Abstract
Clostridium perfringens enterotoxin (CPE) is a cause of food poisoning and is considered a pore-forming toxin, which damages target cells by disrupting the selective permeability of the plasma membrane. However, the pore-forming mechanism and the structural characteristics of the pores are not well documented. Here, we present the structure of CPE determined by x-ray crystallography at 2.0 Å. The overall structure of CPE displays an elongated shape, composed of three distinct domains, I, II, and III. Domain I corresponds to the region that was formerly referred to as C-CPE, which is responsible for binding to the specific receptor claudin. Domains II and III comprise a characteristic module, which resembles those of β-pore-forming toxins such as aerolysin, C. perfringens ϵ-toxin, and Laetiporus sulfureus hemolytic pore-forming lectin. The module is mainly made up of β-strands, two of which span its entire length. Domain II and domain III have three short β-strands each, by which they are distinguished. In addition, domain II has an α-helix lying on the β-strands. The sequence of amino acids composing the α-helix and preceding β-strand demonstrates an alternating pattern of hydrophobic residues that is characteristic of transmembrane domains forming β-barrel-made pores. These structural features imply that CPE is a β-pore-forming toxin. We also hypothesize that the transmembrane domain is inserted into the membrane upon the buckling of the two long β-strands spanning the module, a mechanism analogous to that of the cholesterol-dependent cytolysins.
Keywords: Bacterial Toxins, Crystallography, Membrane Bilayer, Protein Assembly, Protein Domains, Protein Folding, Protein Structure
Introduction
Clostridium perfringens enterotoxin (CPE),2 which damages intestinal epithelia, is a causative agent of food poisoning. The toxin consists of a single chain polypeptide of 319 amino acids. The C-terminal domain of CPE (C-CPE, residues 184–319) recognizes and binds to certain members of the claudin family, components of tight junctions, as a receptor on target cells (1–4), and the N-terminal region is believed to be involved in forming physiological pores to disrupt the selective permeability of the plasma membrane, resulting in cell death (5–7). It was also reported that the physiological pores are composed of a large complex comprising CPE and cellular components such as claudins (2, 8).
The binding between C-CPE and claudins has been well characterized. The 16–17 C-terminal amino acids of C-CPE were reportedly important for the interaction (9, 10), especially, Tyr306, Tyr310, Tyr312, and Leu315 (11–13). According to the crystal structure of C-CPE (14), these residues organize a cleft space that is considered to interact directly with claudins. Claudins are tetratransmembrane proteins. The region of claudins responsible for binding to CPE was located on the C-terminal side of the second extracellular loop and recently designated CPE-SR for CPE sensitivity-related region (15). The bottom of the cleft space of CPE is negatively charged, whereas the CPE-SRs of CPE-sensitive claudins are positively charged. Therefore, it was proposed that electrostatic attraction at least partly contributes to the mutual interaction. In fact, some members of the claudin family, the CPE-SR of which did not show an apparent negative charge, are insensitive to CPE (15).
In contrast to the binding of CPE to claudins, the complex that makes the physiological pores remains little understood. Some electron microscopic techniques and/or analyses of crystal structures have provided insight into construction of the physiological pores made of aerolysin (16–18), cholesterol-dependent cytolysins (19, 20), and staphylococcal α-toxin (21) and so on (22, 23). In contrast, there is no report regarding microscopic observations of the CPE complex on the plasma membrane and molecular structure of the full-length CPE.
Here, we describe the structure of CPE solved by x-ray crystallography at a resolution of 2.0 Å. Our results show that CPE shares features in common with aerolysin, C. perfringens ϵ-toxin, and Laetiporus sulfureus hemolytic pore-forming lectin, implying that CPE organizes transmembrane pores via the similar mechanism adopted by these toxins.
EXPERIMENTAL PROCEDURES
Plasmid Construction
The CPE gene was amplified by PCR with the following primer pair: NcoCPE 5′-aaaaccatggcgATGCTTAGTAACAATTTAAATC-3′ (including a NcoI site (underlined)) and CPEXho 5′-aaactcgagAAATTTTTGAAATAATATTGAATAA-3′ (including a XhoI site (underlined)) using pET16b-CPE (15) as the template. The amplified DNA fragment was cloned into pCR2.1-TOPO TA (Invitrogen) and sequenced. The CPE gene with the correct sequence was excised with NcoI and XhoI and inserted into the NcoI-XhoI site of pET28a(+) (Merck), so that the gene was fused to a His6 tag sequence on the 3′ side. The resultant plasmid was named pET28a-CPEwt.
