Abstract
HIV-1 p17 contains C- and N-terminal sequences with positively charged residues and a consensus cluster for heparin binding. We have previously demonstrated by affinity chromatography that HIV-1 p17 binds strongly to heparin-agarose at physiological pH and to human activated CD4+ T cells. In this study we demonstrated that the viral protein binds to heparan sulfate side chains of syndecan-2, syndecan-4, and CD44v3 purified from HeLa cells and that these heparan sulfate proteoglycans (HSPGs) co-localize with HIV-1 p17 on activated human CD4+ T cells by confocal fluorescence analysis. Moreover, we observed a stimulatory or inhibitory activity when CD4+ T cells were activated with mitogens together with nanomolar or micromolar concentrations of the matrix protein.
Keywords: AIDS, Cytokine, Heparin-binding Protein, HIV, Viral Protein, Heparan Sulfate, Matrix Protein, p17, Proteoglycans, Syndecans
Introduction
HIV-1 p17 matrix protein (MA) forms the N-terminal domain of the GAG Pr55. MA is a 132-amino acid structural protein that is post-translationally myristilated at the N terminus. The three-dimensional structure of HIV-1 MA has been determined by NMR spectroscopy and x-ray crystallography (1–3). The protein folds into a compact core domain consisting of five α-helices and three stranded β-sheets. α-Helices 1–4 form a compact globular domain whereas the C-terminal helix 5 projects away from the membrane and may serve to connect MA and capsid domains in Gag Pr55. In the crystal structure, HIV-1 p17 is trimeric (1). The formation of higher order MA structures could be important for the process of virus assembly and Env incorporation into virus particle (4, 5). HIV-1 matrix protein is the key regulator of the early and late stages of viral replication.
Well established roles of p17 are the transport of P55GAG protein to the membrane (6) and the incorporation of viral envelope glycoproteins into nascent particles (7–9). We have previously shown that HIV-1 p17 is able to enhance the production of IFN-γ and TNF-α when added to phytohemagglutinin (PHA)2 stimulated peripheral blood mononuclear cells (PBMCs) (10). We demonstrated by immunofluorescence assays that this biological activity was induced by the binding of HIV-1 p17 to mitogen-activated PBMCs (11). More recently, we found that HIV-1 p17 is a heparin-binding protein and binds to heparan sulfate (HS) on activated primary human CD4+ T cells (12).
HS is a linear glycosaminoglycan (GAG) composed of repetitive disaccharide units of glucuronic (iduronic) acid and galactosamine or glucuronic (iduronic) acid and glucosamine. The disaccharide units are sulfated at different positions (13). GAGs are attached by covalent linkage to the core protein, forming proteoglycans (PGs). The detailed structure of PGs and their GAG sugars has been reviewed extensively (13, 14). Different cell surface proteins exist in PG forms. They include the betaglycan (TGF low affinity receptor type III), CD44 (a potent hyaluronan receptor), thrombomodulin, syndecans, and glypicans. These two last families are the major PGs of the cell surface (15). They bind proteins of the extracellular environment via their HS chains, regulating a wide spectrum of biological activities, including cell proliferation and differentiation, morphogenesis, wound repair, and host defense. Syndecans are transmembrane PGs with only one transmembrane domain, whereas glypicans are fixed to plasma membrane via a glycosylphosphatidylinositol anchor. Four human syndecan and six human glypican genes have been identified and cloned so far (16).
Syndecans and glypicans are expressed in a cell-, tissue-, and development-specific fashion. CD44 is a widely expressed family of cell surface molecules with structural heterogeneity generated by the alternative splicing of at least nine exons in humans encoding membrane-proximal domains of the extracellular region (17). The v3 exon product of the CD44 molecule is special in the sense that it is the only exon that has a HS assembly site (18, 19). V3 exon-containing variants carrying HS side chains are able to bind and present heparin-binding growth factors and cytokines, like basic FGF, heparin-binding epidermal growth factor (EGF), macrophage inflammatory protein 1β, and hepatocyte growth factor/scatter factor, which may have various functional consequences (19–22). Activated human CD4+ T cells express CD44v3, syndecan-2 (Sy-2), and syndecan-4 (Sy-4) (15, 22–24).
In this study, we investigated whether HIV-1 p17 matrix protein was able to bind to these HSPGs. We found that the viral protein binds HS side chains of Sy-2, Sy-4, and CD44v3 purified from HeLa cells and that these HSPGs co-localize with HIV-1 p17 on activated human CD4+ T cells by confocal fluorescence analysis. Moreover, we observed a stimulatory or inhibitory activity when CD4+ T cells were activated with mitogens together with nanomolar or micromolar concentrations of the matrix protein, respectively.
EXPERIMENTAL PROCEDURES
Reagents
Unless otherwise stated all chemicals were purchased from Sigma-Aldrich. Recombinant HIV-1 p17 matrix protein was purchased from Austral Biologicals (distributed by AMS Biotechnology, Oxon, UK). The most of the recombinant protein (>90%, molecular mass 14,900) is present in a monomer conformation (as determined by SDS-PAGE and gel filtration chromatography; data not shown). Anti-HIV-1 p17 mouse monoclonal antibody MK-1 was generated in our laboratory (11). Unfractioned heparin (UFH) was obtained from Parker-Davis (Milan, Italy). Rabbit polyclonal anti-human Sy-2 and anti-human Sy-4 antibodies were obtained from Santa Cruz Biotechnologies. Rabbit polyclonal anti-human CD44v3 and anti-human CD41 antibodies were provided by ABCAM (Cambridge, UK).
