Abstract
Daxx is involved in transcriptional control and apoptosis. It comprises several domains, including a regulatory C terminus that is responsible for the interaction with numerous proteins such as p53, promyelocytic leukemia protein (PML), and Hsp27. Here, we describe the identification and characterization of two novel variants of Daxx termed Daxx-β and Daxx-γ, which are generated by alternative splicing. Alternative splicing results in a truncated regulatory C terminus in both proteins. As a consequence, Daxx-β and Daxx-γ show a markedly decreased affinity to PML, which in turn is associated with a different subnuclear localization of these proteins compared with Daxx. Although Daxx is localized mainly in PML-oncogenic domains (PODs) Daxx-β and Daxx-γ display a distinct distribution pattern. Furthermore, Daxx-β and Daxx-γ show a decreased affinity to p53 also due to the truncated C terminus. We provide evidence that the p53 recruitment into PODs is Daxx isoform-dependent. The decreased affinity of Daxx-β/-γ to p53 and PML results in a diffuse localization of p53 throughout the nucleus. In contrast to Daxx, Daxx-β and Daxx-γ are unable to repress p53-mediated transcription. Therefore, alternative splicing of Daxx might indicate an additional level in the cellular apoptosis network.
Keywords: Apoptosis, Nuclear Organization, p53, Signal Transduction, Transcription
Introduction
Deregulation of apoptosis is involved in several diseases, including cancer, characterized by an abnormally prolonged cell survival. The ubiquitously expressed protein Daxx (1) is implicated in apoptosis, but whether its function is pro- or anti-apoptotic is still controversially discussed. Daxx contains several domains that are essential for interaction with a growing number of proteins (1–3). Originally, Daxx was isolated as a CD95-binding protein that activates c-Jun-NH2-terminal kinase (JNK) via directly binding to and activating of the apoptosis signal regulating kinase-1 (ASK-1), thereby enhancing CD95-mediated apoptosis in a FADD/procaspase-8-independent manner (1, 4–6). Daxx is located mainly in the nucleus where it associates with the promyelocytic leukemia protein (PML)3 in speckled subnuclear structures called PML oncogenic domains (PODs) or PML nuclear bodies (7–10). It was shown that co-localization of Daxx and PML to PODs correlates with Daxx-mediated enhancement of apoptosis. The C-terminal part of the protein is responsible for binding to PML and also exerts its apoptosis-promoting potential (8–10). Depletion of Daxx by siRNA is associated with an elevated level of apoptosis as well as an increased sensitivity to CD95 and stress-induced apoptosis (11, 12). Similarly, targeted disruption of the daxx gene in mice results in embryonic lethality accompanied by global apoptosis in the entire embryo, pointing to a rather anti-apoptotic than pro-apoptotic role of Daxx (13, 14).
Consistent with its nuclear localization, Daxx is also involved in transcriptional control. It interacts directly with several transcription factors, including ETS1 (15), Pax3, and Pax5 (2, 16, 17), androgen receptor, p53 family proteins (18–20), Smad4, and glucocorticoid receptor (21), thereby acting as a transcriptional co-repressor. The capacity of Daxx to repress transcription is in part controlled by modified PML and homeodomain-interacting protein kinase-1 (HIPK1) (7, 8, 22). HIPK1 was shown to be able to relocate Daxx to chromatin, facilitating the Daxx-dependent recruitment of histone acetylases, which leads to transcriptional repression (8, 22). Recent data suggest that this dual subnuclear localization of Daxx is also controlled in a cell cycle-dependent manner, revealing an S phase-specific heterochromatic accumulation of Daxx (13). By interacting with p53 and suppressing its transcriptional activity, Daxx is involved directly in the regulation of one of the most important cellular tumor suppressors and beside apoptosis thereby potentially implicated in cellular processes such as cell cycle arrest, cellular senescence, genome stability, and angiogenesis (18–20).
Recent findings show that Daxx cooperates with the Axin-HIPK2-p53 complex to induce cell death (23) and that the MDM2-Daxx-HAUSP complex could be disrupted by the tumor suppressor protein RASSF1A-mediated self-ubiquitination of MDM2 (24). An another important role for Daxx is its function as a transcriptional repressor of CCAAT/enhancer-binding protein β (25), which is involved in tissue-specific gene expression and thereby takes part in fundamental cellular processes such as proliferation and differentiation.
In the present study we report on the identification and characterization of two novel Daxx splice variants, Daxx-β and Daxx-γ, which both have a truncated and modified regulatory C terminus affecting the localization, binding to PML and p53, and associates with the incapability to repress p53-mediated transcription.
EXPERIMENTAL PROCEDURES
Cell Culture and Transient Transfection
All renal cell carcinoma (RCC) cell lines were derived from typical representatives of the clear cell and chromophilic/papillary types of RCC, as established in our laboratory. HEK293 and HeLa cells were grown in DMEM containing 10% (v/v) FBS, 2 mm glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. Cell lines HepG2, OST, HCT-15, HaCaT, Molt-3, Raji, DU145 and MCF-7 were cultured in RPMI 1640 medium (Invitrogen) containing 10% (v/v) FBS, 2 mm glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. CH-LA-90, SK-N-H, DTC-1 and TC-71 cells were grown in the same medium, but dishes were precoated with 0.1% gelatin. All cells were incubated at 37 °C in an atmosphere containing 5% CO2. For transfections HeLa, HepG2, and HEK293 cells were seeded out in 6-well dishes or 100-mm plates, and 24 h later transfection of the respective plasmids was performed using Polyfect transfection reagent following the manufacturer's guide (Qiagen).
Generation of Stably Transfected RCC Cell Lines by Retroviral Infection
AmphoPackTM packaging cells (Clontech) were seeded out in 100-mm plates at a density of 5,000 cells/dish and were maintained in DMEM containing 10% (v/v) FBS, 200 mg/liter arginine, 72 mg/liter asparagine (Serva Electrophoresis), 10 mm HEPES, 2 mm glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). Twenty-four hours later, cells were transfected with 20 μg of empty pLEGFP-C1 vector (Clontech) or GFP-Daxx, GFP-Daxx-β, and GFP-Daxx-γ expression constructs (see below) using FuGENE transfection reagent (Roche Diagnostics). The supernatants (after 48 h) containing infectious viral particles were collected, sterile-filtered, and used to infect subconfluent cells of the RCC cell line clearCa-6. Additionally, 10 μg/ml protamine sulfate (Sigma) was added to increase transduction efficiency. After 48 h cells were washed twice with PBS, and fresh medium was added containing 0.8 μg/ml G418 (Invitrogen) to select out uninfected cells. After 3 weeks of culturing with selection medium cell populations with up to 95% of stably overexpressing clearCa-6 cells were obtained as demonstrated by FACS analysis (data not shown). Each transfected cell line represented a pool of multiple stable transfectants thereby avoiding clonal bias. Generally, for further analysis (all methods used in this work) all cells were collected (including dead cells in the supernatant).
Assessment of Cell Viability by the 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium Bromide (MTT) Assay
Cells were seeded out onto 96-well plates at a density of 7,500 cells/well. After 24 h, 2 μg/ml topotecan (Smith Kline Beecham, München, Germany), 1 μg/ml doxorubicin (Sigma), 10 μg/ml etoposide (Sigma), or 1 μg/ml paclitaxel (Taxol; Bristol-Meyers Squibb) was added to the cells or left untreated serving as control. At the indicated time points, cell number was analyzed using the colorimetric MTT assay.