Expression and Purification of CPE
Ten milliliters of an overnight culture of Escherichia coli BL21-CodonPlus (DE3)-RIL (Stratagene) transformed with pET28a-CPEwt was inoculated into 1 liter of LB medium containing 50 μg/ml kanamycin and 50 μg/ml chloramphenicol. The culture was grown at 37 °C with vigorous shaking until the absorbance at 600 nm reached 0.6. Expression of the recombinant protein was induced by adding isopropyl-1-thio-β-d-galactopyranoside (Nacalai tesque, Kyoto, Japan) to give a final concentration of 1 mm. Six hours later, the cells were harvested by centrifugation at 5,000 rpm for 20 min at 4 °C, washed with ice-cold water, suspended in 50 mm sodium phosphate buffer, pH 8.0, including 0.3 m NaCl and 1 mm imidazole, and disrupted by sonication using Sonifier S-350 (Branson, Danbury, CT) at 4 °C. The suspension of the disrupted cells was centrifuged at 12,000 rpm for 20 min at 4 °C. The supernatant was subjected to nickel-nitrilotriacetic acid affinity gel (HIS-Select Nickel Affinity Gel; Sigma-Aldrich) chromatography, and the soluble recombinant protein was eluted with an imidazole concentration gradient (1–500 mm). The fraction containing CPE was collected and dialyzed against 10 mm Tris-HCl, pH 8.0. The homogeneity of the purified preparation was confirmed by 12.5% PAGE without SDS.
Crystallization
An 8 mg/ml solution of the recombinant C-terminally His-tagged CPE in 0.01 m Tris-HCl, pH 8.0, was employed for crystallization. Crystallization trials were set up at room temperature as sitting-drop vapor-diffusion experiments on Linbro crystallization plates. Initial screening was performed using the sparse-matrix method with commercial crystal screening kits (Hampton Research). The crystallization droplets were made of 1 μl of the CPE solution and 1 μl of a reservoir solution containing 25% (w/v) polyethylene glycol 3350, 0.2 m sodium chloride, and 0.1 m Tris-HCl, pH 8.0, at 293 K, and equilibrated with 500 μl of the reservoir solution. Triangle-shaped tabular crystals appeared in a few weeks and grew to maximum dimensions of about 0.3 × 0.3 × 0.2 mm.
Data Collection, Phasing, and Refinement
X-ray diffraction data on CPE crystals were collected at 100 K in a nitrogen stream, after the crystals were soaked with the reservoir solution containing 20% glycerol as a cryoprotectant. Native data at 1.98 Å were collected from a single crystal with a Rayonix MX225HE CCD detector on the beamline BL44XU at SPring-8 (Table 1). The x-ray wavelength was 0.9 Å, the angle oscillation range was 0.5°, and the crystal-to-detector distance was 230 mm.
TABLE 1.
Data collection, structural solution, and refinement statistics for CPEs
| Diffraction data | CPE |
|
|---|---|---|
| Native1 | SmCl2 | |
| X-ray source | SPring-8/BL44XU | CuKα |
| Wavelength (Å) | 0.900 | 1.54178 |
| Resolution (Å) | 1.98 | 3.1 |
| Space group | P213 | P213 |
| Unique reflections | 22,192 | 6,058 |
| Rmerge (%)a | 6.9(31.9) | 12.0(48.8) |
| Completeness (%)b | 99.3(100) | 100(100) |
| Redundancy | 10.5 | 21.3 |
| Refinement | Refmac5 | |
| Resolution range (Å) | 69.6–1.98 | |
| R-factora/Rfreea (%) | 19.3/25.2 | |
| No. of protein atoms | 2,236 | |
| No. of solvent atoms | 136 | |
| Ramachandran distribution (% favored, allowed, outlier) | 98.0, 1.4, 0 | |
| Root mean square bonds (Å), angles (°) | 0.023, 1.95 | |
| Average B value (Å2) | 43.3 | |
a Rmerge = Σ|Ii − <Ii>|/Σ<Ii>, where Ii is the observed intensity and <Ii> is the average intensity over symmetry equivalent measurements.
R-factor = Σ‖Fobs| −|Fcalc‖/Σ|Fobs|.
Rfree is calculated as R-factor, but on 5% of all reflections that were not used in the crystallographic refinement.
b Completeness for all reflections and for the highest resolution shell in parentheses.
More than 10 heavy-atom compounds were tested to collect diffraction data. Suitable derivatives were used such as SmCl2 (soaking concentration, 5 mm), which yielded an overall figure of merit of 0.66 at 3.1 Å by a combination of single anomalous dispersion and molecular replacement phasing (Table 1). Crystallographic phases were calculated and refined by Phaser (24). The resulting electron density maps modified by Parrot (25) automatically allowed the tracing of about 80% of one independent molecule by Buccaneer (26). Model building and inspection were based on COOT (27). Atomic coordinates and structural factors for CPE have been deposited with the Protein Data Bank (PDB), with accession code 3AM2. All images of the molecular structure were prepared with PyMOL.
