Abstract
Transmembrane adaptor proteins (TRAPs) are important organizers and regulators of immunoreceptor-mediated signaling. A bioinformatic search revealed several potential novel TRAPs, including a highly conserved protein, proline rich 7 (PRR7), previously described as a component of the PSD-95/N-methyl-d-aspartate receptor protein complex in postsynaptic densities (PSD) of rat neurons. Our data demonstrate that PRR7 is weakly expressed in other tissues but is readily up-regulated in activated human peripheral blood lymphocytes. Transient overexpression of PRR7 in Jurkat T cell line led to gradual apoptotic death dependent on the WW domain binding motif surrounding Tyr-166 in the intracellular part of PRR7. To circumvent the pro-apoptotic effect of PRR7, we generated Jurkat clones with inducible expression of PRR7 (J-iPRR7). In these cells acute induction of PRR7 expression had a dual effect. It resulted in up-regulation of the transcription factor c-Jun and the activation marker CD69 as well as enhanced production of IL-2 after phorbol 12-myristate 13-acetate (PMA) and ionomycin treatment. On the other hand, expression of PRR7 inhibited general tyrosine phosphorylation and calcium influx after T cell receptor cross-linking by antibodies. Moreover, we found PRR7 constitutively tyrosine-phosphorylated and associated with Src. Collectively, these data indicate that PRR7 is a potential regulator of signaling and apoptosis in activated T cells.
Keywords: Adaptor Proteins, Cell Death, Signal Transduction, Src, T Cell Receptor
Introduction
Activation of leukocytes through multichain immunoreceptors (TCR,3 BCR, FcRs) involves a complex array of membrane-linked and cytoplasmic proteins, including transmembrane and cytoplasmic adaptor proteins (1). Several of the transmembrane adaptors (TRAPs) are associated with lipid rafts, namely LAT (2, 3), PAG (4, 5), NTAL (6, 7), and LIME (8, 9), whereas others (LAX, SIT, TRIM, GAPT) are present in non-raft membrane (10–13). All of these proteins are composed of a short N-terminal extracellular peptide, a single transmembrane segment, and a cytoplasmic part containing multiple tyrosine and other protein interaction motifs. In the case of the raft-associated proteins, a palmitoylation motif (CXXC or CXC) lies between the transmembrane segment and the intracellular part (14). TRAPs play positive and negative regulatory roles in immunoreceptor signaling. The most prominent role is played by LAT, which is essential for several aspects of TCR signaling and plays an important role in the immunoreceptor signaling in other leukocyte subsets (15). In addition, experiments on genetic models have assigned important regulatory functions to other members of TRAP family, including NTAL, LAX, TRIM, and SIT (16, 17).
In an effort to discover additional TRAPs of functional importance, we performed an in silico search for proteins possessing the features characteristic of known TRAPs. Among 149 candidate genes, one of the best hits was PRR7. Previously, PRR7 was identified in the PSD fraction of rat forebrain tissue by a proteomic approach (18). It was shown to associate with PSD-95 (via a PDZ domain binding motif) and was found in a protein complex containing the N-methyl-d-aspartate receptor subunits NR1 and NR2B. These biochemical data suggested that PRR7 could be potentially involved in modulation of neural activities.
In the present study we found that PRR7 is weakly expressed in many other tissues, including immune cells, and is up-regulated during T cell activation. Moreover, we show that PRR7 overexpression in Jurkat cells substantially affected TCR signaling and cell survival.
EXPERIMENTAL PROCEDURES
Bioinformatic Search
The in silico search in human genome (USCS build hg16) for proteins possessing the features characteristic of known TRAPs was done with following parameters: a short extracellular N-terminal sequence, a single hydrophobic sequence starting at amino acids (aa) 5–50 (prediction using TMHMM Version 2.0) (19), a palmitoylation motif at aa 20–60 (CXC or CXXC), tyrosine-based phosphorylation motifs (YXX(I/L/V/A)) and/or C-terminal Group I PDZ binding motifs ((S/T)X(L/V)).
RNA and RT-qPCR
RNA was isolated using a Mini RNA purification kit (Zymo Research, Orange, CA) or TRI Reagent RT (MRC, Cincinnati, OH). Contaminating DNA was removed using a DNAfree kit (Ambion, Austin, TX). A human normal tissue FirstChoice RNA Survey Panel and a lymph node FirstChoice Total RNA were purchased from Ambion. Human hippocampal RNA was purchased from BioChain Institute (Hayward, CA). Total RNA (1 μg) was transcribed using SuperScript III RT (Invitrogen) with a combination of random pentadecamer and anchored oligo(dT)20 primers. RT-qPCR was performed using a LightCycler 480 SYBR Green I Master chemistry (Roche Applied Science) and an amount of cDNA equivalent to 20 ng of total RNA. Primers specific for human PRR7 were 5′-tgtccagtgaagcgtctgag-3′ (forward) and 5′-aagcacgtgaggaacgtgta-3′ (reverse). Raw RT-qPCR data (Ct values) were preprocessed (normalization to optimal reference genes, GAPDH or β-microglobulin, relative expression calculation) and analyzed using GenEx Software (MultiD, Göteborg, Sweden).
Northern Blotting
Total RNA was isolated from 1 × 107 cells using TRI Reagent RT. RNA was resolved (15 μg/sample) by formaldehyde-agarose gel electrophoresis, transferred to positively charged nylon membrane, and hybridized to radioactively ([α-32P]dCTP)-labeled probe in Super Hyb solution (MRC). The probe was prepared from cloned PRR7 cDNA using DecaLabel DNA labeling kit (Fermentas, Ontario, Canada).
