid- and forskolin-stimulated complexes of retinoic acid response elements and CREB1 (cAMP response element-binding protein 1) have also been reported (25).
Post-transcriptional regulation of HAS2 expression by a tetraexonic RNA, transcribed from the opposite genomic DNA strand at the HAS2 locus, has been described by Chao and Spicer (26). This transcript was originally called HASNT, for HA synthase 2 antisense (26), but has since been annotated formally with the NCBI Entrez Gene definition HAS2-AS1 HAS2 antisense RNA (non-protein coding) (Homo sapiens). The most recent HAS2-AS1 reference sequence has NCBI accession number NR_002835.2 and is located on the plus strand of human chromosome 8 from positions 122651586 to 122657564. HAS2-AS1 exon 2 shares sequence complementarity with the first exon of the HAS2 gene (see Fig. 1A). Overexpression of this complementary antisense region has been shown to down-regulate HAS2 transcription and HA synthesis in osteosarcoma cells, but the antisense transcript was not detected in kidney (26).
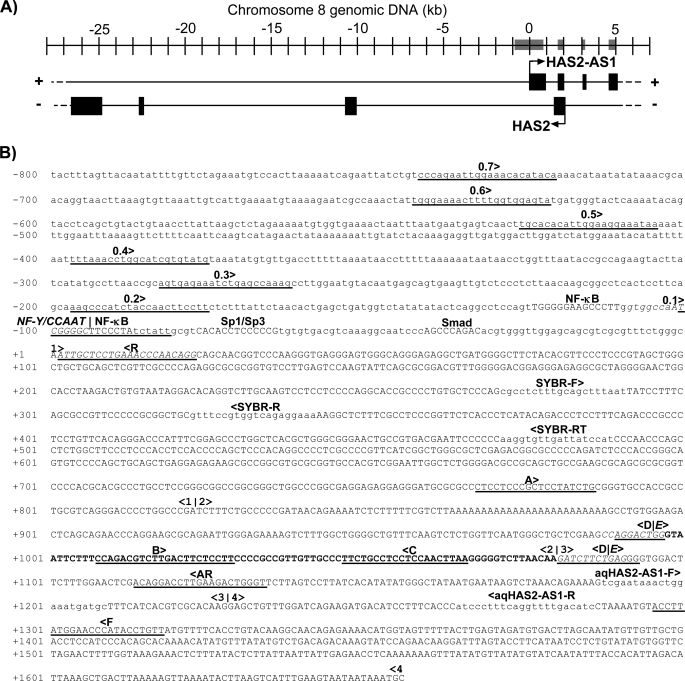
FIGURE 1.
Genomic organization of HAS2-AS1 and HAS2 at locus 8q24.13 and locations of TFBS consensus motifs and primer-binding sites for luciferase reporter, end-point RT-PCR, and qRT-PCR analyses of the human HAS2-AS1 gene. A, the four exons of HAS2-AS1 on the upper (+) strand of chromosome 8 at locus 8q24.13 are shown as black boxes, with the transcription start site indicated by an arrow. HAS2 on the lower (−) strand is depicted similarly, and the overlap of HAS2-AS1 exon 2 and HAS2 exon 1 is evident. The sequences displayed in B are highlighted in A by four gray boxes on the scale bar. B, the complete HAS2-AS1 cDNA sequence NR_002835 (26) is shown from the 5′ terminus at nucleotide +1 to the 3′ terminus at nucleotide +1656 in uppercase letters, with the following exceptions. Exon boundaries are numbered and delineated by vertical lines and arrows, e.g. <2|3> for the boundary between HAS2-AS1 exons 2 and 3. Oligonucleotide primer sequences and primer-binding site positions are displayed with orientation and labeled as described below. Primer positions for SYBR Green qRT-PCR analyses are shown in lowercase letters and are indicated as SYBR-F for the qRT-PCR sense-strand primer, SYBR-R for the qRT-PCR antisense-strand primer-binding site, SYBR-RT for the RT antisense-strand primer-binding site, aqHAS2-AS1-F for the absolute quantification qRT-PCR sense-strand primer, and aqHAS2-AS1-R for the absolute quantification qRT-PCR antisense-strand primer-binding site. End-point RT-PCR primer positions are underlined and designated A–F (including AR), with the exception of primer E, which overlaps for some of its length with primer D, and the relevant sequence is italicized. The alternatively spliced nucleotides in HAS2-AS1 exon 2 are shown in boldface type. Upstream of NR_002835 is genomic sequence from nucleotides −1 to −800. Selected discrete putative TFBSs identified by in silico analysis (30, 31) are shown in uppercase letters. The NF-Y/CCAAT site around nucleotide −100 is shown in italic type, and the overlapping NF-κB motif is shown wholly in uppercase letters. Sense-strand PCR primer sequences for the amplification of luciferase reporter construct inserts spanning ∼0.1–0.7 kb of putative HAS2-AS1 promoter sequence are underlined and designated accordingly, and the common antisense-strand primer-binding site is designated R.
The aim of this study was to identify the expression of HAS2-AS1 in PTCs and then to investigate the role of this RNA in the post-transcriptional regulation of HAS2 expression in these cells. Having established that HAS2-AS1 was transcribed in PTCs, subsequent time course analyses showed coordinated up-regulation of HAS2 and HAS2-AS1 transcription in response to stimulation with the pro-inflammatory cytokine IL-1β or the key fibrotic regulator TGF-β1. We then set out to identify the factors regulating this simultaneous sense and antisense expression and to analyze the effects on HAS2 expression of modulating HAS2-AS1 transcription in these cells. Finally, we investigated the thermodynamic feasibility of HAS2-AS1/HAS2 heterodimer formation in silico and detected locus-specific ribonuclease-resistant double-stranded RNA, providing evidence for the formation of RNA/mRNA duplexes in vitro.