RESULTS
Structural Determination
The crystal data are shown in Table 1. Analysis of the diffraction pattern and systematic absences led to the CPE crystals being assigned to the cubic space group P213, with unit-cell parameters a = b = c = 98.5 Å, α = β = γ = 90°. A total of 22,192 unique reflections were obtained using the program packages of HKL2000 (28). The intensity data in the resolution range of 69.6–1.98 Å were processed with an Rmerge value of 6.9%. Assuming a molecular mass of 35 kDa for the expressed CPE, packing density calculations indicate the most probable value for VM to be 2.27 Å3 Da−1, with one CPE chain per asymmetric unit. This corresponds to a solvent fraction of about 45.9%, a typical value for protein crystals.
The crystal structure of the full-length CPE was solved by a combination of single anomalous dispersion methods using samarium derivatives and molecular replacement techniques with the Lys197 to Phe319 region of C-CPE (PDB accession code 2QUO). The experimental maps calculated by the Phaser and Parrot/Bucaneer procedures were of excellent quality and allowed unequivocal tracing of the chain (residues 36–319) and subsequent refinement at a resolution of 1.98 Å. The structure was refined using REFMAC (29); 5% of the unique reflections were used to monitor the free R-factor. The final values for the general R-factor and free R-factor were 19.3% and 25.2%, respectively (all reflections in the 69.6–1.98 Å range). The refined model consists of 286 amino acids with 136 solvent molecules and two unknown ions. Stereochemistry checks indicated that the refined model was in quite good agreement with expectations for models within this resolution range (Table 1). For N-terminal residues 1–35, electron density was not observed, implying a disordered structure. The asymmetric unit contained one CPE molecule (Fig. 1), and its trimers were generated by crystallographic symmetry (Fig. 2).
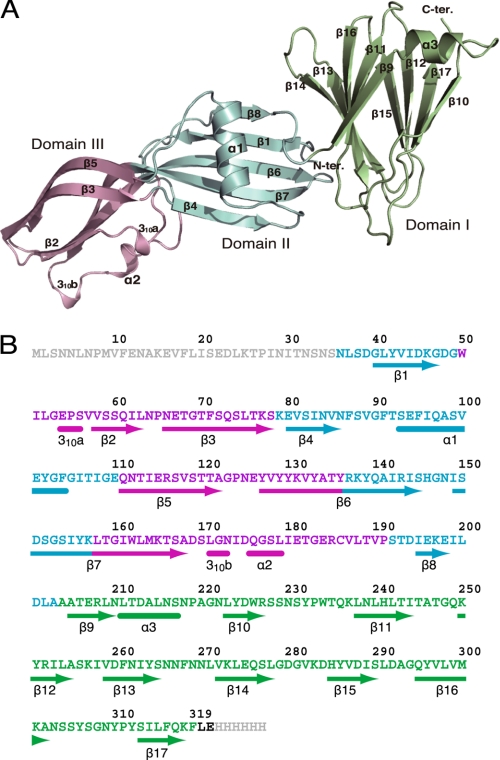
FIGURE 1.
Three-dimensional structure of the full-length CPE monomer. A, ribbon diagram of the CPE monomer. The N-terminal pore-forming domain II, domain III, and the C-terminal claudin-binding domain I are shown in blue, light pink, and lime green, respectively. B, amino acid sequence and secondary structure of CPE. Arrows and end-rounded bars below the sequence indicate β-strand and helical conformations, respectively. Domains I, II, and III are represented by green, blue, and magenta, respectively.
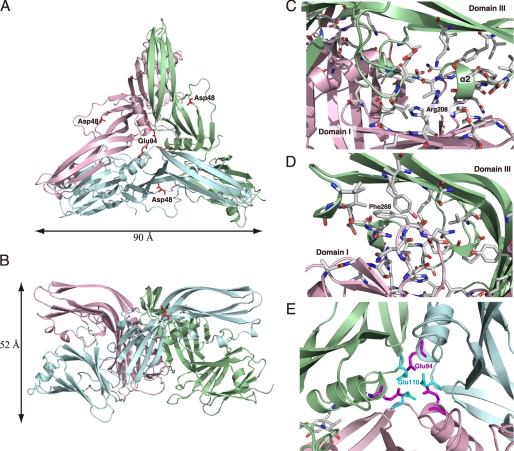
FIGURE 2.
Trimeric structure of CPE. A, view from the top of the trimer and parallel to the 3-fold axis. B, view perpendicular to the 3-fold axis. Each monomer is defined by a different color, blue, green, or pink. Asp48 is rendered as red sticks in each monomer. C–E, views of the interface between monomers. Residues directly involved in contact between the monomers are shown as stick models. The calculations of interfaces between monomers were performed by PISA. C and D, dimeric interface between domains I and III. E, trimeric interfacing center of CPE. Glu94 and Glu110 of each monomer are featured as magenta and light blue sticks, respectively.