Cells and Media
Cell line MEG-01 was provided by S. Watson (University of Birmingham, Birmingham, UK), MOLT-4 and HPB-ALL were from the IMG cell line collection (Institute of Molecular Genetics, Prague, Czech Republic), and Caco-2, U937, and THP-1 were from DSMZ (Braunschweig, Germany). Jurkat, Ramos, and COS-7 cell lines were from ATCC (Manassas, VA), and HEK293FT cells were obtained from Invitrogen. Caco-2, HEK293FT, and COS-7 cells were cultured in DMEM supplemented with 10% FCS (Biochrom AG, Berlin, Germany) and antibiotics at 37 °C in 5% CO2. All other cells were cultured in RPMI 1640 supplemented with 10% FCS and antibiotics at 37 °C in 5% CO2. Human peripheral blood lymphocytes (PBL) were isolated from whole blood of healthy donors by Ficoll-Paque Plus (GE Healthcare) gradient centrifugation. For depletion of adherent cells, PBL were subjected to plastic adherence for 2 h at 37 °C. PBL were cultured as stated above for Jurkat cells.
DNA Constructs
The coding region of human PRR7 was amplified from human T cell line MOLT-4 cDNA and inserted into mammalian expression plasmid pEFIRES-P, provided by Dr. S. Hobbs (Institute of Cancer Research, London, UK) (20). For inducible expression, PRR7 was subcloned into the pZX-LR vector in-frame with the hemagglutinin (HA) tag. The pZX-LR vector was a kind gift of Dr. Claude Labrie (CHUL Research Center, Quebec, Canada) (21). Deletion mutants of PRR7 (Δ2–39, Δ44–274, Δ114–274, Δ151–274, Δ159–274, Δ171–274, Δ207–274, ΔTAV) were generated by PCR and fused to enhanced green fluorescent protein (EGFP) in the pEGFP vector (Clontech, Palo Alto, CA). A tyrosine mutant of PRR7 (all Tyr residues mutated to Phe) was generated by sequential PCR oligonucleotide-directed site-specific mutagenesis and subcloned into the pEGFP vector (Clontech). For co-transfection experiments in COS-7 cells, the following cDNA constructs were used: Src in the pSM vector provided by Dr. A. Weiss (University of California, San Francisco, CA), FLAG-tagged Lck inserted into the pcDNA3 vector provided by Dr. R. Abraham (Mayo Clinic, Rochester, MN), FynT and FynF (constitutively active Fyn) in the pEF-BOS vector provided by Dr. B. Schraven (Otto-von-Guericke-University, Magdeburg, Germany), Yes in the pSG5 vector provided by Dr. C. Benistant (CRBM, CNRS, Universities of Montpellier I and II, France), Myc-tagged Lyn in the pcDNA3.1 vector provided by Dr. S. Watson (University of Birmingham, Birmingham, UK), FLAG-tagged Hck in the pcDNA1 vector provided by Dr. G. Langsley (Institut Pasteur, Paris, France).
Antibodies
Hybridomas producing monoclonal antibodies (mAbs, TRAP-3/01–13) to human PRR7 were prepared by standard hybridoma techniques using mice immunized with the recombinant protein corresponding to aa 147–274. Using the deletion mutants, the epitope recognized by the mAbs was mapped to the stretch of aa 147–170. The mAbs were cross-reactive with murine and rat PRR7 in immunoblots (not shown). The phosphotyrosine antibody (P-TYR-02) used for detection of phospho-LAT in Jurkat whole cell lysates was developed in our laboratory as well. For cell stimulation, the following non-commercial antibodies were used: C305 IgM mAb to Jurkat cell TCR provided by Dr. A. Weiss and 248.23.2 IgM mAb to CD28 (22). Antibodies to the following antigens were obtained from the indicated commercial sources: LAT (LAT-01), PAG (MEM-255), CD3 (MEM-57), CD5 (MEM-32), Lck (LCK-01), ZAP-70 (ZAP-03), CD69 (FN50, Alexa Fluor 647), tubulin (TU-01), and LAMP-1 (CD107a, B-T47) from Exbio (Vestec, Czech Republic); GAPDH from Sigma; Src (N-16), c-Jun (N), ERK2 (C-14), TCRζ (6B10.2) from Santa Cruz Biotechnology (Santa Cruz, CA); IL-2 (MQ1–17H12, APC) from eBioscience (San Diego, CA); phosphotyrosine (4G10) from Upstate Biotechnology (Lake Placid, NY); HA tag (6E2), pZAP-70 Tyr-319, pPLCγ1 Tyr-783, PLCγ1, pp42/44 ERK1/2 (Thr-202/Tyr-204), pp38 MAPK (T180/Y182), p38 MAPK, pJNK (T183/Y185), JNK, pc-Jun (Ser-63/Ser-73), pSFK (Y416), and pLck (Y505) from Cell Signaling Technology (Beverly, MA).
Inhibitors and Inhibitor Assay
The following inhibitors were used: the inhibitor of the Src family kinases, PP2 (Calbiochem), final concentration 10 μm and the pan-caspase inhibitor, Z-VAD-FMK (Alexis Biochemicals), final concentration 10 μm. Z-VAD-FMK inhibitor assay was performed using 2.5 × 105 cells in 96-well tissue culture plates in triplicates. Viability was assessed by flow cytometry (propidium iodide exclusion and side plus forward scatter gating).
Transfections and Inducible Expression
Jurkat cells (107) were transfected with 15 μg of the corresponding plasmid DNA in 300 μl of RPMI medium supplemented with 10% FCS by electroporation (250 V, 950 microfarads) using a GenePulser electroporator (Bio-Rad). For the pZRD-inducible system (21), based on the RheoSwitch Mammalian Inducible Expression System (New England Biolabs, Ipswich, MA), transfectants were selected in the culture medium containing Zeocin (200 μg/ml, Invitrogen). Stably transfected GFP-positive cells were sorted and further subcloned by limiting dilution. The clones (denoted J-iPRR7) that expressed PRR7 only after induction with RSL1 (500 nm for maximal induction, RheoSwitch Ligand 1, New England Biolabs) were used in further experiments. COS-7 cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Cell Stimulation
To analyze PRR7 up-regulation, freshly isolated PBL were stimulated using 1) anti-CD3 IgG (MEM-57) immobilized on tissue culture plastic (10 μg/ml) and soluble anti-CD28 IgM (100× diluted hybridoma supernatant), 2) PHA-L (2.5 μg/ml, Sigma), and 3) a combination of PMA (10 ng/ml, Sigma) and ionomycin (500 ng/ml, Sigma).