Cells
PBMCs were isolated by Ficoll-Hypaque gradient centrifugation of leukopacks obtained by apheresis of healthy donors. CD4+ T cells were purified by positive selection using magnetic beads (Invitrogen) as described previously (11). The preparations were >96% pure as judged by flow cytometry. The HeLa cell line was obtained from the European Collection of Animal Cells. Cells were cultured in complete RPMI 1640 medium supplemented with 2 mm l-glutamine and 10% fetal calf serum (FCS).
DEAE-Sephacel Chromatography
Adherent HeLa cells cultured on plastic substrata were rinsed with cold phosphate-buffered saline (PBS) and scraped in the same buffer. After centrifugation (6000 × g; 20 min) the pellet was extracted in 25 mm Tris buffer, pH 8.5, containing 1% Triton X-100, 8 m urea, and 0.1 mm phenylmethylsulfonyl fluoride (TTU buffer) and kept overnight at 4 °C. After centrifugation (10,000 × g; 30 min). this cleared extract was applied to a DEAE-Sephacel column (1.5 × 5 cm) previously equilibrated with TTU buffer and eluted by increasing concentrations of NaCl (0.1, 0.2, 0.3, 0.4, 0.5, 1.0, and 2.0 m) in TTU buffer. Two fractions of 5 ml were collected for each molarity.
Preparation of Microtiter Solid Phase
Fractions collected from DEAE-Sephacel column were diluted 1:2 with water (to reduce saline concentration), and then 50 μl of each dilution was coated onto the wells of a microtiter plate (Nunc, Maxisorp, Denmark) for 18 h at 4 °C. After washing with PBS, wells were blocked for 2 h at 37 °C with 100 μl/well PBS containing 2% FCS (PBSF). This solid phase was used to perform two distinct enzyme-linked immunosorbent assays (ELISAs). The first had the purpose of detecting the presence of Sy-2, Sy-4, and CD44v3 proteoglycans (PG-ELISA) in the eluted fractions and the second was to evaluate whether any interaction occurred with the matrix protein (p17-ELISA).
PG-ELISA
To each well 50 μl of anti Sy-2, Sy-4, CD44v3 or unrelated rabbit polyclonal antibody (all at 2 μg/ml in PBSF) was added and incubated for 2 h at room temperature. After washing three times with PBS, 50 μl of polyclonal goat anti-rabbit IgG-horseradish peroxidase (HRP) conjugate, diluted 1:1000 with PBSF, was added to each well and incubated for 1 h at room temperature. After three washes with PBS, HRP activity was detected by adding 100 μl/well 3,3′,5,5′-tetramethylbenzidine liquid substrate. The enzymatic reaction was blocked after 10 min with 100 μl/well 2 n H2SO4. Absorbances at 450 nm were measured in each well using a microtiter plate reader.
p17-ELISA
To detect an interaction between p17 and molecules eluted from a DEAE-Sephacel column, 0 or 0.25 μg/ml HIV-1 p17 in PBSF was added (50 μl) to each fraction, adsorbed on the microtiter wells and incubated for 2 h at room temperature. After three washes with PBS, 50 μl of anti-p17 monoclonal antibody (MK-1) was added to each well and incubated for further 2 h at room temperature. After the washing step, 50 μl of polyclonal rabbit anti-mouse IgG-HRP conjugate, diluted 1:1000 with PBSF, was added. After washing the plate three times, peroxidase activity was detected by adding 100 μl of 3,3′,5,5′-tetramethylbenzidine and blocking the reaction after 10 min with 100 μl of 2 n H2SO4. Absorbances were measured as described above. In some experiments, HIV-1 p17 (0.25 μg/ml) was preincubated for 1 h at room temperature with 10 μg/ml UFH.
HSPG/HIV-1 p17 Complex Capture ELISA
Fractions interacting with the matrix protein in p17-ELISA were pooled and dialyzed with PBS. No reacting fractions were pooled and dialyzed with PBS and used as negative control.
Four different solid phases were prepared using rabbit polyclonal antibodies to human Sy-2, Sy-4, CD44v3, and CD41 (used as unrelated antibody). Each antibody was coated in triplicate by adding 100 μl/well antibody solution at 2 μg/ml in 50 mm sodium carbonate buffer, pH 9.6. After overnight incubation at room temperature, the wells were saturated with 200 μl of PBSF for 18 h at room temperature. After two washing steps with PBS, 50 μl of pooled fractions and 50 μl of PBSF were mixed into each well of the four solid phases. After 2 h of incubation at room temperature, the wells were washed three times with PBS. To each well of the four solid phases, 100 μl of HIV-1 p17 at 0 μg/ml or 0.25 μg/ml in PBSF was added in triplicate and incubated for 2 h at room temperature. After three washes, 100 μl of mouse monoclonal antibody MK-1 (5 μg/ml in PBSF) was added to all wells and incubated for 2 h at room temperature. After three washing steps, 100 μl of polyclonal rabbit anti-mouse IgG-HRP conjugate (1:1000) was added to each well. After the washing procedure, the enzymatic activity was measured as described above. Ιn some experiments HIV-1 p17 (0.25 μg/ml) was preincubated for 1 h at room temperature with increasing amounts of heparin (0, 0.2, 1, 5, and 20 μg/ml UFH). The assay was then carried out as already described.