EXPERIMENTAL PROCEDURES
Cell Culture
The culture of the human kidney PTC line HK-2 was as described previously (22). Recombinant IL-1β and TGF-β1 were obtained from R&D Systems Europe Ltd. (Abingdon, Oxfordshire, United Kingdom) and were used in accordance with the recommendations of the manufacturer.
Reverse Transcription and End-point RT-PCR
Extraction of total (22) as well as nuclear and cytoplasmic (27) RNAs was carried out as described previously. Nuclear and cytoplasmic RNA extracts were treated with 20 units/ml DNase (Promega UK Ltd., Southampton, Hampshire, United Kingdom) and, where appropriate, 100 μg/ml RNase A (Roche Diagnostics Ltd., Burgess Hill, West Sussex, United Kingdom) at 37 °C for 2 h. First-strand cDNA synthesis from 0.5–1 μg of total RNA was performed using the cDNA reverse transcriptase high capacity kit from Applied Biosystems (Warrington, Cheshire, United Kingdom). End-point RT-PCR primers were designed to span intron-exon boundaries to obviate amplification from contaminating genomic DNA, with the exception of the oligonucleotides used in heteroduplex detection, sequence data for which are given in the legend to Fig. 8B. Details of all other HAS2-AS1-specific primer sequences/binding sites are shown in Fig. 1B. All oligonucleotides were obtained from Invitrogen Ltd. (Paisley, Renfrewshire, United Kingdom) or Metabion (Martinsried, Bavaria, Germany). As we have detailed previously (22), end-point RT-PCRs were carried out; their products were visualized; and where appropriate, gel fragments were purified and sequenced to confirm the identity of both HAS2-AS1-related products and products of nonspecific amplification.
FIGURE 8.
RNA duplex modeling and detection. A, computed minimum free energy secondary structure of the heterodimer of L-HAS2-AS1 RNA (green) and HAS2 mRNA (red) showing the duplex formed by regions of perfect complementarity. B, detection of ribonuclease- and deoxyribonuclease-resistant cytoplasmic RNA duplex by RT-PCR with primers designed to amplify transcription outside (a, 5′-cttcgagcagcccattgaac-3′ and 5′-agcctgtggaagactcagca-3′) or within (b, 5′-aaacagttgccctttgcatc-3′ and 5′-tgatttgtctctgcccatga-3′) the predicted region of L-HAS2-AS1/HAS2 heteroduplex formation.
Reverse Transcription and SYBR Green Quantitative RT (qRT)-PCR Analysis of HAS2-AS1
Our SYBR Green qRT-PCR assay for HAS2-AS1 was optimized for linear detection with an efficiency of ∼100% using the oligonucleotide primers shown in Fig. 1B. Total RNA was isolated as described above, followed by a HAS2-AS1-specific RT step using 250 ng to 2 μg of total RNA and the SYBR-RT primer at a final concentration of 100 nm, 500 nm, or 1.0 μm. RT was carried out at 45 °C for 5 min, 37 °C for 1 h, and 85 °C for 5 min, followed by chilling at 4 °C. qRT-PCR was then performed in a total reaction volume of 20 μl using 1.3 μl of cDNA, the Power SYBR Green PCR Master Mix (Applied Biosystems), and HAS2-AS1-specific PCR primers SYBR-F and SYBR-R at a final concentration of 300 nm. Samples were processed in Applied Biosystems fast optical 96-well reaction plates in a 7900HT fast real-time PCR system (Applied Biosystems) with cycling parameters of 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Melting curve analysis was performed to confirm amplification of a single product. Following optimization, routine HAS2-AS1 analyses were carried out using RT from 1 μg of total RNA, followed by qRT-PCR as described above. Onboard SDS Version 2.3 software was used for the analysis of output data from biological triplicates.
qRT-PCR using TaqMan Assays
TaqMan assays for HAS2 (catalog no. Hs00193435_m1), Smad2 (Hs00183425_m1), Smad3 (Hs00232222_m1), Sp1 (Hs00412720_m1), Sp3 (Hs01595811_m1), and 18 S rRNA (4310893E) were obtained from Applied Biosystems. A custom HAS2-AS1 TaqMan assay (HAS2-AS1_LS) was also purchased from Applied Biosystems and used, as recommended by the manufacturer, to analyze cDNA generated from subsequent siRNA knockdown experiments. For HAS2 and 18 S rRNA qRT-PCR, 1 μl of cDNA was added to 19 μl of a qRT-PCR mixture in 1× TaqMan Fast Universal PCR Master Mix (Applied Biosystems); 2 μl of cDNA was added to 18 μl for the corresponding HAS2-AS1 reaction. Biological triplicates of all TaqMan assays were analyzed as described above.
Absolute Quantification of HAS2 and HAS2-AS1 Copy Numbers Using SYBR Green qRT-PCR
Absolute quantification of HAS2 mRNA was carried out by standard means using the HAS2 open reading frame (a gift from Dr. Andrew P. Spicer, Texas A&M University System Health Science Center, Houston, TX) inserted into an expression vector that we have described previously (28) and oligonucleotide sequences aqHAS2-F (tagaagaagatcccatggttgg) and aqHAS2-R (ggaatgagatccaggaatcgta). HAS2-AS1 RNA was quantified similarly using the full-length antisense sequence in pBluescript (Epoch Biolabs Inc., Missouri City, TX) together with oligonucleotide primer sequence aqHAS2-AS1-F and an oligonucleotide binding at the aqHAS2-AS1-R site shown in Fig. 1B.