For short term activation (Ca2+ flux, phospho-Tyr blots), Jurkat cells were stimulated with anti-TCR IgM (C305) (23) (10 μg/ml) at 37 °C for 2 min. For activation of Jurkat cells in an IL-2 production assay, the combination of PMA and ionomycin was used as described above.
Preparation of Cell Lysates, Immunoprecipitations, Biochemical Methods
Generally, 5 × 107 cells were lysed in 1 ml of lysis buffer (20 mm Tris pH 7.5, 100 mm NaCl, 5 mm iodoacetamide, 10 mm EDTA, 50 mm NaF, 10 mm Na4P2O7, 10% v/v glycerol, and Protease Inhibitor Mixture III, Calbiochem) containing 1% w/v detergent LM (Calbiochem) or Brij-98 (Sigma) for 30 min on ice. To remove nuclei and other insoluble materials, the lysate was spun down at 16,000 × g for 10 min at 4 °C. Density gradient ultracentrifugation and gel filtration on Sepharose 4B were performed as previously described (24). For immunoprecipitation experiments, we coupled the anti-PRR7 IgG (TRAP3-03) to CNBr-Sepharose 4B (Amersham Biosciences) or used soluble antibodies and Protein A/G PLUS-agarose IP Reagent (Santa Cruz). Palmitoylation of PRR7 was examined using the acyl-biotinyl exchange chemistry-based method (25). Briefly, plasma membranes from 5 × 107 cells were isolated, and palmitate protein modifications were removed by hydroxylamine and replaced with biotin. Biotinylated proteins were then immunoprecipitated on streptavidin-agarose beads. Other biochemical methods (SDS-PAGE, immunoblotting) were performed essentially as described before (4).
Microscopy
Jurkat cells were allowed to adhere to polylysine-coated coverslips for 30 min, then fixed with 4% w/v formaldehyde for 15 min at room temperature and permeabilized with 0.1% Triton X-100 (Sigma) for 5 min. Blocking was performed in 2.5% BSA and 10% goat serum (Sigma) in PBS for 30 min. Cells were then incubated with 100×-diluted LAMP-1 antibody followed by 750×-diluted Alexa 647-labeled goat anti-mouse IgG secondary antibody (Molecular Probes, Invitrogen). The DNA dye, Hoechst 33258 (1 μg/ml, Invitrogen), was used to visualize nuclei. Images were captured with a Leica SP5 confocal microscope and a 63× objective lens (Leica Microsystems, Mannheim, Germany).
Flow Cytometry
CD69 surface staining and intracellular IL-2 staining were done according to the standard protocols. To measure apoptosis in cells expressing PRR7, we performed annexin V/Dy-647 (Apronex Biotechnologies, Vestec, Czech Republic) staining in combination with the DNA dye, Hoechst 33258, according to the manufacturer's instructions. To measure calcium response to anti-TCR activation, cells were first loaded with 5 μm Fura Red (Molecular Probes, Invitrogen) in loading buffer (1 × Hanks' balanced salt solution, 2% FCS, without Ca2+ or Mg2+) for 30 min at 37 °C. After washing, cells were resuspended in loading buffer supplemented with Ca2+ and Mg2+ and kept on ice. Cells were warmed up for 10 min at 37 °C and then stimulated with an irrelevant control antibody or the anti-TCR C305 mAb (10 μg/ml). Calcium flux was monitored for 240 s. Flow cytometry was carried out on an LSRII instrument (BD Biosciences), and cells were sorted on FACSVantage (BD Biosciences). Analysis of the data was performed using FlowJo software (Tree Star, Ashland, OR) as specified in the figure legends (Figs. 6 and 8).
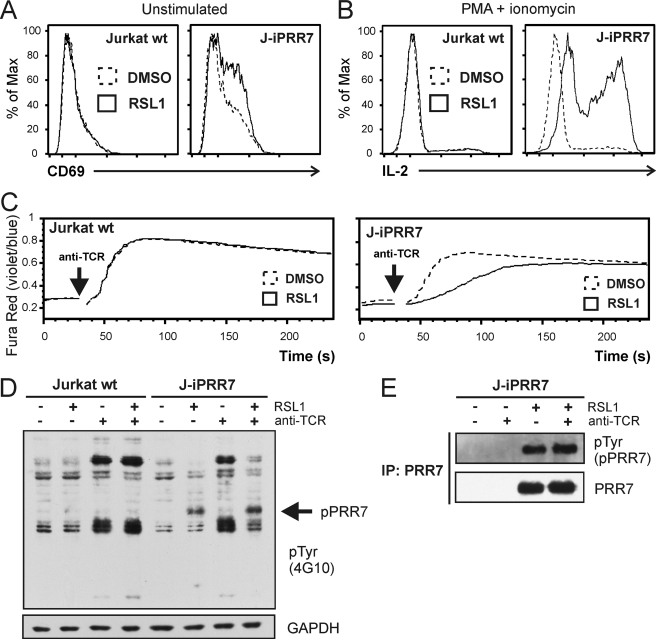
FIGURE 6.