Confocal Immunofluorescence Microscopy
Human primary CD4+ T cells were activated for 72 h with 5 μg/ml PHA and phorbol ester PMA (10 ng/ml). Cells were collected and after a washing step with cold PBS, air-dried on multiwell immunofluorescence slides at room temperature. Cells were fixed for 5 min with cold 70% ethanol in water. Fixed cells were blocked with PBS containing 5% normal human serum. CD4+ T cells were incubated with HIV-1 p17 (2 μg/ml in PBSF) for 1 h at room temperature and after washing reacted with MK-1 monoclonal antibody (5 μg/ml in PBSF). Complexes between HSPGs and the matrix protein were detected by incubating for further 1 h at room temperature with rabbit anti-mouse IgG-Texas Red conjugate diluted 1:50 in PBSF. To co-localize Sy-2, Sy-4, and CD44v3, cells were stained with the related primary antibodies for 1 h at room temperature followed by FITC-labeled secondary antibodies for 30 min each at room temperature. Preparations were mounted in 0.1 m Tris-HCl, pH 8.5, 25% glycerol, 10% (w/v) Mowiol (Calbiochem), and 2.5% 1,4-diazobicyclo[2.2.2]octane and examined with confocal laser scanning microscopy (MCR 1024; Bio-Rad). The system is controlled by Bio-Rad LaserSharp 2000 software. Data acquisition was done in a sequential way to avoid cross-talk between the emission spectra of the fluorochromes, with all acquisition settings kept constant for all images. Cells were examined using an oil immersion 63× objective. Unrelated rabbit and mouse antibodies were used as negative controls. The images were used for the co-localization analysis by the LaserSharp Emulations software (Bio-Rad).
Activation of Primary Human CD4+ T Cells in the Presence or Absence of HIV-1 p17 and Cytokine Measurements
Primary human CD4+ T cells (2 × 106/ml) were stimulated into flat-bottom 24-well plate for 3 days with 5 μg/ml PHA, 10 ng/ml PMA, and increasing amounts of HIV-1 p17. In some experiments we used resting PBMCs. Levels of TNF-α and IL-2 in cell supernatants were measured by ELISA kits according to the manufacturer's instructions (SA Biosciences).
Statistical Analysis
Statistical analysis was performed by using a Student's t test. A p value of < 0.05 was used as the criterion of statistical significance.
RESULTS
Preparation of Proteoglycans from HeLa Cells
We have previously showed that HIV-1 p17 matrix protein binds strongly to heparin/heparan sulfate and to HS side chain of PGs on activated human CD4+ T cells (12). Naive and memory-activated human CD4+ T cells express Sy-2 and Sy-4, and it has been demonstrated that the cross-linking of these HSPGs inhibited T cell proliferation (24). CD44v3 isoform carries chondroitin sulfate and HS side chains, and it is highly expressed on activated CD4+ T cells. Furthermore, this isoform is frequently expressed on tissue-infiltrating lymphocytes and on leukocytes of patients with autoimmune disease. Thus, we focused our attention on these HSPGs to study the interaction between HS and p17 on activated CD4+ T cells.
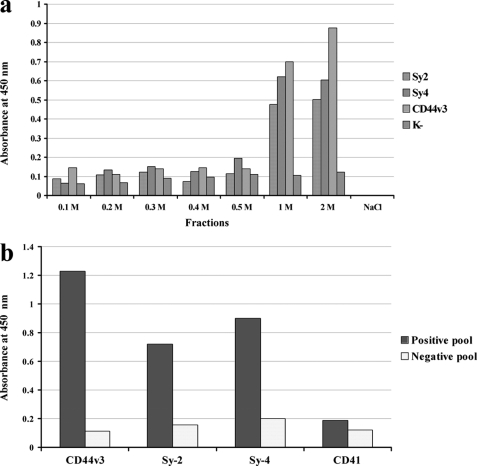
We utilized HeLa cells to isolate Sy-2, Sy-4, and CD44v3 because they are constitutively expressed on this tumor cell line. Previous studies (25) showed that PGs can be extracted efficiently from the plasma membranes by detergent treatment. We used both a chaotropic agent (urea) and Triton X-100 detergent in the extraction buffer. Extracted PGs were subjected to DEAE chromatography using a step gradient elution. Fractions were coated on the wells of a microtiter plate, and to detect the presence of Sy-2, Sy-4, and CD44v3 in the eluates an indirect ELISA (PG-ELISA) was performed. As shown in Fig. 1a only fractions eluted at high NaCl molarity reacted with the anti-PG antibodies. As expected, these proteins bind strongly to anion exchange solid phase for their high negative charge.