RNA/DNA Extraction, RT-PCR, PCR on Genomic DNA, and Sequencing
For RT-PCR first strand cDNA synthesis was carried out using 1 μg of total RNA (extracted with RNeasy; Qiagen), 25 nmol of dNTPs, 0.5 unit of recombinant ribonuclease inhibitor RNasin®, 0.5 μg of random primers, 5 mm MgCl2, and 15 units of avian myeloblastosis virus reverse transcriptase (Promega). The reactions were incubated at 55 °C for 1 h and subsequently terminated by heat inactivation at 95 °C for 5 min. Then, 5 μl was subjected to real-time PCR in a 20-μl PCR mixture containing 2 μl of 10× Fast Start Reaction mix, 3 mm MgCl2 (Roche Diagnostics), and 0.5 μm respective forward and reverse primers. For amplification of Daxx transcripts, the primers 5′-CTT CCT TCA ATG GAG GCG T-3′ (+) and 5′-CCG AGG CTG TGA ATG-3′ (−) were used (GenBank accession no. AF015956) and for GAPDH amplification (GenBank accession no. J04038) PCR was performed with primers 5′-AAC AGC GAC ACC CAC TCC TC-3′ (+) and 5′-GGA GGG GAG ATT CAG TGT GGT-3′ (-), respectively. PCR conditions were as follows: initial denaturation at 95 °C for 10 min and 50 cycles composed of <1 s at 95 °C, 10-s annealing at 66 °C and 20-s primer extension at 72 °C. Corresponding PCR with genomic DNA as template was also performed using up to 300 ng of the respective DNA with an identical PCR setup. Additionally, identities of PCR products were confirmed by DNA sequencing.
Expression Constructs
To create Daxx C-terminally fused to GFP, PCR amplification with human cDNA (obtained from HeLa cells) as template was performed using Pfu polymerase (Promega) with the following primers. The forward primer termed Daxx-I (5′-ACT TCC TCC GTC GAC GGG ATT GGA TCC C-3′) contained a SalI site, and the reverse primer termed Daxx-II (5′-TCC GGT GGA TCG ATG CAG CTA ATC AG-3′) contained a ClaI site (each underlined). The PCR product was subcloned into the SalI and ClaI sites of the pLEGFP-C1 vector (Clontech) to obtain pLEGFP-Daxx. Using pLEGFP-Daxx as template Daxx-β was generated by a two-step PCR method. During the first PCR step within two separate PCRs a Daxx 3′-amplification product and 5′-amplification product, respectively, were generated independently using two different primer pairs of which the respective nonflanking primers were in part complementary. The fist primer pair was composed of the Daxx-I primer and Daxx-β1 (5′-ATG TGG AAA GGC AAA GCC CGG CTG TCC CAA AC-3′) and the second one comprised the primers Daxx-II and Daxx-β2 (5′-GTT TGG GAC AGC CGG GCT TTG CCT TTC CAC AT-3′). In the second step PCR both amplification products were mixed, and fusion of the fragments was achieved by amplification with the respective outer primers Daxx-I and Daxx-II. According to the region that is deleted in Daxx-β by alternative splicing (nucleotides 19–170 of exon 6) due to the design of the primers this region was also deleted in PCR product of the second amplification step. Thereby this product resembles the coding sequence of human Daxx-β. Daxx-γ (lacking nucleotides 1–170 of exon 6) was generated using the same strategy by replacing primers Daxx-β1 and Daxx-β2 with Daxx-γ1 (5′-GGC CAT TAG GAA ACA GCC CGG CTG TCC CAA AC-3′) and Daxx-γ2 (5′-GTT TGG GAC AGC CGG GCT GTT TCC TAA TGG CC-3′). As described for Daxx the sequences encoding Daxx-β and Daxx-γ were subcloned into the SalI and ClaI sites of the pLEGFP-C1 vector (Clontech) to obtain pLEGFP-Daxx-β and pLEGFP-Daxx-γ expressing GFP-fused Daxx-β and Daxx-γ, respectively. The constructs pDSRed2-Daxx, pDSRed2-Daxx-β, and pDSRed2-Daxx-γ were made by exchange of the GFP-encoding sequence of the respective pLEGFP-Daxx vectors by the DNA sequence coding for DSRed2 which was obtained by the pDSRed2-C1 vector (Clontech) and subcloned via the AgeI and XhoI sites. To create HA-tagged Daxx splice variants a HA tag encoding double-stranded DNA cassette was made by annealing the two complementary oligonucleotides 5′-GAT CTA CCG GTC GCC ACC ATG GCT TAC CCA TAC GAT GTT CCA GAT TAC GCG G-3′ and 5′-TCG ACC GCG TAA TCT GGA ACA TCG TAT GGG TAA GCC ATG GTG GCG ACC GGT A-3′ which contain an internal AgeI site (underlined) and flanking XhoI and SalI “sticky ends,” respectively. The latter were used to subclone the HA tag encoding the DNA cassette into the pLEGFP-Daxx vector. At least, GFP was deleted by AgeI digestion, and subsequent relegation led to pLHA-Daxx. The vectors pLHA-Daxx-β and pLHA-Daxx-γ were constructed by exchange of the Daxx-encoding sequence, and the “mock control” pLHA-C1 was made by introducing the HA tag-encoding DNA cassette into pLEGFP-C1 (Clontech) according to the above mentioned procedure.
Western Blot Analysis
Western blot analysis was performed under standard conditions. The following primary antibodies were used in this study: anti-Daxx mAb (Novocastra Laboratories, Newcastle, UK), anti-β-actin mAb (Sigma), anti-PML mAb (Santa Cruz Biotechnology), anti-HA mAb (Cell Signaling), anti-CD95 polyclonal antibody (Santa Cruz Biotechnology), A.v. monoclonal antibody (anti-GFP mAb, Clontech). Of note, this anti-GFP antibody is able to detect blue fluorescent protein too.
Immunoprecipitation
After transfection of HEK293 cells with the respective plasmids as described above, cells were washed with ice-cold PBS buffer, and then cell lysates were prepared by adding ice-cold lysis buffer containing 50 mm HEPES (pH 7.0), 250 mm NaCl, 5 mm EDTA, 0.1% (v/v) Nonidet P-40 supplemented with a complete protease inhibitor mix (Roche). Then, the cell lysate was harvested and centrifuged for 20 min at 20,000 × g and 4 °C to remove intact cells and cell debris. After centrifugation, the supernatant was considered the protein fraction. Either anti-GFP antibody (full-length A.v. polyclonal antibody, dilution 1:150; Clontech) or an anti-Daxx polyclonal antibody (dilution 1:20; Santa Cruz Biotechnology) was added to 100–500 μg of total protein and incubated for 1 h at 4 °C while gently rotating. Subsequently, 40 μl of protein A-Sepharose beads (Sigma) was added, and the mixture was incubated for an additional 16 h. Beads were washed five times with lysis buffer, and the immune complex was released from the beads by boiling in sample buffer for 5 min. Then, immunoprecipitated proteins were analyzed by Western blotting.
Fluorescence Microscopy and Immunostaining
HeLa cells were seeded out on glass coverslips and then transfected with the indicated plasmids. After 48 h cells were washed in PBS, fixed in 2% (w/v) paraformaldehyde for 10 min at room temperature, and permeabilized in PBST (PBS containing 0.1% (v/v) Triton X-100) for 5–10 min at room temperature. Then cells were blocked by incubation with 5% (w/v) BSA in PBST (blocking buffer) for 1 h following incubation with anti-PML mAb antibody (dilution 1:50; Santa Cruz Biotechnology) at room temperature. After washing three times with PBST, for detection TRITC-conjugated goat anti-mouse secondary antibody (Sigma) diluted 1:50 in blocking buffer was added and incubated for 1 h at room temperature followed by washing once in PBST and five times in PBS. According to the experimental design, during the PBST washing step DAPI staining was performed by incubating with 100 ng/ml DAPI solution for 5 min at room temperature. Finally, slides were mounted in 100 mm Tris-HCl (pH 8.5), 10% (w/v) Mowiol 4-88, 2.5% (w/v) 1,4-diazabicyclo[2.2.2]octan (DABCO) and 22% (v/v) glycerol and analyzed by laser scanning confocal microscopy (LSM, Zeiss, Jena, Germany).