CPE Monomer
The monomer of CPE shows an elongated shape (95 × 42 × 32 Å), which is composed of 17 β-strands and 5 helices, three of which are very short, comprising fewer than 6 amino acid residues (Fig. 1). These results are consistent with previous data from circular dichroism spectra showing that CPE contains about 80% β-sheet (30). Three distinct domains could be distinguished in the molecule and were designated domain I, domain II, and domain III as shown in Fig. 1. Domain I almost corresponds to the region referred to as C-CPE, which binds to the specific receptor claudin (2, 4). This domain forms a nine-strand β-sandwich with a short helical element between strands β9 and β10, the fold of which is almost the same as that reported previously (14), except for some loop regions (residues 217–223 and 265–272). The crystal packing is, however, completely different. The major interactions in the previously determined structure mainly involve Arg227 and Ser304, whereas in the structure determined here, marked hydrophobic interactions occur between Val282 in the C-terminal region, and Phe319 in the C-terminal region of a neighboring molecule (14). Domains II and III organize a module that shows an elongated caterpillar shape, 60 Å long and 24–28 Å thick (Fig. 1). The module is composed of 8 β-sheets, β1–β8, two α-helices, and two short 310 helical segments. Two long β-strands, β6 and β7, organize a stem that spans the entire length of the module with a slight distortion between the domains. In addition, domain II contains the largest α-helix and three β-strands that form an antiparallel sheet together with β6 and β7. Domain III contains three β-strands and three small helices in addition to β6 and β7.
Trimeric Assembly of CPE
Although the monomer of CPE is found in the asymmetric unit in this crystal, it does form a crystallographic trimer (Fig. 2, A and B). The trimeric fold is stabilized by a number of specific interactions between each monomer. Analyses with PISA revealed major interactions between domain III and domain I of two symmetry-related molecules. The helical regions at the edges of domain III interact with the β-barrel region of domain I of an adjacent molecule. Arg208 of domain I is inserted into α2, which has negatively charged interfaces (Fig. 2C). On the other side, Phe268 of the loop connecting β13 and β14 of domain I is hydrophobically inserted into helix 310b at the edge of domain III (Fig. 2D). These two specific interactions determine the position of domain I relative to domain III. The trimeric assembly shows an apparent conduit structure in its center. Glu94 and Glu110 face the respective counterpart of each monomer and organize the narrow regions (diameter = 4.4–4.6 Å) of the conduit (Fig. 2E). Although the α1 helices of each monomer are close to each other, directed toward the conduit, there are neither α-helices nor β-sheets forming the conduit walls. Analysis of samarium-derivative datasets shows that three symmetrical side chains of Glu94 and Glu110 keep a samarium ion in each narrow region. To know whether the conduit structure is involved in the toxicity of CPE, we examined CPE mutants, in which Glu94 or Glu110 was independently replaced with Asp, Gly, Gln, Tyr, or Leu, for cytotoxicity in Vero cells. All CPE mutants were found to retain the cytotoxicity (data not shown).
Structural Homologues of CPE
A DALI search for structural homologues of the full-length CPE uncovered a hemagglutinin component, HA3, of Clostridium botulinum toxin, and C. perfringens ϵ-toxin. C. botulinum HA3 is expressed as a single chain and proteolytically cleaved into HA3a and HA3b chains, both of which share homology with the N-terminal module of CPE composed of domains II and III (2ZOE-A chain, Z = 14.0; 2ZOE-B chain, Z = 10.9). BLAST also revealed the amino acid sequences of HA3a (residues 17–173, GenBank accession number D38562) and HA3b (residues 211–423, GenBank accession number D38562) to show 24 and 27% identity with CPE, residues 33–188 and 39–240, respectively. In contrast to HA3, the structural similarity of C. perfringens ϵ-toxin with CPE is low (PDB code 1UYJ, Z = 5.2). When domains II and III of CPE were subjected to the DALI search, aerolysin and L. sulfureus hemolytic pore-forming lectin (LSL) also emerged in addition to C. perfringens ϵ-toxin (Fig. 3A).
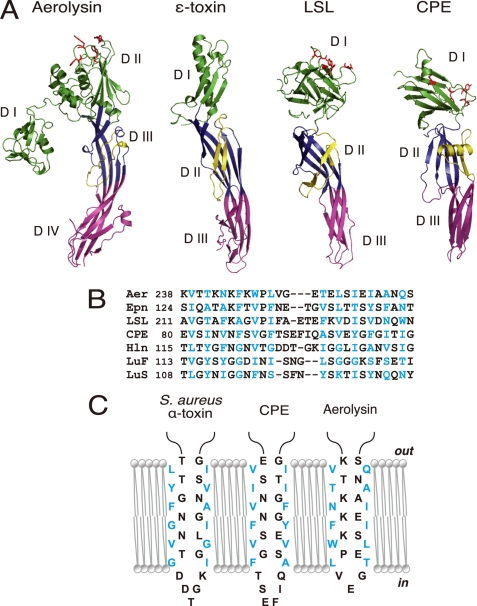
FIGURE 3.