FIGURE 1.
a, DEAE-Sephacel chromatography and PG-ELISA. HSPGs were extracted from HeLa cells with TTU buffer. The extract was chromatographed over a DEAE-Sephacel column equilibrated with TTU buffer. Elution was performed by stepwise salt gradient (NaCl in TTU buffer). Eluted fractions were diluted 1:2 in water and coated on microtiter wells. Fractions were assayed for HSPG presence by an indirect ELISA using anti-CD44v3, anti-Sy-2, anti-Sy-4 antibodies or an unrelated antibody as negative control (K−). The data are from a representative experiment of three independent experiments. b, capture ELISA. To define which HSPG interacts with the matrix protein, specific antibodies to Sy-2, Sy-4, CD44v3, and unrelated antibody (anti-CD41) were adsorbed on microtiter plate wells. Positive and negative fractions were pooled and reacted with the four solid phases. Captured antigens were reacted with the matrix protein. Complexes were detected by MK-1 antibody and by rabbit anti-mouse IgG-HRP conjugate. Data are representative of three independent experiments.
To detect whether eluates interact with HIV-1 p17, each fraction was coated on microtiter wells and then tested for HIV-1 p17 binding. The complexes HSPG/p17 were then detected using a specific anti-p17 mouse monoclonal antibody (MK-1). Fractions eluted at 1.0 m and 2.0 m NaCl strongly reacted with the matrix protein, indicating that HIV-1 p17 was captured by the solid phases containing HSPGs. To best understand the nature of this interaction, the matrix protein was preincubated with heparin which is a strong competitor for HS binding (12) and then tested for the binding to the solid phases. The interaction between p17 and eluates at 1.0 and 2.0 m NaCl was completely abolished (data not shown), indicating the specificity of the reaction.
Capture ELISA
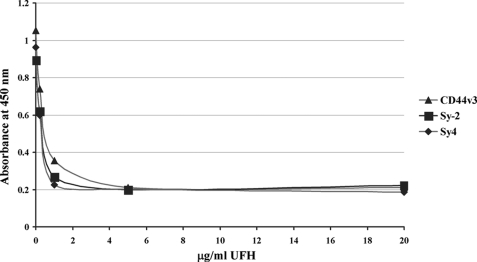
To investigate whether HIV-1 p17 binds to one or more HSPGs a capture ELISA method was employed. Four different solid phases were prepared using rabbit polyclonal antibody anti-Sy-2, anti-Sy-4, anti-CD44v3, and anti-CD41 (as unrelated antibody). Positive or negative (to measure unspecific binding) pooled fractions were allowed to react with the four different solid phases. HIV-1 p17 matrix protein was then incubated with the selectively captured antigens. HSPG/p17 complexes were detected with anti-p17 monoclonal antibody MK-1. As shown in Fig. 1b, all captured HSPGs bind to HIV-1 p17 with comparable signal levels. No significant signals were observed for the unrelated capture antibody and for the negative fractions. To investigate the specificity of this interaction further, a competitive variant of the capture ELISA was employed. A constant amount of HIV-1 p17 (0.25 μg/ml) was preincubated with heparin at different concentrations (0, 0.2, 1, 5, and 20 μg/ml), and then the solutions were tested in the capture ELISA. Heparin at 1 μg/ml completely inhibited the interaction between p17 and HSPGs (Fig. 2). These data demonstrate that HIV-1 p17 matrix protein interacts with the HS side chains of Sy-2, Sy-4, and CD44v3 and not with the protein core of the PGs.
FIGURE 2.
UFH inhibits the interaction between HSPGs and p17. The matrix protein (0.25 μg/ml) was mixed with increasing concentrations of UFH (0, 0.2, 1, 5, 20 μg/ml) for 1 h before the addition to HSPGs on capture ELISA solid phases. An assay was then performed as described for the capture ELISA assay. Data are representative of three independent experiments.
HIV-1 p17 Matrix Protein Co-localized with Sy-2, Sy-4, and CD44v3 on Activated CD4+ T Cells
Because recently published data show that activated CD4+ T cells express Sy-2, Sy-4, and CD44v3 HSPGs (23–26), we performed a co-localization analysis to verify whether the MA binds to these three HSPGs on activated CD4+ T cells as occurs for HeLa cells. In these experiments, slides were coated with activated CD4+ T cells. HSPG/p17 complexes were stained with Texas Red and HSPGs with FITC.
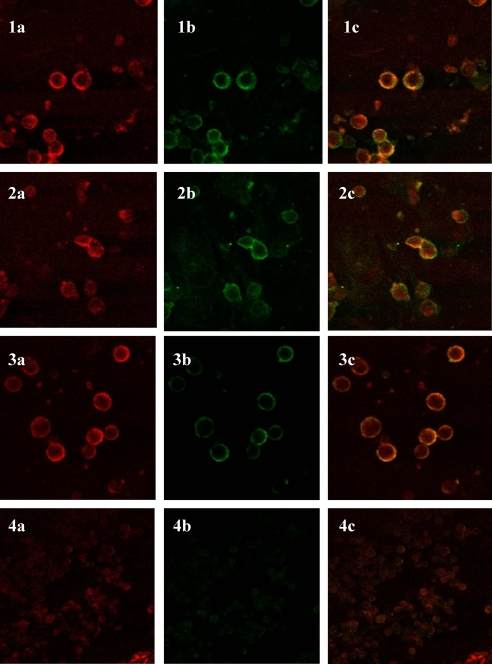
The LaserSharp Emulation software provides quantitative analysis of the degree of co-localization of two fluorophores on a pixel-by-pixel basis. Co-localization can be evaluated using Pearson correlation coefficient. A coefficient between 0 and −1 indicates no co-localization, whereas a value of 1 attests a perfect co-localization, which can be reached in theory (27). Micrographs of stained cells and merged pictures of CD44v3/p17, Sy-2/p17, and Sy-4/p17 are shown in Fig. 3.