Activated phenotype and impaired TCR signaling in J-iPRR7 cells. A, J-iPRR7 cells were induced for 12 h to express PRR7, stained for the surface lymphocyte activation marker, CD69, and analyzed by flow cytometry. B, J-iPRR7 cells were induced to express PRR7. After 12 h, the cells were stimulated for additional 6 h with PMA and ionomycin in the presence of brefeldin A (5 μg/ml), fixed, stained for cytokine IL-2, and analyzed by flow cytometry. C, J-iPRR7 cells were induced for 16 h to express PRR7 and then loaded with a calcium indicator dye Fura Red. Calcium response after the anti-TCR IgM (C305, 10 μg/ml) stimulation was measured by flow cytometry. Collected data were analyzed in FlowJo software and plotted as mean fluorescence values. D, J-iPRR7 cells were induced to express PRR7 (16 h). Cells were stimulated with the anti-TCR IgM (C305, 10 μg/ml, 2 min) or left unstimulated. Whole cell lysates were analyzed by immunoblotting with the anti-phospho-Tyr mAb (4G10). Membrane was reprobed with GAPDH antibody as a loading control. E, J-iPRR7 cells were treated as in panel D, and lysate was subjected to immunoprecipitation (IP) with the anti-PRR7 mAb. Immunoprecipitated material was analyzed by immunoblotting with the anti-phospho-Tyr mAb (4G10) and the anti-PRR7 mAb.
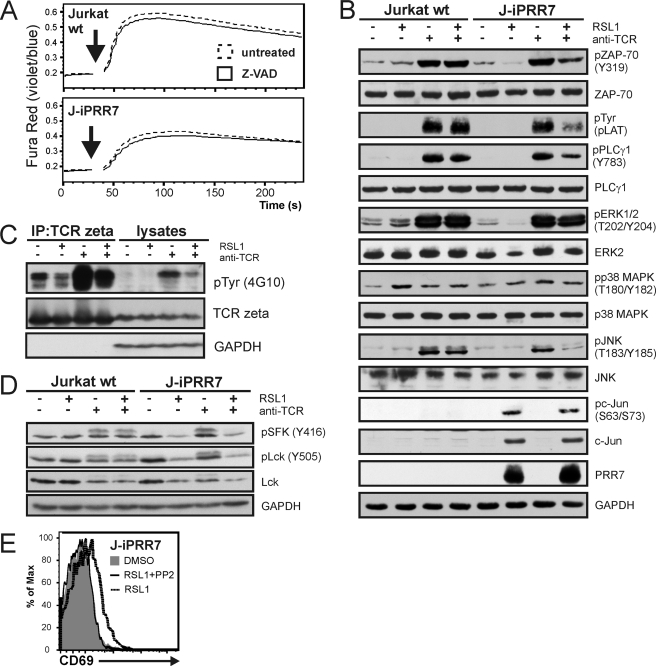
FIGURE 8.
Mechanisms of PRR7 dependent regulation of J-iPRR7 signaling. A, J-iPRR7 cells were induced to express PRR7 in the presence of the pan-caspase inhibitor Z-VAD-FMK (10 μm, 16 h) and then loaded with a calcium indicator dye Fura Red. Calcium response after the anti-TCR IgM (C305, 10 μg/ml, black arrows) stimulation was measured by flow cytometry. Collected data were analyzed in FlowJo software and plotted as mean fluorescence values. B, J-iPRR7 cells induced to express PRR7 (16 h) were stimulated with the anti-TCR IgM (C305, 10 μg/ml, 2 min) or left unstimulated. Whole cell lysates were analyzed by immunoblotting with antibodies specific for proteins involved in TCR signaling. Phosphorylation status of LAT was detected using anti-Tyr(P) antibody (clone P-TYR-02) developed in our laboratory. Staining of GAPDH was used as a sample loading control. C, J-iPRR7 cells were treated as stated above (panel B), and lysates were subjected to immunoprecipitation (IC) with the anti-TCRζ mAb. Immunoprecipitates and whole cell lysates were immunoblotted with the anti-phospho-Tyr mAb (4G10) and Abs specific for TCRζ and GAPDH (sample loading control). D, J-iPRR7 cells were treated as stated above (panel B). Whole cell lysates were analyzed by immunoblotting with antibodies specific for phosphorylated forms of Lck, total Lck, and GAPDH (sample loading control). E, J-iPRR7 cells were induced to express PRR7 in the presence or absence of PP2 (10 μm, 18 h), stained for CD69, and analyzed by flow cytometry. In all the above assays, Jurkat wt cells and DMSO treatment were used as controls (not shown in panels C and E).
RESULTS
PRR7 Sequence Analysis
Human PRR7 (18) is a 274-aa protein with a sequence typical for TRAPs. It has a short N-terminal extracellular peptide, a single transmembrane segment, and a cytoplasmic part containing several conserved binding motifs (Fig. 1A). These motifs include multiple SH2 domain binding and/or endocytic tyrosine-based motifs (YXX(I/L/V/A)), multiple proline-rich SH3 binding motifs (PXXP), group I WW domain binding motifs (PPXY), and a C-terminal class I PDZ domain binding motif (TTAV). In addition, PRR7 contains a potential submembrane palmitoylation motif (CCXC, Fig. 1A).
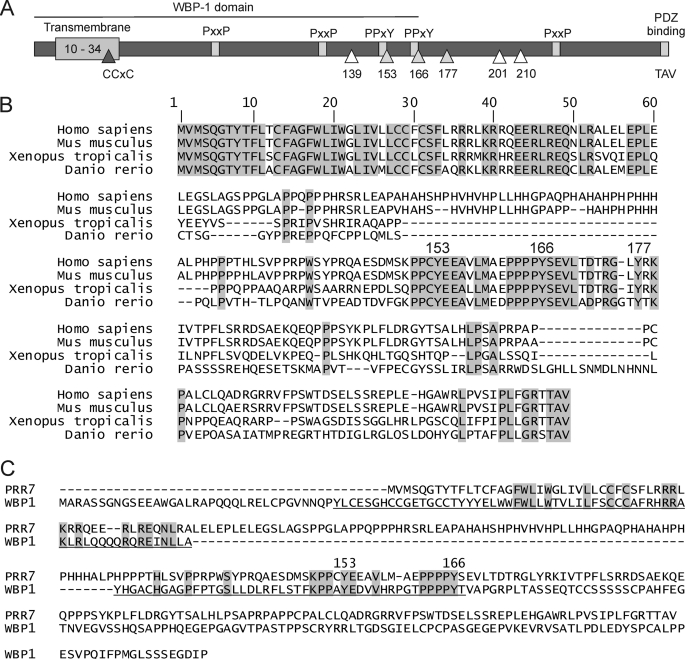
FIGURE 1.