HAS2-AS1-Luciferase Reporter Analysis
Putative HAS2-AS1 promoter sequences were PCR-amplified using the primers/primer-binding sites highlighted in Fig. 1B, high fidelity Platinum Pfx DNA polymerase (Invitrogen Ltd.), and the cycling parameters recommended by the manufacturer. Sense-strand primers 0.1–0.7 (KpnI) and antisense-strand primer R (NotI) bore tails suitable for restriction endonuclease digestion and subsequent ligation into a modified pGL3 luciferase reporter vector, after which they were sequenced to ensure fidelity of amplification as described previously (29). Transient transfection into HK-2 cells cultured in 6-well plates (BD Biosciences, Oxford, Oxfordshire, United Kingdom) was carried out using Lipofectamine LTX and Plus Reagent (Invitrogen Ltd.) in accordance with the manufacturer's recommendations. The ability of each HAS2-AS1 promoter fragment to drive transcription of the firefly luciferase gene was tested, according to the manufacturer's instructions, using the Dual-Luciferase reporter assay system (Promega UK Ltd.). In each test sample, in addition to the above firefly luciferase activity, this assay also detected the Renilla reniformis luciferase activity of a second vector that was cotransfected to control for variation in transfection efficiency. The relative luciferase activity of the test constructs was then expressed as the ratio of firefly to Renilla luciferase activities. We have described the growth medium to maximize luciferase output previously (22).
siRNA Knockdown of Gene Expression
Annealed oligonucleotide siRNA reagents for the knockdown of Smad2 (catalog no. 107875), Smad3 (115717), Sp1 (143158), and Sp3 (115337) and scrambled negative control transfection (4611) were purchased from Applied Biosystems and used in accordance with the manufacturer's guidelines. Three custom siRNAs specific for HAS2-AS1, and not for HAS2, were purchased from Applied Biosystems (catalog no. 295471, 295472, and 295473). A final concentration of 30 nm each siRNA was transfected into HK-2 cells cultured in 12-well plates (BD Biosciences) using siPORT amine transfection reagent as described previously (23). The previously determined time points of 3 h for IL-1β and 48 h for TGF-β1 were used for subsequent cytokine stimulations.
Sequence Data Base Analysis
HAS2-AS1 locus sequences were retrieved from the genome browser at the UCSC Genome Bioinformatics site and analyzed for the presence of putative transcription factor-binding sites (TFBSs) using the CISTER program (30) and the updated MatInspector program from the Genomatix suite (31). Selected putative TFBSs are shown in Fig. 1B.
Secondary Structure Investigations
The secondary structures of HAS2 mRNA and HAS2-AS1 RNA were investigated in silico to verify the thermodynamic feasibility of interaction to form a heterodimer. Minimum free energy and partition function calculations were performed using the Vienna suite of programs to probe the relative stabilities of folded RNA monomers, homodimers, and heterodimers (32, 33).
Statistical Analysis of Promoter Activity Assay Data
Data were calculated as the ratio of the firefly luciferase activity for each HAS2-AS1 promoter-reporter construct to the corresponding value for the cotransfected R. reniformis luciferase vector. Where appropriate, statistical analysis was performed using the Wilcoxon signed rank test, and p < 0.05 was considered statistically significant.
Statistical Analysis of qRT-PCR Data
Fold changes in expression were calculated using 2−(ΔCt1−ΔCt2), where ΔCt represents the difference between the threshold cycle (Ct) for each target gene and 18 S rRNA (34). Values for p were calculated by the t test using Microsoft Excel and were considered significant below 0.05.
RESULTS
PCR Analysis of HAS2-AS1
Fig. 2A shows that, following RT from total PTC RNA, HAS2-AS1 RNA-specific amplification was detected by end-point RT-PCR using primers spanning two intron-exon junctions. We then optimized a SYBR Green qRT-PCR assay from which the typical amplification data shown in Fig. 2B were obtained, and the melting curve of the products generated is shown in Fig. 2C. Both panels demonstrate that the amplification from negative control reactions, including any contaminating genomic DNA remaining in the RNA extracts, was negligible. Fig. 2D illustrates that, using this assay, the detection of HAS2-AS1 was quantitative within the range of 250 ng to 2 μg of total PTC input RNA for the three RT primer concentrations shown. The absolute quantification of HAS2 and HAS2-AS1 RNAs shown in Fig. 2E showed that the ratio of mRNA to RNA was 4.7:1 in quiescent HK-2 cells and 4.4:1 in IL-1β-stimulated cells.
FIGURE 2.