Apoptosis Assay and Flow Cytometry
For induction of apoptosis, HEK293 cells were seeded out in 6-well dishes and co-transfected with pKex-Apo-1 expressing the CD95 receptor together with plasmids encoding the HA-tagged Daxx-splice variants or the corresponding mock control pLHA-C1 (see above). Forty-eight hours after transfection cells were either incubated with 2 μg/ml CD95-agonistic antibody CH11 (Immunotech, Hamburg; Germany) or left untreated for an additional 12 h followed by assessment of apoptotic cells, which was performed by Annexin V staining. In brief, cells were washed twice with cold PBS and detached by incubation with PBS containing 0.05% (v/v) EDTA for 5 min at 37 °C. Then, cells were divided and 50% were collected by centrifugation and resuspended in binding buffer containing 10 mm HEPES (pH 7.4), 140 mm NaCl, and 2.5 mm CaCl2 (BD Bioscience) at a concentration of 1 × 106 cells/ml. A total of 1 × 105 cells in a final volume of 100 μl was mixed with 5 μl of Annexin V-phycoerythrin and 5 μl of 7-AAD staining solution (BD Bioscience) and incubated in the dark for 15 min at room temperature. Afterward, 400 μl of binding buffer was added, and cells were analyzed by flow cytometry using a FACSCalibur flow cytometer (BD Bioscience). Cells positively stained for Annexin V and 7-AAD were considered as apoptotic (late apoptosis). Data acquisition was performed with CellQuest software and WinMDI, respectively. In every experiment, a quantified percentage of apoptosis was referred to the corresponding transfection efficiency, which was determined separately by staining of the overexpressed CD95 receptor. Therefore, the remaining 50% of total cell amount was collected by centrifugation and resuspended in PBS containing 3% (v/v) FBS (FACS-buffer) at a concentration of 1 × 106 cells/ml. A total of 1 × 105 cells in a final volume of 100 μl was incubated with an FITC-conjugated anti-CD95 mAb antibody (dilution 1:100; Serotec, Düsseldorf, Germany) or the respective FITC-conjugated isotypic control (dilution 1:100; DakoCytomation, Hamburg; Germany) for 30 min at 4 °C in the dark. Then, cells were washed with FACS-buffer and analyzed by flow cytometry. Cells transfected with pLHA-C1 and without pKex-Apo-1 served as control to determine the endogenous level of CD95 receptor expression which was set to zero to detect percentage of overexpressed CD95 receptor in co-expression experiments.
Luciferase Reporter Assays
HeLa cells were seeded out in 6-well dishes and co-transfected with the 200 ng of p53-Luc, 50 ng of pcDNA-p53-GFP, and 400 ng of plasmids encoding HA-Daxx, HA-Daxx-β, HA-Daxx-γ, or the corresponding empty vector pLHA-C1, respectively, as described above. In all samples, 20 ng of Renilla luciferase-expressing reporter plasmid pRL-EF-1a was included to normalize transfection efficiencies. The total DNA amount for transfection was kept constant by adding pLHA-C1 to a final amount of 2 μg. After 48 h, cells were lysed and Renilla and firefly luciferase activity was monitored using the Dual Luciferase kit (Promega) according to the manufacturer's instructions.
RESULTS
Identification of Two Novel Daxx Splice Variants Daxx-β and Daxx-γ in Various Cell Lines
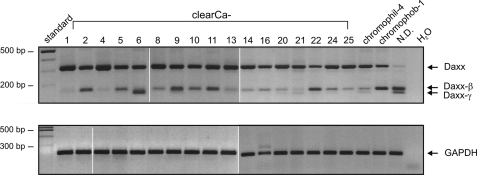
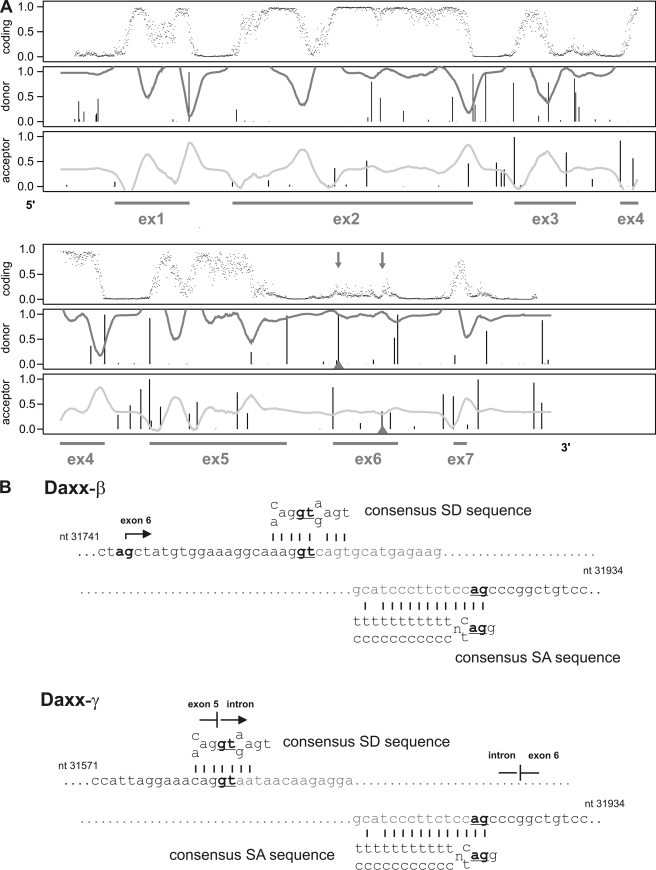
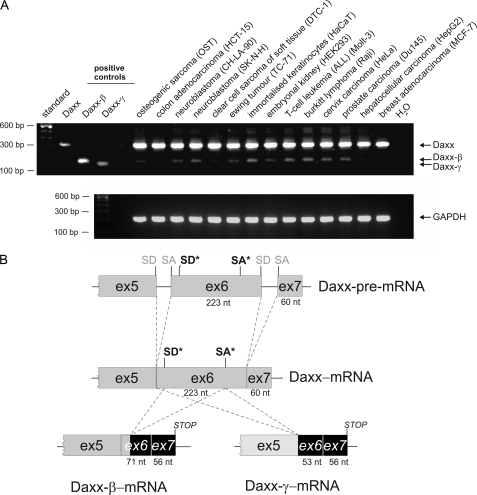
We investigated the role of Daxx in human carcinomas (Fig. 1A) and found that Daxx mRNA expression could be detected in all tested RCC cell lines. After DNA sequencing, all three amplification products could be identified as Daxx mRNA sequences. Comparing the respective PCR product sequence with genomic data the largest 316-bp amplification product could be referred to the regularly spliced form of Daxx. In contrast, the 164-bp fragment and the 146-bp fragment showed partial deletion of exon 6, with the smaller fragment lacking nucleotides 1–170 of exon 6 and the 164-bp fragment missing nucleotides 19–170 (GenBank accession no. AF015956). NetGene2 analysis of genomic Daxx sequence (GenBank accession no. Z97183) revealed that the deleted sequence in the 164-bp fragment is flanked by potential splice donor (SD) and splice acceptor (SA) sequences (Fig. 2A) that match to the consensus sequences of common SD sites {C/A}AG|GT{A/G}AGT and SA sites (T/C)11N{C/T}AG|G as shown in Fig. 2B. Thus, the mRNA sequence corresponding to the 164-bp fragment was identified as an alternatively spliced form of Daxx and termed Daxx-β. The smaller fragment was designated as Daxx-γ whereas usage of the regular SD site at the exon 5/intron boundary with the alternative SA in exon 6 leads to the lack of the entire first 170 nucleotides of exon 6. As shown in Fig. 3A many tested cell lines displayed expression of Daxx-β (6 of 14) whereas Daxx-γ mRNA could not be detected in any cell line. Because only four of 20 RCC cell lines revealed a Daxx-γ expression, this reflects the distribution pattern of the Daxx isoforms and identifies Daxx-β as more abundantly expressed than Daxx-γ.
FIGURE 1.
Expression of different Daxx mRNA variants in various RCC cell lines of all major histological types. Besides the expected 316-bp PCR amplification product two additional smaller fragments were generated. GAPDH PCRs were performed as control. Abbreviations: clearCa, clear cell renal carcinomas; chromophil, papillary/chromophilic renal cell carcinoma; chromophob, chromophobic renal cell carcinoma; N.D., the histological subtype of this RCC line is not determined.