Human PRR7 protein scheme and conservation. A, PRR7 features a short extracellular part (predicted aa 1–9) followed by a single transmembrane segment (predicted aa 10–34), a submembrane palmitoylation motif CCXC (aa 28–31), six intracellular tyrosine residues (gray triangles for fully conserved residues, white triangles for less conserved residues), multiple SH3 domain binding proline-rich motifs (PXXP, only conserved motifs are shown, aa 74–77, 123–126, 222–225), group I WW domain binding motifs (PPXY, only conserved motifs shown, aa 150–153, 163–166), and a C-terminal PDZ-binding motif (TTAV, aa 271–274). The N-terminal part of PRR7 (aa 1–167) corresponds to the predicted WBP-1 domain as indicated. This scheme is based on GenBankTM protein sequence NP_085044. B, shown is alignment of PRR7 protein sequences from four vertebrate species: Homo sapiens (human), Mus musculus (mouse), Xenopus tropicalis (frog), and Danio rerio (zebrafish). The sequences were aligned using ClustalW program from the EMBL-EBI website. Absolutely conserved residues are shaded in gray. Tyrosine residues conserved in all vertebrates (Tyr-153, Tyr-166, Tyr-177 in humans) are numbered above the alignment. C, a comparison of human PRR7 and WBP-1 protein sequences is shown. The sequence corresponding to the predicted WBP-1 domain is underlined. Identical residues are shaded in gray. Two conserved tyrosine residues (Tyr-153 and Tyr-166 in human PRR7) in the predicted WW domain binding motifs are indicated above the sequences.
PRR7 is a highly conserved protein. The percentage of aa identity is more than 94% among available sequences from placental mammals, and a substantial level of aa identity is observed even if lower vertebrate species (amphibian, fish) are included (Table 1). The highest degree of conservation of PRR7 protein sequence was observed in the extracellular, transmembrane, and submembrane parts, including the palmitoylation motif. In addition, the region containing three tyrosine residues, Tyr-153, Tyr-166, and Tyr-177, and the C terminus with the PDZ domain binding motif also displayed a very high degree of homology (Fig. 1B). Interestingly, N-terminal part of PRR7 as well as sequences surrounding tyrosines 153 and 166 form a putative domain identified in Pfam data base (26) as the WBP-1 domain (PF11669). Among other proteins, this domain is present in WW domain-binding protein 1 (WBP-1) where it mediates the interaction with WW domains of Yes-associated protein (YAP) via tandem PPXY motifs in its C terminus (27). Similar motifs encompass tyrosines 153 and 166 in PRR7, suggesting possible involvement of this region in an interaction with a WW domain containing protein (Fig. 1C).
TABLE 1.
Conservation of PRR7 protein in vertebrates
Orthologous protein sequences were aligned and compared to human PRR7 using ClustalW program. Species in this table are sorted according to the calculated protein identity with human PRR7.
| Species | Length | Identity |
|---|---|---|
| aa | % | |
| Pongo pygmaeus (orangutan) | 273 | 99 |
| Pan troglodytes (chimpanzee) | 259 | 98 |
| Macaca mulatta (macaque) | 276 | 98 |
| Rattus norvegicus (rat) | 269 | 98 |
| M. musculus (mouse) | 269 | 97 |
| Spermophilus tridecemlineatus (squirrel) | 269 | 97 |
| Canis familiaris (dog) | 272 | 97 |
| Bos taurus (cattle) | 269 | 97 |
| Echinops telfairi (tenrec) | 270 | 95 |
| Sus scrofa (pig) | 269 | 94 |
| Monodelphis domestica (opossum) | 280 | 81 |
| X. tropicalis (frog) | 227 | 56 |
| D. rerio (fish) | 246 | 48 |
| Gasterosteus aculeatus (fish) | 279 | 44 |
PRR7 Expression Profile
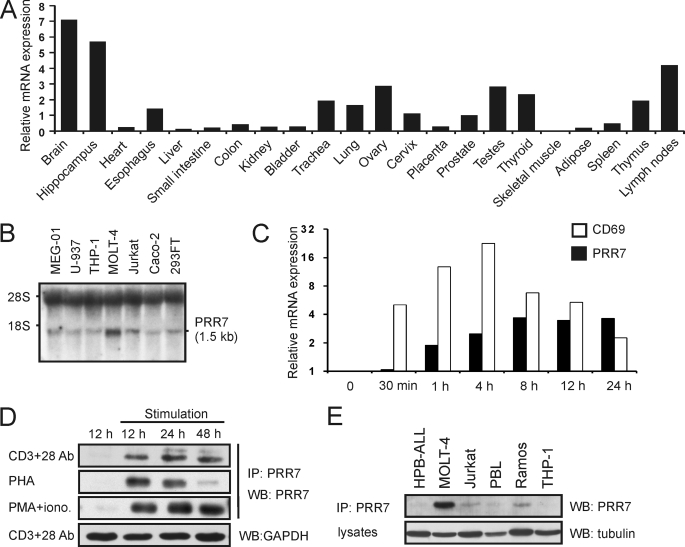
To determine the PRR7 expression pattern in human tissues, we performed RT-qPCR on a panel of multiple tissue RNAs. As shown in Fig. 2A, PRR7 mRNA is expressed most strongly in brain tissue and moderately in several other tissues: esophagus, trachea, lung, ovary, cervix, prostate, testes, thyroid, including the immune organs thymus and lymph nodes. We also found low levels of PRR7 mRNA in a number of human cell lines (Jurkat, U937, THP-1, MEG-01, CaCo2, HEK293FT); the only cell line exhibiting relatively high expression was T cell line MOLT-4 (Fig. 2B). We next analyzed whether the expression level of PRR7 mRNA in PBL changes after activation. Interestingly, we observed rapid up-regulation of PRR7 mRNA in PHA-stimulated PBL. It was detectable at 1 h and peaked at 8 h after stimulation (Fig. 2C).