End-point RT-PCR detection of HAS2-AS1 transcription, optimization of SYBR Green qRT-PCR, and absolute quantification of HAS2 and HAS2-AS1 RNAs. A, detection of HAS2-AS1 expression in PTCs by end-point RT-PCR using primers A and AR (see Fig. 1B for details of all HAS2-AS1-specific primers not used in duplex detection). B (from left to right), pairs of HAS2-AS1 qRT-PCR profiles are shown following amplification using primers SYBR-F and SYBR-R from 2 μg, 1 μg, 500 ng, and 250 ng of input RT product, respectively. Each pair comprises amplifications from RT reactions carried out using either 0.1 or 1.0 μm primer SYBR-RT. Profiles for negative control reactions with no reverse transcriptase, no RNA, and no cDNA are contained within a box that is highlighted with an asterisk. C, melting curve of HAS2-AS1 SYBR Green amplification products shown in B. The negative control reaction range is illustrated by a bracket highlighted with an asterisk. D, variation in Ct in the presence of different quantities of input RNA (see B above) and the different concentrations of HAS2-AS1-specific RT primer displayed in the key. E, absolute quantification of the copy number of HAS2 mRNA using primers aqHAS2-F and aqHAS2-R (see “Experimental Procedures” for details) and of HAS2-AS1 RNA using primers aqHAS2-AS1-F and aqHAS2-AS1-R in unstimulated cells (white bars) and in response to stimulation with IL-1β (1 ng/ml) for 3 h (black bars). Data are displayed from one of two replicate experiments, each carried out in quadruplicate, and error bars are S.D.
Expression of HAS2-AS1 and HAS2 in Response to Cytokine Stimulation
Time courses following the expression of HAS2-AS1 and HAS2 by qRT-PCR in response to stimulation of PTCs with IL-1β are shown in Fig. 3 (A and B, respectively). An initial peak of HAS2-AS1 and HAS2 expression was seen at ∼2–3 h. The expression of both RNAs then declined rapidly after 6 h before rising steadily through 48 h. Fig. 3 (C and D) shows that the addition of TGF-β1 also led to the up-regulation of both transcripts in a coordinated fashion, increasing over time points to 72 h.
FIGURE 3.
Cytokine-stimulated up-regulation of HAS2-AS1 and HAS2 expression is coordinated. Shown is the relative expression of HAS2-AS1 RNA (A) and HAS2 mRNA (B) in response to IL-1β (1 ng/ml) stimulation and of HAS2-AS1 RNA (C) and HAS2 mRNA (D) following the addition of TGF-β1 (10 ng/ml). Data are shown from one of two replicate experiments, each carried out in triplicate, and error bars are S.D. *, p < 0.05.
Promoter Activity Analysis of Genomic DNA Sequences Upstream of HAS2-AS1 NR_002835
Each luciferase reporter construct containing an insert of putative HAS2-AS1 promoter sequence showed significant luciferase activity in comparison with the promoterless pGL3-Basic control vector, as shown in Fig. 4. The magnitude of activity varied from ∼5-fold greater than pGL3 for the 0.1-kb vector to >10-fold greater for the 0.7-kb construct. The ability of these inserts to drive the transcription of the luciferase gene demonstrated the constitutive promoter activity of each and implied the presence of the HAS2-AS1 proximal promoter.
FIGURE 4.
HAS2-AS1 proximal promoter constructs exhibit constitutive luciferase activity. The ratio of firefly to Renilla luciferase activities for each promoter construct is normalized relative to the promoterless pGL3-Basic construct (C). The promoter constructs are labeled in accordance with the approximate length of the putative HAS2-AS1 promoter that they amplify, as shown in Fig. 1B. Data are shown from one of three replicate experiments, each carried out in triplicate, and error bars are S.D. *, p < 0.05.
All insert sequences spanned a putative TFBS for both Sp1 and Sp3, mediators of constitutive HAS2 transcription (23), and a binding element for Smad proteins, the transcription factors that mediate response to TGF-β. These motifs were identified in silico within the first 100 nucleotides upstream of HAS2-AS1 NR_002835 (see Fig. 1B).
Stimulated HAS2-AS1 and HAS2 Expression following siRNA Knockdown of Transcription Factors
Combined transcription factor siRNAs for either Sp1/Sp3 or Smad2/Smad3 were administered prior to the addition of IL-1β or TGF-β1, respectively. The knockdown of Sp1 and Sp3 mRNAs inhibited IL-1β-stimulated up-regulation of HAS2 (Fig. 5A) and HAS2-AS1 (Fig. 5B). This effect was also seen when Smad2 and Smad3 mRNAs were knocked down and the induction of HAS2 by TGF-β1 was blunted (Fig. 5, C and D).
FIGURE 5.
IL-1β-stimulated up-regulation of HAS2-AS1 and HAS2 is mediated by Sp1 and Sp3, and TGF-β1-stimulated up-regulation of HAS2-AS1 and HAS2 is mediated by Smad2 and Smad3. Shown is the relative expression of HAS2 mRNA (A) and HAS2-AS1 RNA (B) in response to IL-1β (1 ng/ml) stimulation for 3 h and of HAS2 mRNA (C) and HAS2-AS1 RNA (D) following the addition of TGF-β1 (10 ng/ml) for 48 h. White bars, unstimulated PTCs, and black bars, cytokine-treated cells. Target gene siRNAs were used at 30 nm, and scrambled siRNA (SCR) was used at 60 nm. Data are shown from one of two replicate experiments, each carried out in triplicate, and error bars are S.D. *, p < 0.05.
Stimulated HAS2-AS1 and HAS2 Expression following siRNA Knockdown of HAS2-AS1
Low abundance HAS2-AS1 expression was detected in quiescent PTCs. Therefore, the effectiveness of HAS2-AS1 siRNA knockdown was evaluated following transcriptional induction by IL-1β, which repeatedly induced a greater magnitude of up-regulation of HAS2-AS1 transcription than TGF-β1 (see Figs. 3 and 5). Each of the three siRNAs analyzed significantly inhibited IL-1β induction of HAS2-AS1 RNA, as shown in Fig. 6A. In addition, Fig. 6B shows that each siRNA also blunted the up-regulation of HAS2 mRNA synthesis, but this inhibitory effect was statistically significant only in two of three cases.