FIGURE 2.
Detection of potential alternative splice donor (SD) and splice acceptor (SA) sites in exon 6 of the Daxx gene. A, NetGene2 analysis of the genomic Daxx sequence. The upper panel of the output predicts the coding region whereas values close to 0 indicate an intron region, and values close to 1 indicate exons. Prediction of SD and SA is given below with a confidence level of 90% (30, 31). The underlying exon progression of the Daxx gene is marked by the string (GenBank accession no. Z97183). B, comparison of the alternative SA and SD sites of exon 6 with the consensus sequences for SA and SD sites.
FIGURE 3.
Detection of the different Daxx mRNA transcripts in various human cell lines of different tissue origin. A, amplifications of plasmids coding for Daxx, Daxx-β, and Daxx-γ used as positive controls. GAPDH PCRs were performed for normalization. B, exon organization of the three Daxx splice variants. Regular SD and SA sites are marked in gray; alternative SD and SA sites used to generate Daxx-β and Daxx-γ are illustrated in black and marked with an asterisk.
Daxx-β and Daxx-γ Display a Truncated and Modified Regulatory C Terminus
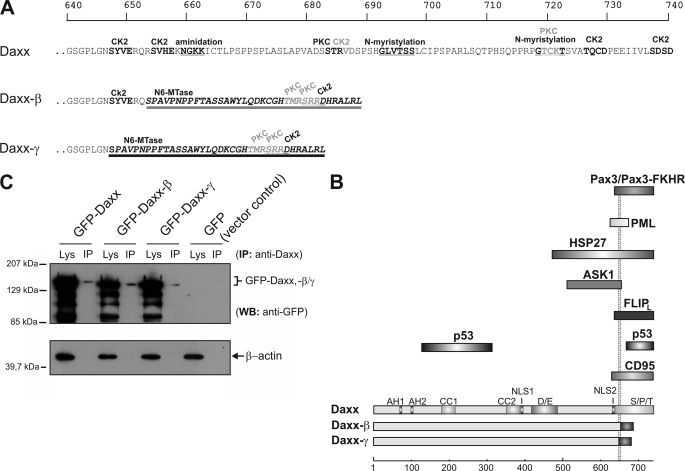
Alternative splicing of Daxx results in a frameshift in both transcripts. This is associated with the generation of a new stop codon that is located 4 nucleotides ahead of the regularly one used in Daxx translation (Fig. 3B). Together with the partial lack of exon 6, at the protein level in Daxx-β and Daxx-γ this leads to truncation and modification of the regulatory C terminus from amino acids 653 and 647, respectively. Compared with Daxx (740 amino acids) (2), Daxx-β consists of 688 amino acids with a calculated molecular mass of 76.3 kDa, and Daxx-γ consists of only 682 amino acid residues (75.6 kDa). According to the identical underlying frameshift, Daxx-β and Daxx-γ only differ in six amino acids at position 647–653. These are deleted in Daxx-γ due to the lack of the first 18 nucleotides of exon 6 in the corresponding mRNA transcript (Fig. 4A). Nevertheless, computational analysis of Daxx-β and Daxx-γ C-terminal protein sequences reveals differences in prediction of secondary structure elements. A PROSITE scan analysis of Daxx isoforms (34) showed that C-terminal modification of Daxx-β and Daxx-γ leads to new binding domains (cf. Fig. 4A). Because this region of Daxx is known to be responsible for binding to numerous proteins such as PML, CD95 receptor, Hsp27 or p53 (1, 7–10, 18, 19, 26) it was reasonable to assume that the modification/truncation of Daxx-β and Daxx-γ has functional consequences (Fig. 4B).
FIGURE 4.
Protein alterations of the novel Daxx isoforms. A, because of a frameshift as a consequence of the splicing event, C-terminal amino acids of the new splice variants are different from those of Daxx from amino acid residues at positions 647 (Daxx-γ, dark underlining) and 653 (Daxx-β, light underlining), respectively. Protein sequences were analyzed by PROSITE scan (34), and potential domains and modification sites are marked. Top scale resembles amino acid progression. B, schematic represents structural characteristics of the three Daxx isoforms. The respective domains are depicted in different shades of gray. Different C termini of Daxx-β/-γ are marked by dark shaded boxes. A subset of Daxx-interacting proteins together with the respective binding domain of Daxx is shown, and the two dotted lines indicate the different C termini of Daxx-β/-γ. Bottom scale represents amino acid progression. Abbreviations: CK2, casein kinase II phosphorylation site; PKC, protein kinase C phosphorylation site; N6-Mtase, N6-methyltransferase signature; Lys, crude cell extract lysate; AH, paired amphipathic helices; CC, coiled-coil regions; D/E, acid-rich domain; NLS, nuclear localization signal. C, recombinant expression of GFP-fused Daxx isoforms in HepG2 cells. GFP immunoprecipitation from vector control was performed and ran at 28 kDa (data not shown). WB, Western blotting.
Decreased Affinity to PML Affects Subcellular Localization of Daxx-β and Daxx-γ
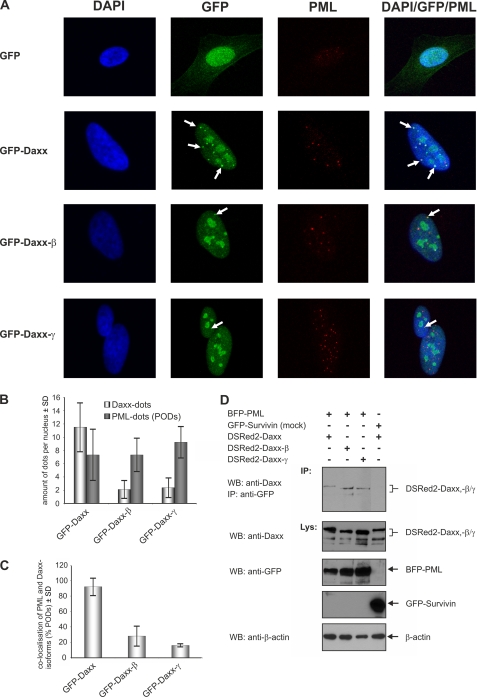
All Daxx isoforms displayed a marked expression of different products with the major ones being identified by immunoprecipitations (cf. Fig. 4C). In contrast to GFP-Daxx, which was located mainly in discrete speckled nuclear dots that are presumably the PODs (7–10), in GFP-Daxx-β- and GFP-Daxx-γ-transfected cells comparable structures were rather underrepresented, and both proteins accumulated predominantly in more patchy nuclear structures (Fig. 5A). These structures resemble the distribution pattern observed for Daxx in PML−/− cells, which is assumed to be the consequence of POD disruption (8). Because in Daxx-β and Daxx-γ the C-terminal 625–740 amino acids of Daxx essential for PML binding are partly truncated/modified, we asked whether an impaired PML interaction is responsible for the different distribution pattern of Daxx-β and Daxx-γ compared with Daxx. Therefore, overexpression of GFP-fused Daxx isoforms combined with immunostaining of endogenous PML was performed in HeLa cells. As shown in Fig. 5A, the majority of GFP-Daxx dots could be identified as PODs because they co-localize perfectly with endogenous PML. Although not totally interrupted, such co-localization was reduced dramatically between PML and GFP-Daxx-β as well as PML and GFP-Daxx-γ. Determination of overall POD quantity revealed that POD formation was not influenced by overexpression of either of the respective Daxx isoforms, thereby excluding minor POD assembling as being responsible for the reduced co-localization between PML and GFP-Daxx-β and GFP-Daxx-γ (Fig. 5B). Consequently, the percentage of PML co-localization of GFP-Daxx-β and GFP-Daxx-γ was markedly decreased (Fig. 5C). To confirm the results obtained by confocal microscopy, co-immunoprecipitation analyses were performed (cf. Fig. 5D). Interestingly, the pure physical interaction of Daxx variants by immunoprecipitation shows increasing binding signals. This fact indicates that co-localization-stabilizing factors in POD complexes are more determining of the binding ability from Daxx to PML in vivo. Together, these findings indicate that the C-terminal truncation/modification of Daxx-β and Daxx-γ leads to a reduced in vivo affinity to PML which is associated with a decreased recruitment of Daxx-β and Daxx-γ into PODs.