Structural similarities between aerolysin, C. perfringens ϵ-toxin, LSL, and CPE. A, ribbon diagrams drawn from PDB data under codes 1PRE for aerolysin (17), 1UYJ for C. perfringens ϵ-toxin (35), 1W3F for LSL (36), and 3AM2 for CPE (this study). Domains corresponding to domains I, II, and III of CPE are defined by green, blue, and pink, respectively. Domains I and II of aerolysin are shown in green. Amphipathic loops (flanking elements) are indicated as yellow ribbons. The residues that participate in receptor binding are depicted as red sticks (11–13, 15, 36, 40). Note that each toxin possesses at least two long antiparallel β-strands spanning domains II and III (domains III and IV for aerolysin). B, sequence alignment of the putative transmembrane domains of different β-PFTs; the aerolysin-like family, aerolysin (Aer), ϵ-toxin (Epn), and LSL, three staphylococcal pore-forming toxins, Staphylococcal aureus α-toxin (Hln, PDB code 7AHL), LukF (PDB code 1PVL), and LukS (PDB code 1T5R), and CPE. The numbers after the toxin names indicate the N-terminal amino acid positions. Alignments were generated based on the alternating pattern of polar and hydrophobic residues. Residues that are putatively facing the lipid bilayer are shown in blue. C, schematic representation of the antiparallel strands forming the β-barrel of staphylococcal α-toxin and the corresponding residues of CPE and aerolysin. The alignment is based on previous reports (21, 37, 41) and sequence similarity. The residues are portrayed as either facing the lipid bilayer or lining the lumen of the pore.
DISCUSSION
The pore-forming toxins exert their toxic effects by elaborating physiological pores on the plasma membrane of target cells, resulting in changes to the membrane permeability and eventually cell lysis. These toxins are divided into two groups, α-pore-forming toxins (α-PFTs) and β-pore-forming toxins (β-PFTs), based on structural properties of transmembrane pores. The former group employs α-helices to insert into the lipid membrane, and the latter uses β-strands to fold amphipathic β-barrels in the membrane (32, 33). Salmonella cytolysin A and staphylococcal δ-toxin exemplify the former (23, 32), and many other PFTs belong to the latter. Although CPE has long been believed to be a PFT, the architecture of the pore it creates has been unknown. Briggs et al. reported the preliminary crystallographic data of CPE but did not described the details (31). In this study, we first determined the overall structure of CPE, information that then enabled us to speculate on the mechanism by which it forms pores.
The structure of CPE provided several points of note. First, CPE shows a trimeric form with an apparent conduit/channel in crystal packing, which may suggest a physiological channel made by CPE as a PFT. However, it is unlikely that this conduit functions as actual transmembrane pores for the following reasons. (i) Unlike in the case of other PFTs, the conduit is not surrounded by a rigid structure such as an α-helix or β-barrel. (ii) The vertical size of the trimer containing the conduit is estimated at 52 Å at most, which is not enough to span the lipid bilayer membrane. (iii) In gel permeation chromatography, CPE eluted at a position estimated at 35 kDa, indicating the toxin to be in a monomeric state in solution (data not shown). In addition, there have been no reports to show that CPE is oligomerized in solution. (iv) Glu94 and Glu110 face the respective counterpart of each monomer and organize the narrow points in the conduit (Fig. 2E). This structure implies that these residues are critical in the function of any pores organized by the trimer. However, substitutions of these residues with Asp, Gln, Gly, Tyr, or Leu did not affect the toxicity (data not shown). (v) The conduit is organized above the receptor binding site, which is located at the bottom of domain I in the trimeric state shown in Fig. 2B. Therefore, the conduit should be positioned above the plasma membrane surface and is not able to make a transmembrane pore. (vi) Asp48, which was reported to be important in the oligomerization of the toxin (7), is located outside of the interface of the trimer (Fig. 2A). These findings indicate that the trimeric assembly with the conduit-like structure emerges only in the crystal.