FIGURE 6.
siRNA knockdown of HAS2-AS1 down-regulates HAS2 transcription. Shown is the relative expression of HAS2-AS1 RNA (A) and HAS2 mRNA (B) in response to the presence of IL-1β (1 ng/ml) for 3 h following transfection with three different siRNAs designed to knockdown HAS2-AS1 RNA. White bars, unstimulated PTCs; black bars, cytokine-treated cells. HAS2-AS1 siRNAs were used at 30 nm, and scrambled siRNA (SCR) was used at 30 nm. Data are shown from one of four replicate experiments, each carried out in triplicate, and error bars are S.D. *, p < 0.05.
End-point RT-PCR Analysis of HAS2-AS1 Splice Variants
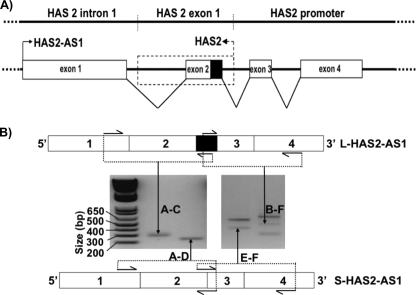
The original description of HAS2-AS1 reported alternative splicing of HAS2-AS1 exon 2, resulting in “long” (L) and “short” (S) HAS2-AS1 isoforms varying in length by 83 nucleotides and sharing either 257 nucleotides (L-HAS2-AS1) or 174 nucleotides (S-HAS2-AS1) of complementary sequence with the first exon of HAS2 (Fig. 7A) (26). Fig. 7B shows the detection of both L- and S-HAS2-AS1 expression in PTCs by agarose gel electrophoresis of end-point RT-PCR-amplified fragments. For L-HAS2-AS1-specific reactions A–C and B–F, oligonucleotides were designed to prime amplification from the 83-nucleotide alternatively spliced region of exon 2. S-HAS2-AS1-specific products were amplified using primers A–D and E–F.
FIGURE 7.
Organization of HAS2-AS1 and HAS2 exon 1 at locus 8q24. 13 and end-point RT-PCR amplification of L- and S-HAS2-AS1-specific fragments from total RNA. A, exons of HAS2-AS1 sequence NR_002835 are depicted as white boxes, with the exception of HAS2-AS1 exon 2, for which the alternatively spliced section is shown as a black box. The HAS2-AS1 exon 1 arrow shows the direction of transcription. Exon 1 of HAS2 NM_005328 plus AJ_604570 (22) is shown as a dashed box, and the dashed arrow shows the direction of HAS2 transcription. The location of HAS2 genomic regions is labeled on the line above. B, end-point RT-PCR showed that L- and S-HAS2-AS1 are expressed in PTCs. L- and S-HAS2-AS1 RNAs are represented as labeled boxes above and below the RT-PCR data. End-point RT-PCR products are displayed as bands on agarose gels following staining with ethidium bromide and are labeled according to the primers used for their amplification, which are displayed in Fig. 1B. A double-stranded DNA size marker is also shown.
Thermodynamic Feasibility of HAS2-AS1/HAS2 Heterodimer Formation and Heteroduplex Detection
HAS2 mRNA and HAS2-AS1 RNA were predicted to interact primarily through duplex formation of their perfectly complementary regions (174 nucleotides for S-HAS2-AS1 and 257 nucleotides for L-HAS2-AS1), with little other intermolecular hybridization predicted between the sequences (Fig. 8A). Based on the ensemble free energies of the species, at equilibrium at 37 °C, an equimolar mixture of HAS2 mRNA and L-HAS2-AS1 was predicted to exist overwhelmingly as a heterodimer (−2180 kcal/mol) without competition from homodimers (−2055 and −1696 kcal/mol for HAS2 mRNA and L-HAS2-AS1, respectively) or intramolecularly folded monomer forms (−1022 and −841 kcal/mol for HAS2 mRNA and L-HAS2-AS1, respectively). Comparable results were obtained for the interaction of HAS2 mRNA with S-HAS2-AS1 (data not shown). The calculations therefore support the existence of a dimer of HAS2 mRNA and HAS2-AS1 RNA in the absence of sequestration of the RNAs in kinetic or thermodynamic traps, which were not considered by this analysis. The RT-PCR assay data shown in Fig. 8B depict the detection of a ribonuclease- and deoxyribonuclease-resistant HAS2-AS1/HAS2 heteroduplex in PTC cytoplasmic RNA. Very little duplex signal was detected in nuclear RNA extracts (data not shown).
DISCUSSION
Understanding the mechanisms that regulate the expression of the human HAS genes is part of an ongoing series of studies in our laboratory that aim to determine the pathogenic mechanisms underlying renal fibrosis. In this investigation, we analyzed the expression of the HAS2 natural antisense RNA, HAS2-AS1, and investigated its role in the regulation of HAS2 expression.
Since the prescient identification of HAS2-AS1 as a post-transcriptional regulator of HAS2 expression and HA synthesis (26), a more detailed knowledge of the transcriptional output of the human genome has been revealed, including the pan-genomic significance of noncoding RNAs in gene expression (35, 36). Indeed, numerous subsequent reports have suggested that cell phenotype is a function of the total transcriptome (35–38). There is also a growing appreciation of the widespread occurrence of long noncoding RNAs, including antisense transcripts like HAS2-AS1, and that their impact on gene expression is likely to vary depending on genomic locus and cellular context (38–40). Therefore, following previous data describing the down-regulation of HAS2 and HA synthesis by HAS2-AS1 in osteosarcoma cells (26), in this investigation, we analyzed the expression of this natural antisense RNA in PTCs.