FIGURE 3.
Co-localization analysis on activated CD4+ T cells. The matrix protein was incubated on fixed cells and detected with anti-p17 monoclonal antibody (MK-1). Complexes were stained with rabbit anti-mouse IgG Texas Red conjugate (red, 1a, 2a, and 3a). CD44v3 (1b), Sy-2 (2b), and Sy-4 (3b) were detected with a primary rabbit polyclonal antibody and stained with FITC-labeled secondary antibody (green). 1c, 2c, and 3c are merged red and green channels. Where the red and green of the image overlap, the pixels appear yellow. 4a, 4b, and 4c images are from unmatched antibodies. Data are representative of three individual experiments.
A co-localization coefficient of 0.67 ± 0.09 was determined for the CD44v3/p17 complex. Similar micrographs were obtained for Sy-4/p17 and Sy-2/p17 with Pearson coefficients of 0.44 ± 0.08 and 0.34 ± 0.09, respectively. Based on these data we conclude that HIV-1 p17 matrix protein co-localizes at varying degrees with HS side chains of Sy-2, Sy-4, and CD44v3. Differences between calculated Pearson coefficients might reflect a different up-regulation degree of HSPGs on the plasma membrane.
Cytokine Production by Activated Primary Human CD4+ T Cells in the Presence of HIV-1 p17
Our previous findings showed that HIV-1 p17 matrix protein enhanced activation and proliferative capacity of activated PBMCs and increased HIV-1 infection. Moreover, the matrix protein enhanced the production of proinflammatory cytokine of stimulated PBMCs (11).
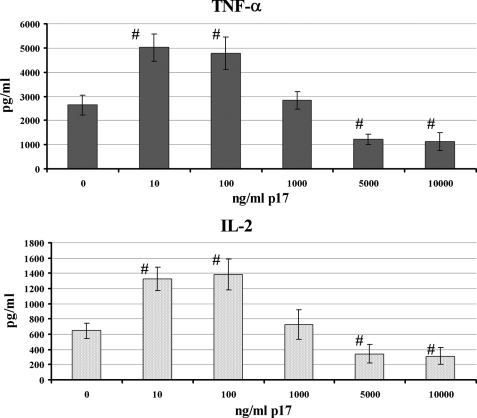
In this study we analyzed the production of TNF-α and IL-2 by mitogen-activated CD4+ T cells in the presence of increasing amounts of HIV-1 p17, using a larger range of concentrations with respect to previous published data (11). As already observed, the addition of nanomolar concentrations of HIV-1 p17 to mitogen-stimulated CD4+ T cells induced a statistically significant (p < 0.05) increase of the secretion of both cytokines (Fig. 4). On the contrary, the viral protein exerted a statistically significant (p < 0.05) inhibitory effect on TNF-α and IL-2 secretion when it was used at micromolar concentrations (Fig. 4). As previously observed (11) in the absence of mitogen activation, p17 had no capacity to induce TNF-α and IL-2 production (data not shown).
FIGURE 4.
Effect of HIV-1 p17 on TNF-α and IL-2 production by activated CD4+ T cells. The cells were activated for 72 h with PHA and PMA in the presence or absence of the matrix protein. Supernatants were collected, and TNF-α and IL-2 production was measured by ELISA. The data are representative of three independent experiments with the error bars indicating the ± S.D. #, p < 0.05 compared with PHA/PMA cell activation.
DISCUSSION
HIV-1 p17 matrix protein is the N-terminal protease cleavage product of the viral precursor protein P55GAG. Oligomerization of the matrix protein forming trimers or hexameric structures seems to be a crucial step in the virus assembly (1, 7, 28). The N-terminal portion of MA, which is myristilated, facilitates the binding of P55GAG to membranes (5) and regulates the targeting of P55GAG to the site of virus particle assembly (9). We have recently showed that the basic regions of the matrix protein also resemble heparin/heparan sulfate binding domains, demonstrating that HIV-1 p17 interacts at physiological ionic strength and pH with heparin and heparan sulfate. Moreover, we found that the matrix protein binds to HSPGs on activated human primary CD4+ T cells (12).
In this report, we further determine the nature of the interaction of the matrix protein with HS side chains of PGs, focusing our attention on the characterization of the protein cores. Previous published data showed that the HSPGs expressed on activated CD4+ T cells are represented by Sy-2, Sy-4, and CD44v3 PGs (23, 24, 26). Thus, to assess whether the matrix protein binds to one or more of these PGs, we extracted HSPGs from HeLa cells, which are constitutively expressed on their plasma membrane, and then we partially purified them by DEAE-Sephacel chromatography. HSPG-containing fractions were pooled and then tested with specific antibodies in a capture ELISA assay. All HSPGs reacted with the matrix protein. The specificity of this interaction was demonstrated by the complete inhibition obtained with the soluble heparin, indicating that the binding occurs solely through association with their HS side chains.