FIGURE 5.
Subcellular localization of the Daxx isoforms. A, vectors encoding GFP-Daxx/-β/-γ as well as the corresponding empty vector transfected into HeLa cells. Representative confocal pictures are shown as single and overlay fluorescence images. White arrows indicate co-localization of the respective GFP-fused Daxx isoform with endogenous PML in the PODs. B, quantitative analysis of the amount of Daxx dots and PODs per nucleus of the transfected cells. C, percentage of co-localization between the Daxx isoforms and PML which was calculated as the ratio of co-localization signals and total number of PODs per nucleus. The fact that the cellular number of PODs may vary during the cell cycle (43) is presumably responsible for the relatively high SD level. D, co-immunoprecipitation of Daxx isoforms with PML. HEK293 cells were transfected with expression plasmids coding for the respective DSRed2-fused Daxx isoform and blue fluorescent protein-PML. Co-transfection of vectors encoding GFP-tagged Survivin and DSRed2-Daxx was serving as control.
Neither Daxx nor Daxx-β or Daxx-γ Is an Enhancer of CD95-mediated Apoptosis in HEK293 Cells
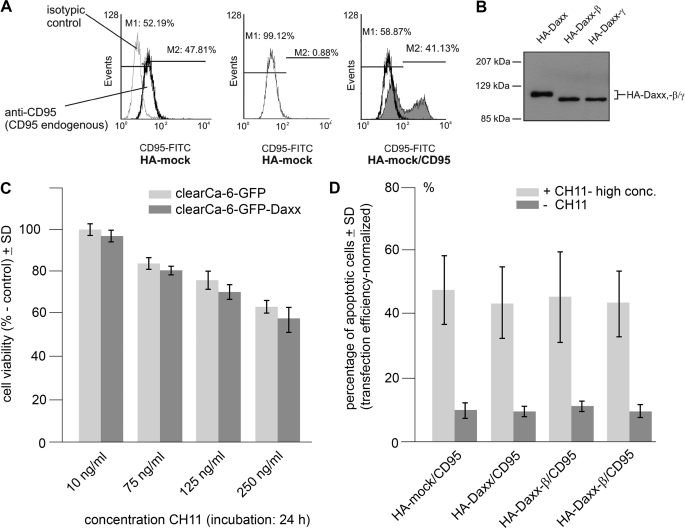
To investigate the apoptosis-promoting potential of Daxx-β and Daxx-γ, co-expression of CD95 receptor with the HA-tagged Daxx, -Daxx-β, or -Daxx-γ was performed in HEK293 cells. Activation of CD95 in the absence of the corresponding ligand occurs due to the overexpression-based close proximity of receptor molecules. This leads to the multimerization of death domains followed by apoptosis induction (1, 27). Apoptosis was monitored by Annexin-V staining, whereas comparable expression was determined by staining of overexpressed CD95 receptor and Western blotting (Fig. 6, A and B). As shown, neither co-expression of HA-Daxx nor the co-expression of HA-Daxx-β or HA-Daxx-γ significantly enhances CD95-mediated apoptosis (Fig. 6C). Nevertheless, no modulation of CD95-mediated apoptosis by either of the Daxx isoforms became obvious (Fig. 6D). We tested different concentrations of CH-11 (up to 2 μg/ml to overexcite the stimulation, shown in Fig. 6D) for 12 h without any significant alteration.
FIGURE 6.
Daxx isoforms do not enhance CD95-mediated apoptosis. HEK293 cells were co-transfected with vectors encoding CD95 and HA-tagged Daxx, Daxx-β, or Daxx-γ. Co-expression of CD95 with empty HA-vector was used as control. A, detection of CD95 overexpression by flow cytometry analysis. Endogenous CD95 expression level of HA-mock-transfected HEK293 cells was analyzed using FITC-labeled anti-CD95 antibody and the corresponding FITC-labeled isotypic control (left panel). Then, the endogenous level of CD95 expression was set to 0 (center panel) to detect the overexpressed amount of CD95 (right panel) in the following experiments, thereby reflecting the respective transfection efficiency. Representative images are shown. B, Western blot analysis demonstrating overexpression of HA-tagged Daxx isoforms. C, cell viability after CD95 induction of GFP and GFP-Daxx-expressing cells at low CH11 concentrations (10–250 ng/ml) in 24 h (mock, light gray; GFP-Daxx, dark gray). D, percentage of apoptotic cells after co-expression of CD95 with HA-Daxx, HA-Daxx-β, HA-Daxx-γ, or empty vector serving as control (dark gray, ± S.D. (error bars); n = 5), and additional stimulation of expressed CD95 receptor by incubation with 2 μg/ml CD95 agonistic antibody CH11 (light gray, ± S.D.; n = 4).
Moreover, by co-immunoprecipitations we were unable to detect any binding of Daxx to the CD95 receptor (data not shown), thereby corroborating previous results made by Hollenbach et al. (2), who could not detect an association between Daxx and CD95 receptor nor any CD95-apoptosis enhancing potential of Daxx. In conclusion, these data suggest that Daxx, Daxx-β, and Daxx-γ overexpression does not sensitize HEK293 cells to CD95-mediated apoptosis.
Daxx-β and Daxx-γ Display a Markedly Reduced Affinity to p53 and Are Unable to Recruit p53 into PODs
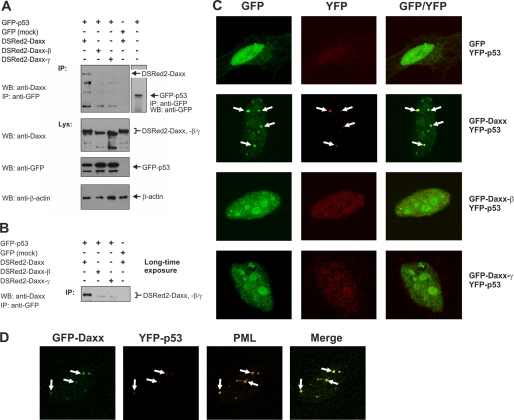
In addition to its already reported function as apoptosis regulator, Daxx is implicated in transcriptional control (2, 15–17). Via the C-terminal S/P/T domain Daxx physically interacts with p53, thereby repressing transcriptional activity of p53 (18–20). Because this domain is modified in Daxx-β and Daxx-γ (Fig. 4B), we tested whether Daxx-β and Daxx-γ are still able to bind p53. Thus, HEK293 cells were transiently co-transfected with plasmids encoding p53-GFP and DSRed2-fused Daxx variants. As expected, immunoprecipitation with an anti-GFP antibody and subsequent Western blot analysis with an anti-Daxx antibody revealed a direct interaction between DSRed2-Daxx and GFP-p53 (Fig. 7A). In contrast, DSRed2-Daxx-β and DSRed2-Daxx-γ could not be detected in the corresponding anti-GFP precipitates, suggesting that the C-terminal modification of these Daxx isoforms is associated with an abrogated p53 interaction (Fig. 7A). However, after a long exposure of the respective membrane, a weak Daxx-β and Daxx-γ signal became visible, indicating that a p53 interaction of Daxx-β and Daxx-γ, although dramatically reduced, was not totally absent (Fig. 7B). This is consistent with recent observations that N-terminal parts of Daxx could also be implicated in binding to p53 (19, 20). After co-expression of Daxx with empty GFP vector, which served as negative control, even with longer exposure no Daxx protein could be detected in the corresponding precipitate. Therefore, these differential p53 interactions of the Daxx splice variants were considered to be specific. To confirm this observation further, HeLa cells co-expressing YFP-p53- and GFP-tagged Daxx isoforms were analyzed by immunofluorescence and confocal microscopy. Consistent with the results obtained by immunoprecipitation, GFP-Daxx and YFP-p53 displayed a co-localization pattern. Here, YFP-p53 is recruited to the Daxx dots, which in the majority were identified as PODs (Fig. 7C). In contrast, after co-expression with GFP-Daxx-β or GFP-Daxx-γ as well as together with unfused GFP serving as control, YFP-p53 yielded a diffuse nuclear distribution pattern and no comparable dot-like accumulation as seen for Daxx co-expression (Fig. 7C). Moreover, co-localization of YFP-p53 with the patchy-like accumulations of GFP-Daxx-β and GFP-Daxx-γ was totally underrepresented, thereby corroborating with the co-immunoprecipitation data. Using co-expression of YFP-p53 and GFP-Daxx combined with immunostaining of endogenous PML, the dot-like co-accumulations of YFP-p53 and GFP-Daxx clearly could be identified as PODs containing PML, p53, and Daxx (Fig. 7D). This further supports previous reports showing that PML targets p53 into PODs (28, 29). Taken together, these results suggest that the C-terminal truncation/modification of Daxx-β/-γ leads to a strongly impaired p53 interaction of these proteins. Furthermore, the differences in subnuclear p53 localization indicate a Daxx isoform-dependent recruitment of p53 into PODs: The indirect binding of p53 to PML is probably mediated by a piggy-back mechanism through the direct binding of Daxx to PML. As expected, Daxx-β and Daxx-γ failed to recruit p53 to the PODs.