Second, the CPE monomer shares structural homology with C. botulinum HA3 and C. perfringens ϵ-toxin, which also show structural homology to each other. HA3 is a component of a hemagglutinin and known to bind to sialyloligosaccharides (34). However, because the sugar binding ability is attributable to a region of HA3 that is not homologous to CPE, it is difficult to predict the structure-function relation of CPE, based on the homology with HA3. Instead, we focus on characteristic features of CPE that are common among the PFTs such as C. perfringens ϵ-toxin, aerolysin, and LSL. These toxins have an elongated architecture with β-sheets and an arrangement of three (or four) domains (17, 35, 36) (Fig. 3A). In this study, the domains of CPE were numbered after those of the other toxins: domains I, II, and III (Figs. 1 and 3). Domain I is composed of the C-terminal region of CPE, whereas domains II and III are formed by the N-terminal region. As a result, the amino acid sequence and the domain arrangement of CPE were in the opposite order, whereas C. perfringens ϵ-toxin, aerolysin, and LSL consist of domains I, II, III (and IV for aerolysin) from the N to C terminus.
As shown in Fig. 3A, these domains are aligned along the long axis of the toxins, except domain I of aerolysin. Domain I (or domains I and II of aerolysin) is responsible for binding to respective receptors. Aerolysin is known to bind a certain set of glycosylphosphatidylinositol-anchored proteins. Domain I of LSL is defined as a lectin binding to carbohydrates. The receptor for ϵ-toxin is not known. Although these toxins similarly have the receptor binding site/residues at the top (Fig. 3A, red sticks), they show less structural similarity in domain I than other domains. It is plausible that the distinct structure of domain I (and domain II for aerolysin) reflects the different receptors recognized by these domains. In contrast to domain I, the module composed of domains II and III shares several structural features in common with these toxins, notably in terms of the stem of two β-strands and a flanking element laying on the major β-sheet in domain II (or domain III for aerolysin) (Fig. 3A, yellow ribbons). The flanking element of CPE shows alternating patterns of polar and hydrophobic residues, which imply amphipathic characteristics. Amphipathic strands have been found in similar flanking elements of aerolysin (16), C. perfringens ϵ-toxin (35), LSL (36), staphylococcal α-toxin (21) and leukocidin (37) (Fig. 3B), all of which are known as β-PFTs. Biochemical analyses of Clostridium septicum α-toxin, which shows clear homology with aerolysin, identified the membrane-spanning domain as an amphipathic region corresponding to the flanking region of aerolysin (38). These strands are believed to be inserted into the lipid bilayer and form the transmembrane β-barrel, the hydrophobic residues of which face the lipid bilayer and the polar residues of which constitute the hydrophilic lumen of pores (Fig. 3C). The amphipathic strand of CPE stretches from Val81 to Ile106 and comprises β4, α1, and the intervening loop (Figs. 1 and 3). A previous report demonstrated that this region participates in the formation of pores after organizing a prepore oligomeric complex (7). Taken together, it is likely that CPE belongs to the β-PFT family, and the Val81-Ile106 region forms the β-hairpin to organize the β-barrel structure spanning the lipid membrane (Fig. 3C).
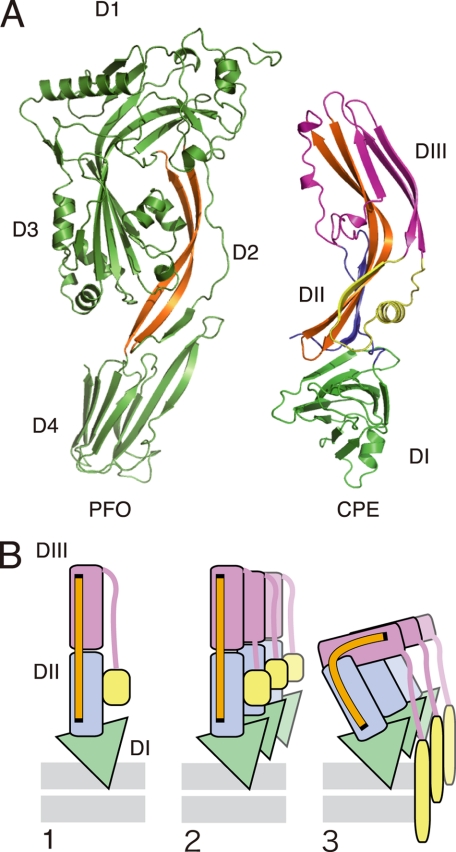
The amphipathic flanking strands reside in the middle of the toxin molecule. Therefore, if this strand takes part in the formation of the transmembrane pore, a certain structural alteration of the toxin must occur. Previous reports demonstrate that cholesterol-dependent cytolysins, a group of β-PFTs including perfringolysin O and pneumolysin, undergo vertical collapse that brings the amphipathic strand to the membrane surface and forms the membrane pore composed of the β-barrel (19, 39). We consider that similar structural changes occur for CPE to accomplish the β-barrel insertion into the membrane. The cholesterol-dependent cytolysins consist of four domains, of which domain 2 is considered to be folded over, bringing the amphipathic strand close enough to the membrane to span the bilayer. Domain 2 of cholesterol-dependent cytolysins and domains II and III of CPE possess at least two long antiparallel β-strands (Fig. 4A, orange strands). Thus, we presume that CPE, like the cholesterol-dependent cytolysins, inserts the amphipathic strand from β4 to α1 into the membrane upon the buckling of domains II and III, which may be conducted by the long antiparallel β-strands as a “hinge module” (Fig. 4B). This mechanism might be applicable to aerolysin, C. perfringens ϵ-toxin, and LSL because they also have similar long antiparallel β-strands.