Co-localization analysis on activated human primary CD4+ T cells confirmed the interaction of the matrix protein with all the three HSPGs. Pearson coefficients were all positive, indicating a co-localization between the matrix protein and the three HSPGs. The Pearson coefficient was higher for CD44v3 and lesser for Sy-2. This effect might be due to differences in the density of the three HSPGs expressed on the plasma membrane.
Sy-2, Sy-4, and CD44v3 PGs are expressed on activated human primary CD4+ T cells and are involved in positive or negative regulation of T cell proliferation and TNF-α production (26, 29). It has been published that the engagement of the GAG moiety of CD44 by RANTES (regulated on activation normal T cell expressed and secreted) induces cellular activation and enhances HIV-1 infectivity, triggering the activation of the p44/p42 MAPK pathway (29). Activated MAPKs are known to phosphorylate various cytoplasmic and membrane-bound cellular substrate (30). Similarly, our previous studies (11) showed that the addition of picomolar amounts of the matrix protein to activated PBMCs enhanced proinflammatory cytokine production, cell proliferation, and HIV replication.
However, it has also been shown that the cross-linking of HS side chains of Sy-2 and Sy-4 on activated naive and memory CD4+ T cells inhibited both cell proliferation and TNF-α production (25). Recently, it has been reported that the binding of the dendritic cell-associated HSPG-dependent integrin ligand to HS chains of Sy-4 on activated T cells induced a down-regulation of proinflammatory cytokines and inhibited cell proliferation (31). These studies indicate that Sy-2 and Sy-4, through an anti-inflammatory action, play a considerable role in the immune regulation.
In this work, we revisited the up-regulation effect of the matrix protein using activated CD4+ T cells instead of PBMCs. CD4+ T cells were activated with PHA, PMA, and increasing amounts of p17. After 3 days supernatants were collected and analyzed for TNF-α and IL-2 production. Interestingly, we found that nanomolar concentrations of the matrix protein enhanced production of both TNF-α and IL-2, confirming our previous findings (11), whereas micromolar amounts produced a significant down-regulation of both cytokines.
We hypothesized that this double effect obtained in vitro might be due to the structure of recombinant HIV p17 protein. This commercial protein is monomeric for >90%. Therefore, at low concentrations monomeric HIV-p17 binds HS side chains of CD44v3, Sy-2, and Sy-4, inducing an overall up-regulation of the activated CD4+ T cells. The interaction between the HS side chains of CD44v3 and p17 could activate Src family kinases as occurs for RANTES (29).
On the contrary, the binding between HS side chains of Sy-2 and Sy-4 and monomeric p17 is ineffective because to transduce a signal, GAG chains must be cross-linked (24). Using high p17 concentrations during cell activation, GAG chains can promote the formation of p17 oligomers on the cell surface as described for some chemokines (33–35). It was, in fact, shown that the receptor clustering is a key element in the syndecans biology (32).
Oligomeric p17 can interact with Sy-2 and Sy-4 cross-linking HS side chains decreasing cell activation and reducing TNF-α and IL-2 production (24). Why in this case the overall effect is a down-regulation even if multimeric p17 binds to CD44V3, at the moment is unknown. In our in vitro experiments, inhibition is only observed at p17 concentrations >100 ng/ml, whereas it was demonstrated that sera of HIV-1 patients contained p17 at concentrations ranging from 0.19 to 13.9 ng/ml (47).
In vivo, local concentrations of the matrix protein are unknown. It was already shown (36) that the matrix protein and other viral proteins persist in the germinal centers of lymph nodes of HIV-1 chronically infected patients. We can only speculate that HSPGs in lymphoid tissues could increase p17 local concentration.
However, in vivo the matrix protein is myristylated and it is thought to form trimers (1) or, as suggested by a recent report (28), hexameric structures. Thus, the multimeric matrix protein might be able to cross-link syndecans even at low molar concentrations. Therefore, the matrix protein through the interaction with Sy-4 that is expressed on activated naive and memory CD4+ T cells might down-regulate cell proliferation of these T cell subpopulations during all stages of the infection without the need to reach high local concentrations to achieve a multimeric structure. Extrapolating our data to the situation in vivo, we can postulate that HIV p17 might have different activities during HIV infection.
In particular, in the early stages of HIV infection, in the context of low levels of immune activation and a moderate decrease of numbers of CD4+ T cells, the matrix protein could fuel the cellular activation and the virus infectivity through CD44v3 interaction, expressed at higher level on all activated CD4+ T cell subsets. Further, in the late stages of the disease characterized by a progressive depletion of CD4+ T cells, an expansion of memory CD4+ T cells and a high viral load, it might participate in immune failure by down-regulating naive and memory CD4+ T cells through the interaction with Sy-2 and Sy-4.
It is well recognized that T cell depletion during the course of HIV-1 infection is not restricted to infected cells; a disruption of T cell homeostasis (37, 38), diminished thymic function (39, 40), peripheral naive T cell expansion failure (41), and distorted lymphoid architecture (42), as well as heightened cellular turnover (43, 44), have been implicated as underlying these depletions.