End-point RT-PCR data confirmed the presence of HAS2-AS1 expression in PTCs. Subsequent absolute quantification revealed that constitutive HAS2-AS1 was expressed in low copy number, particularly in quiescent HK-2 cells, and that the HAS2/HAS2-AS1 ratio in both quiescent and IL-β-stimulated cells was ∼4.5:1.
Our qRT-PCR assay showed that the induction of HAS2 sense and HAS2-AS1 antisense transcription occurred simultaneously in response to stimulation with IL-1β or TGF-β1. Correlated expression of overlapping sense/antisense transcripts has been reported in the expression of rat inducible NOS, where inducible NOS mRNA is stabilized by interaction with a natural antisense inducible NOS RNA (41). The biphasic response to IL-1β seen in Fig. 3 (A and B) has been shown previously in the PTC line HK-2 (42). Luo et al. (42) demonstrated that short-term IL-1β treatment inhibited Smad2/3-mediated TGF-β signaling via rapid and transient NF-κB activation but that longer IL-1β exposure augmented the endogenous/exogenous TGF-β-stimulated Smad response, following a switch in NF-κB nuclear subunit composition.
To analyze further the mechanisms regulating coexpression of HAS2-AS1 and HAS2, we created a nested series of putative promoter-luciferase reporter constructs amplified from the genomic DNA upstream of the 5′-end of the NR_002835 HAS2-AS1 RNA sequence. As demonstrated previously in our identification of the HAS2 promoter (22), our data suggested that sufficient HAS2-AS1 proximal promoter was contained in all vectors to drive luciferase gene transcription.
Analysis in silico showed that all inserts spanned Sp1/Sp3 and Smad consensus motifs. The suppression of induction of both HAS2 and HAS2-AS1 by IL-1β or TGF-β1 following siRNA knockdown of Sp1/Sp3 or Smad2/Smad3, respectively, was consistent with these in silico data. This suggested that cytokine-stimulated up-regulation of HAS2-AS1 and HAS2 transcription involved simultaneous binding of these transcription factors at both proximal promoters at the HAS2-AS1/HAS2 locus.
These data supported our previous findings that Sp1 and Sp3 mediate constitutive transcription of HAS2, which is down-regulated following siRNA knockdown of these transcription factors (23). The additional observation from the present study that IL-1β induction of HAS2 transcription was also suppressed by the mRNA knockdown of these ubiquitous transcription factors implied that Sp1 and Sp3 might perform a gatekeeper role in the transcriptional induction of HAS2 mRNA. However, because HAS2-AS1 was expressed constitutively at low abundance, the same conclusion could not be drawn for the antisense RNA in these cells.
The simultaneous abrogation of induction of both HAS2-AS1 and HAS2 by HAS2-AS1-specific siRNAs suggested that the antisense RNA stabilized or augmented HAS2 expression in PTCs. Very similar siRNA data have been reported in neuroblastoma cells from a study on BACE-1 (beta-site amyloid precursor protein-cleaving enzyme 1), a candidate gene for Alzheimer disease that may drive disease-associated pathology (43). Furthermore, BACE-1 mRNA expression is coordinately up-regulated in vitro by increased transcription of the natural antisense RNA BACE-1-AS in response to cell stressors (43). In contrast to the above, Chao and Spicer (26) showed that overexpression of HAS2-AS1 in osteosarcoma cells down-regulates both HAS2 mRNA synthesis and subsequent HA synthesis, underlining the importance of cellular context in sense/antisense interactions (38–40).
We identified in PTCs both known HAS2-AS1 splice variants (26). L-HAS2-AS1 shares 257 nucleotides of complementary sequence with HAS2 exon 1; the corresponding region for S-HAS2-AS1 is 183 nucleotides (26). Recent data show that the opposite genomic DNA strand at the Wilms tumor suppressor gene (WT1) locus encodes conserved multiple antisense RNAs that may regulate WT1 expression via RNA/mRNA interactions and that are deregulated by epigenetic defects and are abnormal in carcinogenesis (44).
In this investigation, we have provided evidence from both in silico and in vitro analyses that HAS2 mRNA and HAS2-AS1 RNA interact by heteroduplex formation. The predominantly cytoplasmic localization of the RNA/mRNA duplex that we observed has also been reported at the WT1 locus (44). The formation of HAS2-AS1/HAS2 duplexes raises the possibility that HAS2 expression might be affected differently due to the sequence variation in L- and S-HAS2-AS1 transcripts. We intend to pursue this line of investigation further in future studies of the RNA/mRNA interaction while bearing in mind that additional levels of regulation of gene expression by noncoding RNAs continue to come to light (e.g. Refs. 45 and 46).
In summary, we have identified expression in PTCs of the previously described natural antisense RNA for the human HAS2 gene, HAS2-AS1. Coordinated temporal profiles of transcriptional induction of HAS2-AS1 and HAS2 were observed in response to stimulation with IL-1β or TGF-β1. In each case, IL-1β induction was blunted following siRNA knockdown of Sp1 and Sp3, whereas pretreatment with siRNAs for Smad2 and Smad3 suppressed TGF-β1-stimulated up-regulation of both RNAs. The use of HAS2-AS1-specific siRNAs inhibited the up-regulation of both antisense RNA and HAS2 mRNA by IL-1β. End-point RT-PCR revealed the expression of L- and S-HAS2-AS1 splice variants that differ only in their length of sequence complementarity with HAS2 and provided evidence of HAS2-AS1 RNA/HAS2 mRNA duplex formation. We suggest that these data infer a role for HAS2-AS1 in the stabilization or augmentation of HAS2 expression in PTCs. Furthermore, the presence of the above-mentioned HAS2-AS1 splice variants provides evidence of a potential additional level of regulation of HAS2 expression in these cells by formation of heteroduplexes of different lengths.