FIGURE 2.
Expression of PRR7 in primary human cells and tissues. A, shown is RT-qPCR analysis of PRR7 mRNA expression in normal human tissues. PCR reactions were done in triplicate, and mean Ct values were used for further data processing in GenEx software. First, data were normalized to GAPDH mRNA expression, and then relative expression values were calculated. An average expression in all tissues corresponds to value 1 on the vertical axis. B, Northern blot analysis of PRR7 mRNA in human cell lines is shown. Total RNA (15 μg) was resolved on 1% agarose gel and, after transfer to nylon membrane, probed for PRR7 mRNA. The positions of ribosomal RNA bands 18 S and 28 S are indicated. C, shown is an RT-qPCR analysis of kinetics of PRR7 mRNA up-regulation in human PBL after PHA treatment. PCR reactions were done in triplicate, and mean Ct values were used for further data processing in GenEx software. Data were normalized to the optimal reference gene, β2-microglobulin, and relative expression values were calculated. Value 1 on the vertical axis corresponds to basal PRR7 mRNA expression in unstimulated PBL. Expression of the activation marker CD69 was used as a positive control. D, human PBL were stimulated with plate-bound anti-CD3 IgG and soluble anti-CD28 IgM, PHA, or PMA and ionomycin. Cell lysates were concentrated by immunoprecipitation (IP) with the anti-PRR7 mAb (TRAP3-03). Expression of PRR7 was analyzed by immunoblotting (WB) using the anti-PRR7 mAb (TRAP3-10). In addition, equivalence of sample sizes was verified via staining of lysates for GAPDH. A representative result for each type of stimulation is provided. E, PRR7 was immunoprecipitated from lysates of human cell lines with the anti-PRR7-Sepharose. Expression of PRR7 was analyzed by immunoblotting using the anti-PRR7 mAb. The equivalence of sample sizes was verified via staining of lysates with the anti-tubulin mAb.
To examine the expression of PRR7 at the protein level, we generated anti-human PRR7 monoclonal antibodies. Unfortunately, none of the generated antibodies could clearly detect PRR7 after immunoblotting of whole primary cell lysates (except for mouse or rat brain tissues; not shown). Moreover, native PRR7 could not be quantitatively immunoprecipitated from cell lysates using these antibodies, suggesting partial blocking of the epitope (not shown). Nevertheless, it was still possible to use immunoprecipitation to concentrate PRR7 protein to the degree sufficient for detection by immunoblotting also in leukocytes. We then used this approach to verify our results from previous experiments. As shown in Fig. 2D, PRR7 protein was detected in PBL activated by various stimuli, including activation with anti-CD3 and anti-CD28 antibodies, PHA, or PMA and ionomycin. We could also detect PRR7 protein in several lymphoid cell lines, including Jurkat, Ramos, and MOLT-4 (Fig. 2E). The highest protein levels were observed in MOLT-4, which correlated well with the higher PRR7 mRNA level in this cell line (Fig. 2, B and E).
Overexpression of PRR7 Results in Apoptosis in the Jurkat T Cell Line
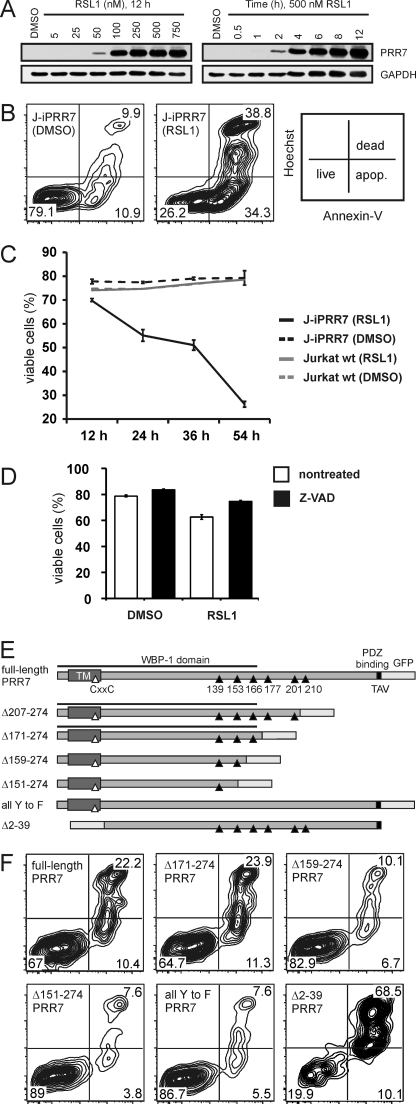
Because the level of PRR7 expression in leukocytes (cell lines and primary PBL) was low and its direct immunoprecipitation was inefficient, we generated multiple Jurkat cell lines transfected with PRR7 constructs containing a C-terminal GFP tag. Surprisingly, we were not able to establish a Jurkat cell line with a sufficiently high expression of PRR7. Direct observation of the PRR7-GFP-transfected Jurkat cells excluded a rapid necrotic cell death of the transfectants. Instead, we consistently observed a gradual loss of the PRR7 transfectants from the culture within a few days (not shown). PRR7high cells were stained with the DNA dye, propidium iodide, and analysis of the staining suggested that overexpression of PRR7 protein caused apoptosis (not shown). To exclude the possibility that the apoptosis induction is at least in part the result of the stress caused by the transfection procedure (electroporation), we generated a stable Jurkat clone (J-iPRR7) inducibly expressing full-length HA-tagged PRR7. In this case a modified RheoSwitch Mammalian Inducible Expression System (21) was utilized. It allowed us to tightly regulate the expression of PRR7 using the synthetic compound RSL1. Without RSL1, PRR7 expression was not detectable; maximal expression was achieved in the presence of RSL1 12 h after induction (Fig. 3A). Strikingly, we observed that in J-iPRR7 cells, apoptosis could be clearly detected within 24 h of induction by RSL1 and gradually increased with time (Fig. 3, B and C), suggesting that apoptosis induction was a direct consequence of PRR7 expression. This process could be to a substantial degree inhibited by caspase inhibitor Z-VAD, demonstrating the involvement of caspase-dependent apoptotic pathways (Fig. 3D).