In this setting, compromised proliferative response of activated T cells in lymphoid organs is a permanent feature limiting the expansion of responding T cells. This “pause” status of activated T cells might intensify the density of productively infected CD4 cells and render activated T cells more susceptible to activation-induced cell death (or apoptosis) through a “bystander” mechanism (45, 46), thereby contributing to the progressive decline of peripheral blood CD4+ T cell pool and to the eventual development of AIDS.
Further studies are necessary for a better understanding of the mechanisms by which HIV p17 might contribute to chronic HIV-1 infection that induces immune perturbations and results in CD4 T-cell depletion. These findings might provide a new insight into HIV-1 pathogenesis and reveal new targets for immune based interventions to slow HIV-1 disease progression.
This work was supported in part by the Ministero dell'Università e della Ricerca Scientifica e Tecnologica.
- PHA
- phytohemagglutinin
- GAG
- glycosaminoglycan
- HS
- heparan sulfate
- HSPG
- heparan sulfate proteoglycan
- PBMC
- peripheral blood mononuclear cell
- PG
- proteoglycan
- PMA
- phorbol 12-myristate 13-acetate
- Sy-2
- syndecan-2
- Sy-4
- syndecan-4
- UFH
- unfractioned heparin.
REFERENCES
- 1. Hill C. P., Worthylake D., Bancroft D. P., Christensen A. M., Sundquist W. I. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 3099–30104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Conte M. R., Matthews S. (1998) Virology 246, 191–198 [DOI] [PubMed] [Google Scholar]
- 3. Verli H., Calazans A., Brindeiro R., Tanuri A., Guimarães J. A. (2007) J. Mol. Graph Model 26, 62–68 [DOI] [PubMed] [Google Scholar]
- 4. Freed E. O. (1998) Virology 251, 1–15 [DOI] [PubMed] [Google Scholar]
- 5. Freed E. O. (2002) J. Virol. 76, 4679–4687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bryant M., Ratner L. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 523–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yu X., Yuan X., Matsuda Z., Lee T. H., Essex M. (1992) J. Virol. 66, 4966–4971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dorfman T., Mammano F., Haseltine W. A., Göttlinger H. G. (1994) J. Virol. 68, 1689–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cannon P. M., Matthews S., Clark N., Byles E. D., Iourin O., Hockley D. J., Kingsman S. M., Kingsman A. J. (1997) J. Virol. 71, 3474–3483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. De Francesco M. A., Caruso A., Fallacara F., Canaris A. D., Dima F., Poiesi C., Licenziati S., Corulli M., Martinelli F., Fiorentini S., Turano A. (1998) AIDS 12, 245–252 [DOI] [PubMed] [Google Scholar]
- 11. De Francesco M. A., Baronio M., Fiorentini S., Signorini C., Bonfanti C., Poiesi C., Popovic M., Grassi M., Garrafa E., Bozzo L., Lewis G. K., Licenziati S., Gallo R. C., Caruso A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 9972–9977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Poiesi C., De Francesco M. A., Baronio M., Manca N. (2008) Virus Res. 132, 25–32 [DOI] [PubMed] [Google Scholar]
- 13. Hardingham T. E., Fosang A. J. (1992) FASEB J. 6, 861–870 [PubMed] [Google Scholar]
- 14. Kjellén L., Lindahl U. (1991) Annu. Rev. Biochem. 60, 443–475 [DOI] [PubMed] [Google Scholar]
- 15. Bernfield M., Götte M., Park P. W., Reizes O., Fitzgerald M. L., Lincecum J., Zako M. (1999) Annu. Rev. Biochem. 68, 729–777 [DOI] [PubMed] [Google Scholar]
- 16. Laskin J. D., Dokidis A., Sirak A. A., Laskin D. L. (1991) Leuk. Res. 15, 515–523 [DOI] [PubMed] [Google Scholar]
- 17. Screaton G. R., Bell M. V., Jackson D. G., Cornelis F. B., Gerth U., Bell J. I. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 12160–12164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jackson D. G., Bell J. I., Dickinson R., Timans J., Shields J., Whittle N. (1995) J. Cell Biol. 128, 673–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bennett K. L., Jackson D. G., Simon J. C., Tanczos E., Peach R., Modrell B., Stamenkovic I., Plowman G., Aruffo A. (1995) J. Cell Biol. 128, 687–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tanaka Y., Adams D. H., Hubscher S., Hirano H., Siebenlist U., Shaw S. (1993) Nature 361, 79–82 [DOI] [PubMed] [Google Scholar]
- 21. van der Voort R., Taher T. E., Wielenga V. J., Spaargaren M., Prevo R., Smit L., David G., Hartmann G., Gherardi E., Pals S. T. (1999) J. Biol. Chem. 274, 6499–6506 [DOI] [PubMed] [Google Scholar]