Acknowledgments
For helpful discussions and advice, we thank Drs. Donald J. Fraser and Robert Steadman (Institute of Nephrology, Cardiff University School of Medicine) and Drs. Keith W. Brown, Anthony R. Dallosso, and Karim Malik (Cancer and Leukaemia in Childhood, Sargent Research Unit, Department of Cellular and Molecular Medicine, School of Medical Sciences, University of Bristol). For very generous help and encouragement on entering the field of HA research, T. B. expresses particular gratitude to Dr. Andrew P. Spicer, discoverer of HAS2-AS1.
This article was selected as a Paper of the Week.
- HA
- hyaluronan (hyaluronic acid)
- HAS
- HA synthase
- PTC
- proximal tubular epithelial cell
- qRT
- quantitative (real-time) RT
- TFBS
- transcription factor-binding site
- L-HAS2-AS1
- long HAS2-AS1
- S-HAS2-AS1
- short HAS2-AS1.
REFERENCES
- 1. Lee J. Y., Spicer A. P. (2000) Curr. Opin. Cell Biol. 12, 581–586 [DOI] [PubMed] [Google Scholar]
- 2. Andhare R. A., Takahashi N., Knudson W., Knudson C. B. (2009) Osteoarthritis Cartilage 17, 892–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bharadwaj A. G., Kovar J. L., Loughman E., Elowsky C., Oakley G. G., Simpson M. A. (2009) Am. J. Pathol. 174, 1027–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Itano N., Kimata K. (2008) Semin. Cancer Biol. 18, 268–274 [DOI] [PubMed] [Google Scholar]
- 5. Lauer M. E., Mukhopadhyay D., Fulop C., de la Motte C. A., Majors A. K., Hascall V. C. (2009) J. Biol. Chem. 284, 5299–5312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Milner C. M., Higman V. A., Day A. J. (2006) Biochem. Soc. Trans. 34, 446–450 [DOI] [PubMed] [Google Scholar]
- 7. Toole B. P. (2004) Nat. Rev. Cancer 4, 528–539 [DOI] [PubMed] [Google Scholar]
- 8. Wight T. N. (2008) Front. Biosci. 13, 4933–4937 [DOI] [PubMed] [Google Scholar]
- 9. Spicer A. P., Seldin M. F., Olsen A. S., Brown N., Wells D. E., Doggett N. A., Itano N., Kimata K., Inazawa J., McDonald J. A. (1997) Genomics 41, 493–497 [DOI] [PubMed] [Google Scholar]
- 10. Sayo T., Sugiyama Y., Takahashi Y., Ozawa N., Sakai S., Ishikawa O., Tamura M., Inoue S. (2002) J. Invest. Dermatol. 118, 43–48 [DOI] [PubMed] [Google Scholar]
- 11. Monslow J., Williams J. D., Norton N., Guy C. A., Price I. K., Coleman S. L., Williams N. M., Buckland P. R., Spicer A. P., Topley N., Davies M., Bowen T. (2003) Int. J. Biochem. Cell Biol. 35, 1272–1283 [DOI] [PubMed] [Google Scholar]
- 12. Gressner O. A., Weiskirchen R., Gressner A. M. (2007) J. Cell. Mol. Med. 11, 1031–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nobili V., Parkes J., Bottazzo G., Marcellini M., Cross R., Newman D., Vizzutti F., Pinzani M., Rosenberg W. M. (2009) Gastroenterology 136, 160–167 [DOI] [PubMed] [Google Scholar]
- 14. Li Y., Rahmanian M., Widström C., Lepperdinger G., Frost G. I., Heldin P. (2000) Am. J. Respir. Cell Mol. Biol. 23, 411–418 [DOI] [PubMed] [Google Scholar]
- 15. El-Chemaly S., Malide D., Zudaire E., Ikeda Y., Weinberg B. A., Pacheco-Rodriguez G., Rosas I. O., Aparicio M., Ren P., MacDonald S. D., Wu H. P., Nathan S. D., Cuttitta F., McCoy J. P., Gochuico B. R., Moss J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 3958–3963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rouschop K. M., Sewnath M. E., Claessen N., Roelofs J. J., Hoedemaeker I., van der Neut R., Aten J., Pals S. T., Weening J. J., Florquin S. (2004) J. Am. Soc. Nephrol. 15, 674–686 [DOI] [PubMed] [Google Scholar]
- 17. Lewis A., Steadman R., Manley P., Craig K., de la Motte C., Hascall V., Phillips A. O. (2008) Histol. Histopathol. 23, 731–739 [DOI] [PubMed] [Google Scholar]
- 18. Jenkins R. H., Thomas G. J., Williams J. D., Steadman R. (2004) J. Biol. Chem. 279, 41453–41460 [DOI] [PubMed] [Google Scholar]
- 19. Meran S., Thomas D. W., Stephens P., Enoch S., Martin J., Steadman R., Phillips A. O. (2008) J. Biol. Chem. 283, 6530–6545 [DOI] [PubMed] [Google Scholar]
- 20. Jones S., Jones S., Phillips A. O. (2001) Kidney Int. 59, 1739–1749 [DOI] [PubMed] [Google Scholar]