FIGURE 4.
Comparison of structure between CPE and perfringolysin O (PFO, accession code 1PFO) (A) and a putative model showing how the toxin molecules are folded to insert the antipathic strands into the membrane (B). Each domain of CPE is represented by the same colors as shown in Fig. 3A. A, long antiparallel strands shown in orange. B, the diagram based on a model presented previously (39). The long antiparallel strands that function as a hinge module are shown as an orange bar. 1, binding to the membrane surface via domain I (DI). 2, prepore complex shown with three monomers. 3, amphipathic strand inserted upon the buckling of the module composed of domains II and III.
Here, we presented an explanation of how CPE forms a transmembrane pore, based on observations of the structural features of the full-length toxin. CPE is considered to be a β-PFT. The amphipathic region from Val81 to Ile106 is inserted into the plasma membrane upon the buckling of the stem module, a mechanism that may be common among some β-PFTs. To test our hypothesis, we are now trying to determine the structure of a CPE-claudin complex or an oligomerized form of CPE, which would enable us to understand better the pore-forming mechanism.
Acknowledgments
We thank Dr. K. Matoba and Prof. S. Harada of the Kyoto Institute Technology and Dr. E. Yamashita, Dr. Y. Umena, and Prof. A. Nakagawa of SPring-8 BL44XU for help with the X-ray data collection. Diffraction data were collected at the Osaka University beamline BL44XU at SPring-8 equipped with MX225-HE (Rayonix), which is supported financially by the Academia Sinica and National Synchrotron Radiation Research Center (Taiwan, China). We also appreciate the secretarial and technical assistance of T. Suzuki and M. Kobayashi.
This work was supported in part by grants-in-aid for scientific research from the Ministry of Education, Culture, Science, and Technology of Japan.
The atomic coordinates and structure factors (code 3AM2) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- CPE
- Clostridium perfringens enterotoxin
- C-CPE
- C-terminal domain of CPE
- LSL
- Laetiporus sulfureus hemolytic pore-forming lectin
- PDB
- Protein Data Bank
- PFT
- pore-forming toxin.
REFERENCES
- 1. Sonoda N., Furuse M., Sasaki H., Yonemura S., Katahira J., Horiguchi Y., Tsukita S. (1999) J. Cell Biol. 147, 195–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Katahira J., Inoue N., Horiguchi Y., Matsuda M., Sugimoto N. (1997) J. Cell Biol. 136, 1239–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hanna P. C., Mietzner T. A., Schoolnik G. K., McClane B. A. (1991) J. Biol. Chem. 266, 11037–11043 [PubMed] [Google Scholar]
- 4. Horiguchi Y., Akai T., Sakaguchi G. (1987) Infect. Immun. 55, 2912–2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kokai-Kun J. F., Benton K., Wieckowski E. U., McClane B. A. (1999) Infect. Immun. 67, 5634–5641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Smedley J. G., 3rd, McClane B. A. (2004) Infect. Immun. 72, 6914–6923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Smedley J. G., 3rd, Uzal F. A., McClane B. A. (2007) Infect. Immun. 75, 2381–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Robertson S. L., Smedley J. G., 3rd, Singh U., Chakrabarti G., Van Itallie C. M., Anderson J. M., McClane B. A. (2007) Cell Microbiol. 9, 2734–2755 [DOI] [PubMed] [Google Scholar]
- 9. Takahashi A., Kondoh M., Masuyama A., Fujii M., Mizuguchi H., Horiguchi Y., Watanabe Y. (2005) J. Control. Release 108, 56–62 [DOI] [PubMed] [Google Scholar]
- 10. Kondoh M., Masuyama A., Takahashi A., Asano N., Mizuguchi H., Koizumi N., Fujii M., Hayakawa T., Horiguchi Y., Watanbe Y. (2005) Mol. Pharmacol. 67, 749–756 [DOI] [PubMed] [Google Scholar]
- 11. Takahashi A., Komiya E., Kakutani H., Yoshida T., Fujii M., Horiguchi Y., Mizuguchi H., Tsutsumi Y., Tsunoda S., Koizumi N., Isoda K., Yagi K., Watanabe Y., Kondoh M. (2008) Biochem. Pharmacol. 75, 1639–1648 [DOI] [PubMed] [Google Scholar]