FIGURE 7.
Daxx isoforms display different p53 interactions. A, co-immunoprecipitation (IP) analysis of DSRed2-fused Daxx isoforms with p53-GFP. HEK293 cells were transiently transfected with equal amounts of expression vectors coding for p53-GFP and DSRed2-Daxx, DSRed2-Daxx-β, or DSRed2-Daxx-γ. The immunoprecipitated GFP-p53 was detected by Western blotting (WB) using anti-GFP antibody (right panel). Co-expression of GFP and DSRed2-Daxx was performed as control. B, long time exposure of respective Western blot membrane described in A. C, confocal fluorescence analysis of YFP-p53 and GFP-tagged Daxx isoforms. HeLa cells were transiently transfected with vectors encoding YFP-p53 and GFP-Daxx, GFP-Daxx-β, or GFP-Daxx-γ, respectively. Co-expression of GFP with YFP-p53 served as control. D, co-localization of GFP-Daxx with YFP-p53 in PODs. HeLa cells were transiently transfected with GFP-Daxx and YFP-p53 expression constructs. Representative images of confocal fluorescence microscopy analyses are show as single and overlay fluorescence signals. Comparing YFP-p53/GFP-Daxx co-localization spots with PML distribution pattern identified these nuclear regions as PODs.
Daxx-β and Daxx-γ Are Unable to Repress p53-mediated Transcription
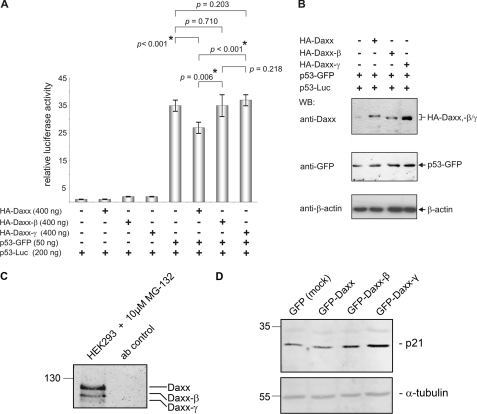
To investigate whether the reduced p53-interacting potential of Daxx-β/-γ also have functional consequences, we performed luciferase assays to analyze repressor activity of Daxx-β and Daxx-γ on p53-dependent transcription. The co-expression of p53-GFP with HA-Daxx leads to a significant decrease of p53-dependent transcription as indicated by a p value of <0.05 (Fig. 8A). In the corresponding co-expression with HA-Daxx-β or HA-Daxx-γ no decrease in transcriptional activity of p53 became obvious, although Western blot analysis demonstrated comparable amounts of overexpressed p53-GFP as well as HA-Daxx, HA-Daxx-β, and HA-Daxx-γ (Fig. 8B). The transcriptional p53 activity detected in these samples was quite similar to that observed by co-expression of the vector control indicating that both Daxx-β and Daxx-γ are unable to repress p53-mediated transcription. Similarly, even in the corresponding controls without ectopically expressed p53-GFP comparable with HA-Daxx overexpression the expression of HA-Daxx-β and HA-Daxx-γ leads to remarkably higher luciferase activity (p53 activity) which was probably mediated by the differential interaction of the Daxx isoforms with endogenous p53 protein. These results therefore suggest that the impaired p53 binding of Daxx-β and Daxx-γ also have functional consequences and is associated with lost repressor activity for p53-dependent transcription. Therefore, we analyzed p21 expression in Daxx variant-overexpressing cells by Western blotting. As shown in Fig. 8D, the p21 protein level increases in Daxx-β/-γ transfected cells, indicating loss or transcriptional repression. Moreover, we observed that the expression of the p53-regulated genes DR5 and Bax is activated by Daxx-β and Daxx-γ at the protein level (data not shown). To highlight the biological significance of our findings, we confirmed the endogenous expression of Daxx-β/-γ (Fig. 8C). Altogether, these data reveal the splicing of Daxx as a new additional level in complexity of p53 regulation.
FIGURE 8.
Daxx-β and Daxx-γ are unable to repress p53-dependent transcription. A, HeLa cells were transiently transfected with p53-GFP expression vector together with a reporter construct expressing luciferase under the control of a p53-responsive promoter and vectors encoding HA-Daxx, HA-Daxx-β, HA-Daxx-γ, or the corresponding empty vector. Equivalent experiments without additionally overexpressing p53-GFP were used as controls. To normalize transfection efficiency, a stable amount of pRL-F1a reporter plasmid was included in every sample (± S.D. (error bars); n = 6). B, to verify comparable expression levels of the respective proteins, aliquots of each sample were analyzed by Western blotting (WB) using anti-Daxx and anti-GFP antibodies. Comparable amounts of protein were confirmed by detection of β-actin. C, confirmation of the existence of the endogenous Daxx variants in cells on protein level. HCT-15 cells were treated with 10 μm MG-132 proteasome inhibitor 2 h before extraction. 1 mg of HCT-15 protein extracts were immunoprecipitated with anti-Daxx monoclonal antibody in the presence of MG-132, separated, and after Western blotting detected with polyclonal anti-Daxx antibody (clone 25C12; Cell Signaling). D, protein expression analysis of p21 as a downstream activated gene of p53 by Western blotting. All three Daxx variants were stably overexpressed in HCT-15 cells. As shown, the Daxx-β- and Daxx-γ-overexpressing cells have p21 up-regulated. Comparable amounts of protein were confirmed by detection of α-tubulin.
DISCUSSION
Alternative splicing has been found to play a key role in the regulation of apoptosis by determining the action of an increasing number of apoptosis-related genes such as coding for CD95 (30), Bcl-X (31), Smac/DIABLO (32), Survivin (33), FLIP (34), as well as some caspases. The role of the basal splicing machinery in regulating alternative splicing remains poorly understood. Recent findings demonstrate that the DNA damage pathway is involved in alternative splicing of Bcl-X (35), whereby the splicing shift requires the activation of certain protein tyrosine phosphatases.
In the present study we report on the identification and characterization of two novel splice variants of the apoptosis-regulator Daxx, designated Daxx-β and Daxx-γ, which are functionally different from Daxx concerning interaction with PML, subcellular localization, and p53-regulating properties. Therefore, Daxx acts as an additional cell death regulator which could also be controlled on level of alternative splicing. Alternative splicing is a tissue-specific phenomenon. The possible causes of different splicing patterns of Daxx-β and Daxx-γ remain unclear; probably they are related to different distribution of tissue-specific splicing factors or the activation state of the cell (35).