- 21. Meran S., Thomas D., Stephens P., Martin J., Bowen T., Phillips A., Steadman R. (2007) J. Biol. Chem. 282, 25687–25697 [DOI] [PubMed] [Google Scholar]
- 22. Monslow J., Williams J. D., Guy C. A., Price I. K., Craig K. J., Williams H. J., Williams N. M., Martin J., Coleman S. L., Topley N., Spicer A. P., Buckland P. R., Davies M., Bowen T. (2004) J. Biol. Chem. 279, 20576–20581 [DOI] [PubMed] [Google Scholar]
- 23. Monslow J., Williams J. D., Fraser D. J., Michael D. R., Foka P., Kift-Morgan A. P., Luo D. D., Fielding C. A., Craig K. J., Topley N., Jones S. A., Ramji D. P., Bowen T. (2006) J. Biol. Chem. 281, 18043–18050 [DOI] [PubMed] [Google Scholar]
- 24. Saavalainen K., Tammi M. I., Bowen T., Schmitz M. L., Carlberg C. (2007) J. Biol. Chem. 282, 11530–11539 [DOI] [PubMed] [Google Scholar]
- 25. Makkonen K. M., Pasonen-Seppänen S., Törrönen K., Tammi M. I., Carlberg C. (2009) J. Biol. Chem. 284, 18270–18281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chao H., Spicer A. P. (2005) J. Biol. Chem. 280, 27513–27522 [DOI] [PubMed] [Google Scholar]
- 27. Hurst S. M., McLoughlin R. M., Monslow J., Owens S., Morgan L., Fuller G. M., Topley N., Jones S. A. (2002) J. Immunol. 169, 5244–5251 [DOI] [PubMed] [Google Scholar]
- 28. Simpson R. M., Meran S., Thomas D., Stephens P., Bowen T., Steadman R., Phillips A. (2009) Am. J. Pathol. 175, 1915–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hoogendoorn B., Coleman S. L., Guy C. A., Smith K., Bowen T., Buckland P. R., O'Donovan M. C. (2003) Hum. Mol. Genet. 12, 2249–2254 [DOI] [PubMed] [Google Scholar]
- 30. Frith M. C., Hansen U., Weng Z. (2001) Bioinformatics 17, 878–889 [DOI] [PubMed] [Google Scholar]
- 31. Cartharius K., Frech K., Grote K., Klocke B., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., Werner T. (2005) Bioinformatics 21, 2933–2942 [DOI] [PubMed] [Google Scholar]
- 32. Hofacker W. F., Fontana W., Stadler P. F., Bonhoeffer L. S., Tacker M., Schuster P. (1994) Monatshefte Chem. Chem. Monthly 125, 167–188 [Google Scholar]
- 33. Bernhart S. H., Tafer H., Mückstein U., Flamm C., Stadler P. F., Hofacker I. L. (2006) Algorithms Mol. Biol. 1, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 35. Carninci P., Yasuda J., Hayashizaki Y. (2008) Curr. Opin. Cell Biol. 20, 274–280 [DOI] [PubMed] [Google Scholar]
- 36. Mattick J. S. (2009) PLoS Genet. 5, e1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mendes Soares L. M., Valcárcel J. (2006) EMBO J. 25, 923–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gingeras T. R. (2007) Genome Res. 17, 682–690 [DOI] [PubMed] [Google Scholar]
- 39. Prasanth K. V., Spector D. L. (2007) Genes Dev. 21, 11–42 [DOI] [PubMed] [Google Scholar]
- 40. Mercer T. R., Dinger M. E., Mattick J. S. (2009) Nat. Rev. Genet. 10, 155–159 [DOI] [PubMed] [Google Scholar]
- 41. Matsui K., Nishizawa M., Ozaki T., Kimura T., Hashimoto I., Yamada M., Kaibori M., Kamiyama Y., Ito S., Okumura T. (2008) Hepatology 47, 686–697 [DOI] [PubMed] [Google Scholar]
- 42. Luo D. D., Fielding C., Phillips A., Fraser D. (2009) Nephrol. Dial. Transplant. 24, 2655–2665 [DOI] [PubMed] [Google Scholar]
- 43. Faghihi M. A., Modarresi F., Khalil A. M., Wood D. E., Sahagan B. G., Morgan T. E., Finch C. E., St Laurent G., 3rd, Kenny P. J., Wahlestedt C. (2008) Nat. Med. 14, 723–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dallosso A. R., Hancock A. L., Malik S., Salpekar A., King-Underwood L., Pritchard-Jones K., Peters J., Moorwood K., Ward A., Malik K. T., Brown K. W. (2007) RNA 13, 2287–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Taft R. J., Glazov E. A., Cloonan N., Simons C., Stephen S., Faulkner G. J., Lassmann T., Forrest A. R., Grimmond S. M., Schroder K., Irvine K., Arakawa T., Nakamura M., Kubosaki A., Hayashida K., Kawazu C., Murata M., Nishiyori H., Fukuda S., Kawai J., Daub C. O., Hume D. A., Suzuki H., Orlando V., Carninci P., Hayashizaki Y., Mattick J. S. (2009) Nat. Genet. 41, 572–578 [DOI] [PubMed] [Google Scholar]
- 46. Taft R. J., Pang K. C., Mercer T. R., Dinger M., Mattick J. S. (2010) J. Pathol. 220, 126–139 [DOI] [PubMed] [Google Scholar]