- 22. Borland G., Ross J. A., Guy K. (2003) Immunology 93, 139–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Roscic-Mrkic B., Fischer M., Leemann C., Manrique A., Gordon C. J., Moore J. P., Proudfoot A. E., Trkola A. (2003) Blood 102, 1169–1177 [DOI] [PubMed] [Google Scholar]
- 24. Teixé T., Nieto-Blanco P., Vilella R., Engel P., Reina M., Espel E. (2008) Mol. Immunol. 45, 2905–2919 [DOI] [PubMed] [Google Scholar]
- 25. Lories V., David G., Cassiman J. J., Van den Berghe H. (1986) Eur. J. Biochem. 158, 351–359 [DOI] [PubMed] [Google Scholar]
- 26. Forster-Horváth C., Bocsi J., Rásó E., Orbán T. I., Olah E., Tímár J., Ladányi A. (2001) Eur. J. Immunol. 31, 600–608 [DOI] [PubMed] [Google Scholar]
- 27. Costes S., Cho E., Catalfamo M., Karpova T., McNally J., Henkart P., Lockett S. (2002) Proc. Microsc. Microanal. 8, S2–1040CD–S2–1048CD [Google Scholar]
- 28. Alfadhli A., Huseby D., Kapit E., Colman D., Barklis E. (2007) J. Virol. 81, 1472–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chang T. L.., Gordon C. J., Roscic-Mrkic B., Power C., Proudfoot A. E., Moore J. P., Trkola A. (2002) J. Virol. 76, 2245–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goldsmith E. J., Akella R., Min X., Zhou T., Humphreys J. M. (2007) Chem. Rev. 107, 5065–5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chung J. S., Bonkobara M., Tomihari M., Cruz P. D., Jr., Ariizumi K. (2009) Eur. J. Immunol. 39, 965–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Carey D. J. (1997) Biochem. J. 327, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Appay V., Dunbar P. R., Cerundolo V., McMichael A., Czaplewski L., Rowland-Jones S. (2000) Int. Immunol. 12, 1173–1182 [DOI] [PubMed] [Google Scholar]
- 34. Proudfoot A. E., Handel T. M., Johnson Z., Lau E. K., LiWang P., Clark-Lewis I., Borlat F., Wells T. N., Kosco-Vilbois M. H. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 1885–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Proudfoot A. E. (2006) Biochem. Soc. Trans. 34, 422–426 [DOI] [PubMed] [Google Scholar]
- 36. Popovic M., Tenner-Racz K., Pelser C., Stellbrink H. J., van Lunzen J., Lewis G., Kalyanaraman V. S., Gallo R. C., Racz P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 14807–14812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Seddon B., Zamoyska R. (2003) Curr. Opin. Immunol. 15, 321–324 [DOI] [PubMed] [Google Scholar]
- 38. Surh C. D., Sprent J. (2005) Semin. Immunol. 17, 183–191 [DOI] [PubMed] [Google Scholar]
- 39. Douek D. C., Betts M. R., Hill B. J., Little S. J., Lempicki R., Metcalf J. A., Casazza J., Yoder C., Adelsberger J. W., Stevens R. A., Baseler M. W., Keiser P., Richman D. D., Davey R. T., Koup R. A. (2001) J. Immunol. 167, 6663–6668 [DOI] [PubMed] [Google Scholar]
- 40. Teixeira L., Valdez H., McCune J. M., Koup R. A., Badley A. D., Hellerstein M. K., Napolitano L. A., Douek D. C., Mbisa G., Deeks S., Harris J. M., Barbour J. D., Gross B. H., Francis I. R., Halvorsen R., Asaad R., Lederman M. M. (2001) AIDS 15, 1749–1756 [DOI] [PubMed] [Google Scholar]
- 41. Sieg S. F., Rodriguez B., Asaad R., Jiang W., Bazdar D. A., Lederman M. M. (2005) J. Infect. Dis. 192, 62–70 [DOI] [PubMed] [Google Scholar]
- 42. Schacker T. W., Brenchley J. M., Beilman G. J., Reilly C., Pambuccian S. E., Taylor J., Skarda D., Larson M., Douek D. C., Haase A. T. (2006) Clin. Vaccine Immunol. 13, 556–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lempicki R. A., Kovacs J. A., Baseler M. W., Adelsberger J. W., Dewar R. L., Natarajan V., Bosche M. C., Metcalf J. A., Stevens R. A., Lambert L. A., Alvord W. G., Polis M. A., Davey R. T., Dimitrov D. S., Lane H. C. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 13778–13783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kovacs J. A., Lempicki R. A., Sidorov I. A., Adelsberger J. W., Herpin B., Metcalf J. A., Sereti I., Polis M. A., Davey R. T., Tavel J., Falloon J., Stevens R., Lambert L., Dewar R., Schwartzentruber D. J., Anver M. R., Baseler M. W., Masur H., Dimitrov D. S., Lane H. C. (2001) J. Exp. Med. 194, 1731–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Finkel T. H., Tudor-Williams G., Banda N. K., Cotton M. F., Curiel T., Monks C., Baba T. W., Ruprecht R. M., Kupfer A. (1995) Nat. Med. 1, 129–134 [DOI] [PubMed] [Google Scholar]
- 46. Lu W., Andrieu J. M. (1995) Nat Med 1, 386–387 [DOI] [PubMed] [Google Scholar]
- 47. Fiorentini S., Riboldi E., Facchetti F., Avolio M., Fabbri M., Tosti G., Becker P. D., Guzman C. A., Sozzani S., Caruso A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3867–3872 [DOI] [PMC free article] [PubMed] [Google Scholar]