Daxx is ubiquitously expressed and involved in several apoptotic pathways, including cell death triggered by CD95 (1) and TGF-β (36). Daxx was initially identified as a pro-apoptotic molecule that interacts with CD95, thereby enhancing CD95-mediated apoptosis. Recent studies, however, reveal that depletion of Daxx by RNAi is associated with an elevated level of basic apoptosis as well as an increased sensitivity to stress and CD95-induced apoptosis (1, 4, 12, 37). Taking this into account, the exact role of Daxx in programmed cell death is still a matter of debate. Moreover, Daxx is a transcriptional co-regulator that binds to several transcription factors including p53, and by repressing p53-dependent transcription Daxx is involved in p53-mediated apoptosis (18–20). In addition, emerging evidence suggests Daxx to be implicated in the pathogenesis of several human malignancies and neurodegenerative disorders (2, 38).
Although Daxx-β and Daxx-γ are also nuclear proteins with both nuclear localization signal-encoding sequence regions being preserved during alternative splicing, Daxx isoforms differ in their subnuclear localization. Whereas Daxx accumulates mainly in PODs, confirming previous observations that Daxx binds to SUMO-1-modified PML via amino acid residues 625–661 (7, 8), Daxx-β and Daxx-γ display a markedly reduced co-localization with PML in PODs. Because the overall number of PODs was comparable in Daxx-, Daxx-β-, or Daxx-γ-overexpressing cells, this could not be related to a reduced POD formation, indicating that the truncated PML-interacting domain of Daxx-β and Daxx-γ strongly affects affinity to PML. However, binding to PML is not totally abrogated in Daxx-β and Daxx-γ. Nevertheless, the reduced PML binding is associated with a drastically decreased sequestration of Daxx-β and Daxx-γ to PODs so that these Daxx isoforms accumulate predominantly in alternative more patchy-like nuclear structures. According to the observations made by Ishov et al. (7) and Li et al. (8) showing that Daxx is located at condensed chromatin in cells lacking detectable PML, our data suggest that the nuclear accumulations of Daxx-β and Daxx-γ could also be referred to sites of heterochromatin where these proteins assemble as a result of the insufficient PML-binding-dependent recruitment to PODs. It was found that Daxx exhibits a cell cycle-dependent distribution pattern. In the late S phase it associates with condensed chromatin in a complex including chromatin-remodeling protein ATRX (13). This presumably explains why comparable patchy-like accumulations of Daxx could also be observed, although to a much lesser extent.
Co-localization with PML appears to correlate with the pro-apoptotic activity of Daxx because mutants that failed to localize to PODs have lost their apoptosis-promoting potential (9, 10). Moreover, the C-terminal domain of Daxx (modified in Daxx-β/-γ) is thought to mediate interaction with CD95, thereby enhancing CD95-dependent apoptosis via activation of the ASK1/JNK pathway (1, 4–6). Therefore, we analyzed whether the CD95 apoptosis-promoting properties of Daxx-β/-γ differed from those mediated by Daxx. Using co-transfection-based experiments, neither Daxx nor Daxx-β or Daxx-γ co-expression with CD95 specifically enhanced or modulated CD95-mediated apoptosis in HEK293 cells. A potential dominant negative effect of Daxx-β or Daxx-γ by interfering with endogenous Daxx was not detected. However, cell type-specific effects, e.g. the expression levels of reported Daxx inhibitors such as Hsp27 (26) and FLIPL (39) as well as yet unknown regulators to be responsible for the discrepancy with previous observations identifying Daxx as a CD95-dependent apoptosis promoting molecule cannot totally be excluded.
The C-terminal domain of Daxx, specifically amino acids 667–740, is essential for binding to p53 with subsequent repression of p53-mediated transcription (18, 19). Consistent with these data we could detect a direct association of Daxx and p53. In contrast, Daxx-β and Daxx-γ lacking this domain displayed a strongly diminished interaction with p53. However, analogous to the reduced interaction with PML, the binding of Daxx-β/-γ to p53 was not totally absent which confirms prior observations that N-terminal parts of Daxx could be also implicated in p53 binding (19, 20). This interaction (Daxx/p53) is accompanied with a recruitment of p53 to PODs, thereby corroborating recent reports that p53 co-localizes with PML in PODs and that Daxx display a co-localization pattern with p53 in subnuclear dot-like structures (19, 28, 29). We clearly demonstrated for the first time that these structures are in effect PODS containing Daxx and p53 co-localized to PML. Interestingly, during co-expression of p53 with Daxx-β or -γ, p53 was dispersed throughout the nucleus, and no dot-like p53 accumulations became obvious. Because this effect was not due to a generally reduced POD formation (see above), this indicates an underlying Daxx isoform-specific effect. In contrast to previous suggestions that PML may directly target p53 into PODs (28, 29) our data therefore point to Daxx as being responsible for this recruitment because Daxx-β and Daxx-γ failed to direct p53 into PODs. According to these observations a piggy-back mechanism for the Daxx-mediated p53 targeting into PODs could be postulated with p53 being indirectly recruited to PODs via directly interacting with Daxx which is bound to PML. Consistently, the reduced binding affinity of Daxx-β and Daxx-γ to p53 and PML could explain the impaired capacity of these isoforms to guide p53 into PODs. The association of Daxx with p53 results in repression of p53 transcriptional activity (18–20), and its localization to PODs has been shown to be crucial for p53 regulation (28, 29, 40–42). Our data indicate that Daxx, Daxx-β, and Daxx-γ may exert different impact on p53 signaling. In fact, using luciferase reporter assays we showed that in agreement with recent findings Daxx was able to repress p53-mediated transcription (18–20). In contrast, the diminished PML/p53 binding of Daxx-β and Daxx-γ was associated with a loss of the potential to suppress p53-dependent transcription. In the same way Daxx-β/-γ-overexpressing cells are not able to attenuate p21 transcription and consecutive p21 protein expression. This demonstrates that Daxx isoforms have different p53-regulating properties. Therefore, it is conceivable that Daxx-β and Daxx-γ may play a role in other p53-regulated biological events such as the regulation of the DNA damage response by inducing cell cycle arrest and DNA repair or when the damage is severe, the activation of death receptors (DR5, CD95) and other apoptotic factors (Bax). Gostissa et al. (18) showed that the Daxx-mediated transcriptional repression of p53 triggers p53-dependent apoptosis by influencing the balance between transcription of cell cycle and pro-apoptotic-related target genes. Indeed, overexpression of Daxx sensitizes U2OS cells to treatment with cisplatin (18), and MCF-7 and Jurkat cells became more sensitive to topotecan and doxorubicin treatment. This further suggests that Daxx-β/-γ may also have lost the p53 apoptosis-promoting potential. Generally, mRNAs of different isoforms originate from the same pre-mRNA precursor pool. This would implicate that splicing of Daxx effectively reduces cellular susceptibility to p53-mediated apoptosis. Thus, it appears reasonably that splicing of Daxx is a putative cellular mechanism to acquire/increase apoptosis resistance.
Analysis of the Daxx variants is accompanied by some difficulties. Rising antibodies against the novel C terminus failed to recognize Daxx variants by Western blotting even in a Daxx-β/-γ-overexpressed control cell line (data not shown), indicating an inappropriate antigenic determinant. Moreover, loss-of-function experiments by siRNA are quite difficult because the truncated Daxx splice variants generated by exon deletion contain the same sequences as the regular Daxx variant. Therefore, an adequate design of oligonucleotide probes to distinguish between the different Daxx variants is not viable.
The findings reported here identify Daxx as an additional apoptosis-related gene that could be regulated on level of alternative splicing thereby generating Daxx isoforms with different potential in repressing p53-mediated transcription. According to the complex p53 regulatory network, splicing of Daxx, as a regulator of the p53-regulated/dependent gene expression, indicates an additional level in the regulation of the cellular signal transduction system.
Acknowledgements
We thank Drs. P. H. Krammer, I. Schmitz, P. P. Pandolfi, F. Essmann, R. H. Stauber and V. Kolb-Bachhofen for providing expression constructs; Drs. C. Poremba, K. L. Schaefer, and V. Kolb-Bachhofen for providing cell lines; Drs. C. Homberg and C. Wiek for retroviral transfection; and M. Krahnke-Schoelzel for excellent technical assistance.