FIGURE 3.
PRR7-inducible system and induction of apoptosis by overexpression of PRR7 in Jurkat cells. A, to determine the optimal concentration of RSL1 for PRR7 induction, J-iPRR7 cells were treated with an increasing amount of RSL1 for 12 h (left panel). J-iPRR7 cells were treated with 500 nm RSL1, and expression of PRR7 in several time intervals was analyzed by immunoblotting with the anti-PRR7 mAb (TRAP3-10, right panel). Staining for GAPDH was used as a sample loading control. B, J-iPRR7 cells were induced to express PRR7 or left uninduced (DMSO control). After 54 h, apoptosis in both cultures was determined by annexin V and Hoechst 33258 staining. C, J-iPRR7 and Jurkat wt cells were treated with 500 nm RSL1 to induce PRR7 expression or left untreated (DMSO control). Cell viability was determined by flow cytometry after 12, 24, 36, and 54 h. Data are presented as the mean of three experiments (±S.D.). D, J-iPRR7 cells were induced to express PRR7 or left uninduced (DMSO control) for 24 h in the presence of the pan-caspase inhibitor, Z-VAD-FMK (10 μm). The percentage of viable cells was determined by propidium iodide (PI) staining and flow cytometry. The experiment was done in triplicate cultures, and data are represented as the mean values ± S.D. E, shown is a scheme of PRR7-GFP mutants used in this study. Tyrosine residues are represented by black triangles. F, Jurkat cells were transfected with corresponding plasmid DNA coding for wt or mutated PRR7 fused to GFP. After 48 h, cells were stained with annexin V/Dy647 and Hoechst 33258 to analyze apoptosis in the GFP-positive cells.
Region Surrounding Tyr-166 Is Critical for Apoptosis Induction
To identify the regions of the PRR7 molecule involved in the apoptosis induction, we generated a set of Jurkat cell lines transiently expressing either full-length PRR7-GFP or its mutants gradually truncated from the C terminus (Fig. 3E). We also generated a mutant where all the tyrosines in the PRR7 sequence were mutated to phenylalanines (all Y to F) and a mutant lacking the N terminus, including extracellular peptide, transmembrane segment, and the palmitoylation motif (Δ2–39-PRR7-GFP). As expected, expression of the full-length PRR7 resulted in a high degree of apoptosis in Jurkat cells (Fig. 3F). Similar results were obtained when aa downstream of Thr-171 were removed, suggesting that the C-terminal region, including the highly conserved Tyr-177 and the PDZ binding motif, are dispensable for apoptosis induction. In contrast, when 12 more aa were removed (Δ159–274-PRR7-GFP), a significant drop in the percentage of apoptotic cells was observed, and only a very mild additional reduction was noticed after further shortening of PRR7 protein by 8 or more aa (Δ151–274-PRR7-GFP) (Fig. 3F and not shown). These data suggested that the region critical for the apoptosis induction is located between aa 159 and 171 of PRR7. Importantly, the tyrosine mutant of PRR7 (all Y to F) also induced only a mild level of apoptosis under the same conditions, indicating that one or more tyrosine residues are involved. The only tyrosine in the critical region identified by the deletion mutants is the highly conserved Tyr-166. Thus, our observations suggest that a tyrosine-based motif surrounding this Tyr-166 is essential for the induction of apoptosis by PRR7, although the involvement of other tyrosine residues cannot be completely excluded by this type of analysis.
Finally, the mutant lacking extracellular and transmembrane parts induced the highest level of apoptosis in Jurkat transfectants (Fig. 3F). This finding was surprising as we rather expected a loss of function when removing the membrane localization sequences.
Intracellular Localization of PRR7 Mutants Partially Correlates with Apoptosis Induction
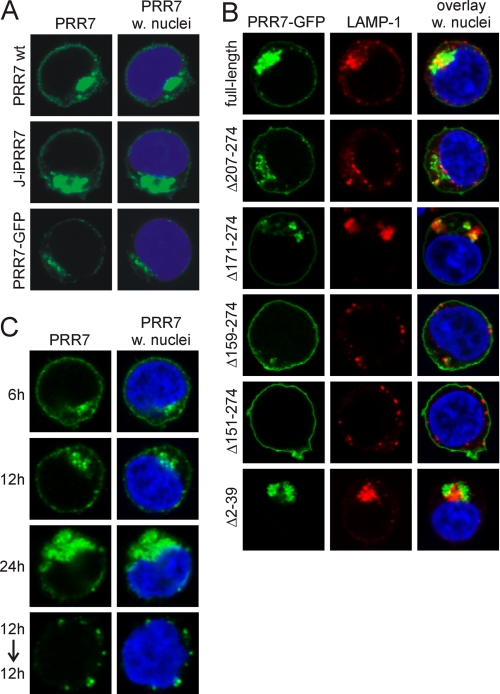
Strong pro-apoptotic effects of the construct lacking putative membrane anchoring sequences prompted us to analyze subcellular localization of PRR7 and its mutants. Full-length PRR7 was localized at the plasma membrane and to large vesicular perinuclear structures, which did not co-localize with an early endosomal marker transferrin (not shown) nor with lysosomal marker LAMP-1 (CD107a, Fig. 4, A and B). The C-terminal tag modifications had no effect on this localization pattern as tagged versions of PRR7 (PRR7-GFP, PRR7-HA) were indistinguishable from non-tagged PRR7 (Fig. 4A), suggesting that the C-terminal PDZ binding motif requiring a free carboxyl group at the C terminus for binding (28) is not involved in this process.