- 12. Harada M., Kondoh M., Ebihara C., Takahashi A., Komiya E., Fujii M., Mizuguchi H., Tsunoda S., Horiguchi Y., Yagi K., Watanabe Y. (2007) Biochem. Pharmacol. 73, 206–214 [DOI] [PubMed] [Google Scholar]
- 13. Ebihara C., Kondoh M., Harada M., Fujii M., Mizuguchi H., Tsunoda S., Horiguchi Y., Yagi K., Watanabe Y. (2007) Biochem. Pharmacol. 73, 824–830 [DOI] [PubMed] [Google Scholar]
- 14. Van Itallie C. M., Betts L., Smedley J. G., 3rd, McClane B. A., Anderson J. M. (2008) J. Biol. Chem. 283, 268–274 [DOI] [PubMed] [Google Scholar]
- 15. Kimura J., Abe H., Kamitani S., Toshima H., Fukui A., Miyake M., Kamata Y., Sugita-Konishi Y., Yamamoto S., Horiguchi Y. (2010) J. Biol. Chem. 285, 401–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Iacovache I., Paumard P., Scheib H., Lesieur C., Sakai N., Matile S., Parker M. W., van der Goot F. G. (2006) EMBO J. 25, 457–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Parker M. W., Buckley J. T., Postma J. P., Tucker A. D., Leonard K., Pattus F., Tsernoglou D. (1994) Nature 367, 292–295 [DOI] [PubMed] [Google Scholar]
- 18. Wilmsen H. U., Leonard K. R., Tichelaar W., Buckley J. T., Pattus F. (1992) EMBO J. 11, 2457–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Czajkowsky D. M., Hotze E. M., Shao Z., Tweten R. K. (2004) EMBO J. 23, 3206–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sekiya K., Danbara H., Yase K., Futaesaku Y. (1996) J. Bacteriol. 178, 6998–7002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Song L., Hobaugh M. R., Shustak C., Cheley S., Bayley H., Gouaux J. E. (1996) Science 274, 1859–1866 [DOI] [PubMed] [Google Scholar]
- 22. Dutta S., Mazumdar B., Banerjee K. K., Ghosh A. N. (2010) J. Bacteriol. 192, 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mueller M., Grauschopf U., Maier T., Glockshuber R., Ban N. (2009) Nature 459, 726–730 [DOI] [PubMed] [Google Scholar]
- 24. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cowtan K. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 470–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cowtan K. (2008) Acta Crystallogr. D Biol. Crystallogr. 64, 83–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 28. Otsinoski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 29. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 30. Granum P. E., Harbitz O. (1985) J. Food Biochem. 9, 137–146 [Google Scholar]
- 31. Briggs D. C., Smedley J. G., 3rd, McClane B. A., Basak A. K. (2010) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66, 794–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Geny B., Popoff M. R. (2006) Biol. Cell 98, 667–678 [DOI] [PubMed] [Google Scholar]
- 33. Gouaux E. (1997) Curr. Opin. Struct. Biol. 7, 566–573 [DOI] [PubMed] [Google Scholar]
- 34. Nakamura T., Kotani M., Tonozuka T., Ide A., Oguma K., Nishikawa A. (2009) J. Mol. Biol. 385, 1193–1206 [DOI] [PubMed] [Google Scholar]
- 35. Cole A. R., Gibert M., Popoff M., Moss D. S., Titball R. W., Basak A. K. (2004) Nat. Struct. Mol. Biol. 11, 797–798 [DOI] [PubMed] [Google Scholar]
- 36. Mancheño J. M., Tateno H., Goldstein I. J., Martínez-Ripoll M., Hermoso J. A. (2005) J. Biol. Chem. 280, 17251–17259 [DOI] [PubMed] [Google Scholar]
- 37. Miles G., Movileanu L., Bayley H. (2002) Protein Sci. 11, 894–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Melton J. A., Parker M. W., Rossjohn J., Buckley J. T., Tweten R. K. (2004) J. Biol. Chem. 279, 14315–14322 [DOI] [PubMed] [Google Scholar]
- 39. Tilley S. J., Orlova E. V., Gilbert R. J., Andrew P. W., Saibil H. R. (2005) Cell 121, 247–256 [DOI] [PubMed] [Google Scholar]
- 40. MacKenzie C. R., Hirama T., Buckley J. T. (1999) J. Biol. Chem. 274, 22604–22609 [DOI] [PubMed] [Google Scholar]
- 41. Rossjohn J., Feil S. C., McKinstry W. J., Tsernoglou D., van der Goot G., Buckley J. T., Parker M. W. (1998) J. Struct. Biol. 121, 92–100 [DOI] [PubMed] [Google Scholar]