This work was supported in part by a grant from the Dr. Mildred-Scheel-Stiftung für Krebsforschung.
- PML
- promyelocytic leukemia protein
- FACS
- fluorescence-activated cell sorter
- HIPK
- homeodomain-interacting protein kinase
- POD
- PML-oncogenic domain
- RCC
- renal cell carcinoma
- SA
- splice acceptor
- SD
- splice donor
- TRITC
- tetramethylrhodamine isothiocyanate.
REFERENCES
- 1. Yang X., Khosravi-Far R., Chang H. Y., Baltimore D. (1997) Cell 89, 1067–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hollenbach A. D., Sublett J. E., McPherson C. J., Grosveld G. (1999) EMBO J. 18, 3702–3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kiriakidou M., Driscoll D. A., Lopez-Guisa J. M., Strauss J. F., 3rd (1997) DNA Cell Biol. 16,1289–1298 [DOI] [PubMed] [Google Scholar]
- 4. Chang H. Y., Nishitoh H., Yang X., Ichijo H., Baltimore D. (1998) Science 281, 1860–1863 [DOI] [PubMed] [Google Scholar]
- 5. Chang H. Y., Yang X., Baltimore D. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 1252–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ko Y. G., Kang Y. S., Park H., Seol W., Kim J., Kim T., Park H. S., Choi E. J., Kim S. (2001) J. Biol. Chem. 276, 39103–39106 [DOI] [PubMed] [Google Scholar]
- 7. Ishov A. M., Sotnikov A. G., Negorev D., Vladimirova O. V., Neff N., Kamitani T., Yeh E. T., Strauss J. F., 3rd, Maul G. G. (1999) J. Cell Biol. 147, 221–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li H., Leo C., Zhu J., Wu X., O'Neil J., Park E. J., Chen J. D. (2000) Mol. Cell. Biol. 20, 1784–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Torii S., Egan D. A., Evans R. A., Reed J. C. (1999) EMBO J. 18, 6037–6049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhong S., Salomoni P., Ronchetti S., Guo A., Ruggero D., Pandolfi P. P. (2000) J. Exp. Med. 191, 631–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen L. Y., Chen J. D. (2003) Mol. Cell. Biol. 23, 7108–7121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Michaelson J. S., Leder P. (2003) J. Cell Sci. 116, 345–352 [DOI] [PubMed] [Google Scholar]
- 13. Ishov A. M., Vladimirova O. V., Maul G. G. (2004) J. Cell Sci. 117, 3807–3820 [DOI] [PubMed] [Google Scholar]
- 14. Michaelson J. S., Bader D., Kuo F., Kozak C., Leder P. (1999) Genes Dev. 13, 1918–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li R., Pei H., Watson D. K., Papas T. S. (2000) Oncogene 19, 745–753 [DOI] [PubMed] [Google Scholar]
- 16. Emelyanov A. V., Kovac C. R., Sepulveda M. A., Birshtein B. K. (2002) J. Biol. Chem. 277, 11156–11164 [DOI] [PubMed] [Google Scholar]
- 17. Lehembre F., Müller S., Pandolfi P. P., Dejean A. (2001) Oncogene 20, 1–9 [DOI] [PubMed] [Google Scholar]
- 18. Gostissa M., Morelli M., Mantovani F., Guida E., Piazza S., Collavin L., Brancolini C., Schneider C., Del Sal G. (2004) J. Biol. Chem. 279, 48013–48023 [DOI] [PubMed] [Google Scholar]
- 19. Kim E. J., Park J. S., Um S. J. (2003) Nucleic Acids Res. 31, 5356–5367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhao L. Y., Liu J., Sidhu G. S., Niu Y., Liu Y., Wang R., Liao D. (2004) J. Biol. Chem. 279, 50566–50579 [DOI] [PubMed] [Google Scholar]
- 21. Lin D. Y., Lai M. Z., Ann D. K., Shih H. M. (2003) J. Biol. Chem. 278, 15958–15965 [DOI] [PubMed] [Google Scholar]
- 22. Ecsedy J. A., Michaelson J. S., Leder P. (2003) Mol. Cell. Biol. 23, 950–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li Q., Wang X., Wu X., Rui Y., Liu W., Wang J., Wang X., Liou Y. C., Ye Z., Lin S. C. (2007) Cancer Res. 67, 66–74 [DOI] [PubMed] [Google Scholar]
- 24. Song M. S., Song S. J., Kim S. Y., Oh H. J., Lim D. S. (2008) EMBO J. 27, 1863–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wethkamp N., Klempnauer K. H. (2009) J. Biol. Chem. 284, 28783–28794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Charette S. J., Lavoie J. N., Lambert H., Landry J. (2000) Mol. Cell. Biol. 20, 7602–7612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boldin M. P., Mett I. L., Varfolomeev E. E., Chumakov I., Shemer-Avni Y., Camonis J. H., Wallach D. (1995) J. Biol. Chem. 270, 387–391 [DOI] [PubMed] [Google Scholar]
- 28. Fogal V., Gostissa M., Sandy P., Zacchi P., Sternsdorf T., Jensen K., Pandolfi P. P., Will H., Schneider C., Del Sal G. (2000) EMBO J. 19, 6185–6195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guo A., Salomoni P., Luo J., Shih A., Zhong S., Gu W., Pandolfi P. P. (2000) Nat. Cell Biol. 2, 730–736 [DOI] [PubMed] [Google Scholar]
- 30. Hughes D. P., Crispe I. N. (1995) J. Exp. Med. 182, 1395–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Akgul C., Moulding D. A., Edwards S. W. (2004) Cell. Mol. Life Sci. 61, 2189–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fu J., Jin Y., Arend L. J. (2003) J. Biol. Chem. 278, 52660–52672 [DOI] [PubMed] [Google Scholar]
- 33. Mahotka C., Wenzel M., Springer E., Gabbert H. E., Gerharz C. D. (1999) Cancer Res. 59, 6097–6102 [PubMed] [Google Scholar]
- 34. Golks A., Brenner D., Fritsch C., Krammer P. H., Lavrik I. N. (2005) J. Biol. Chem. 280, 14507–14513 [DOI] [PubMed] [Google Scholar]
- 35. Shkreta L., Michelle L., Toutant J., Tremblay M. L., Chabot B. (2011) J. Biol. Chem. 286, 331–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Perlman R., Schiemann W. P., Brooks M. W., Lodish H. F., Weinberg R. A. (2001) Nat. Cell Biol. 3, 708–714 [DOI] [PubMed] [Google Scholar]
- 37. Salomoni P., Guernah I., Pandolfi P. P. (2006) Cell Death Differ. 13, 672–675 [DOI] [PubMed] [Google Scholar]
- 38. Tzeng S. L., Cheng Y. W., Li C. H., Lin Y. S., Hsu H. C., Kang J. J. (2006) J. Biol. Chem. 281, 15405–15411 [DOI] [PubMed] [Google Scholar]
- 39. Kim Y. Y., Park B. J., Seo G. J., Lim J. Y., Lee S. M., Kimm K. C., Park C., Kim J., Park S. I. (2003) Biochem. Biophys. Res. Commun. 312, 426–433 [DOI] [PubMed] [Google Scholar]
- 40. Bernardi R., Pandolfi P. P. (2003) Oncogene 22, 9048–9057 [DOI] [PubMed] [Google Scholar]
- 41. Hofmann T. G., Will H. (2003) Cell Death Differ. 10, 1290–1299 [DOI] [PubMed] [Google Scholar]
- 42. Pearson M., Carbone R., Sebastiani C., Cioce M., Fagioli M., Saito S., Higashimoto Y., Appella E., Minucci S., Pandolfi P. P., Pelicci P. G. (2000) Nature 406, 207–210 [DOI] [PubMed] [Google Scholar]
- 43. Doucas V., Evans R. M. (1996) Biochim. Biophys. Acta 1288, M25–29 [DOI] [PubMed] [Google Scholar]