FIGURE 4.
Subcellular localization of full-length PRR7 and deletion mutants in Jurkat cells. A, shown is a comparison of subcellular localization of PRR7 protein with (w.) and without tags. Jurkat cells expressing the PRR7 wt protein (without a tag) were fixed 18 h post-transfection and stained with PRR7 antibody. J-iPRR7 (PRR7 with HA-tag) cells were induced for 18 h to express PRR7 with 500 nm RSL1, fixed, and stained with the anti-HA-tag mAb. Jurkat PRR7-GFP transfectants were fixed 18 h post-transfection. Samples were analyzed by confocal microscopy. B, cells were transfected with corresponding plasmid DNA, and after 18 h cells were allowed to adhere to polylysine-coated coverslips, fixed, stained for lysosomal marker LAMP-1 (CD107a) and DNA dye Hoechst 33258, and analyzed by confocal microscopy. C, J-iPRR7 cells were induced to express PRR7 with 500 nm RSL1. Cells were fixed 6, 12, and 24 h afterward or induced for 12 h, washed, and incubated an additional 12 h in fresh medium without RSL1 before fixation. All samples were stained with the anti-HA-tag mAb and DNA dye Hoechst 33258 and analyzed by confocal microscopy.
Time course analysis of PRR7 in the inducible system revealed that shortly after expression induction by RSL1, PRR7 was found mainly at the plasma membrane and later gradually accumulated in the perinuclear compartment. After RSL1 withdrawal, the plasma membrane staining was lost, and PRR7 could be detected only in the perinuclear cytoplasmic vesicles (Fig. 4C). These data suggest a continuous active process of PRR7 removal from the plasma membrane by endocytosis.
We next analyzed PRR7 deletion mutants. C-terminal deletions up to aa 171 (Δ207–274, Δ171–274-PRR7-GFP) did not have any effect on PRR7 localization. However, further shortening of the cytoplasmic part of PRR7 up to aa 159 and 151 (Δ159–274, Δ151–274-PRR7-GFP) resulted in gradual loss of vesicular accumulation and a shift to an almost exclusive surface localization (Fig. 4B). Additional shortening of PRR7 sequence did not have any further effect on the subcellular localization of this protein (not shown). These results suggest that the region critical for internalization of PRR7 is positioned between aa 151 and 171. Strikingly, this region partially overlaps with the sequence responsible for the apoptosis induction (159–171), suggesting a link between PRR7 internalization and apoptosis. In agreement with this, the PRR7 N-terminal deletion mutant, which caused the highest level of apoptosis (Δ2–39-PRR7-GFP), was localized exclusively in the intracellular vesicular compartment (Fig. 4B). The region found to be critical for PRR7 internalization contains two highly conserved tyrosine motifs, suggesting that they may serve as tyrosine-based internalization motifs. However, the all Y to F mutant of PRR7 showed only a mild reduction in the size of intracellular PRR7 compartment (not shown), displaying at the same time strong defects in the apoptosis induction. These results indicated that tyrosines within the critical conserved region are not a part of the tyrosine-based internalization sequences, but rather, they are involved in pro-apoptotic signaling mediated by PRR7. Importantly, these tyrosines form the putative WW domain binding motif of the PRR7 WBP-1 domain, suggesting possible involvement of a WW-containing protein in this process.
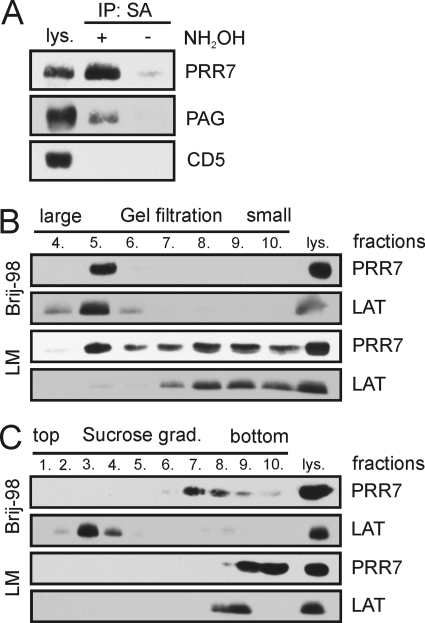
PRR7 Partitions to Large Detergent-resistant Complexes
Similarity to palmitoylated lipid raft-associated TRAPs suggested that PRR7 may employ lipid rafts as signaling platforms. Indeed, we found PRR7 to be palmitoylated in the J-iPRR7 cells (Fig. 5A). However, in contrast to these adaptors, PRR7 was found in a different type of detergent (Brij-98)-resistant complexes. These were large (as determined by gel filtration on Sepharose 4B) but almost non-buoyant (as determined by density gradient ultracentrifugation) and resistant to lipid raft-disrupting detergent laurylmaltoside (Fig. 5, B and C).
FIGURE 5.
Biochemical characterization of the PRR7 protein. J-iPRR7 cells were induced to express PRR7 (16 h) and subjected to several biochemical assays. A, palmitoylation of PRR7 was examined using the acyl-biotinyl exchange chemistry-based method as described under “Experimental Procedures.” A palmitoylated transmembrane adaptor protein, PAG, was used as a positive control; a non-palmitoylated transmembrane protein, CD5, and lysate that was not treated with NH2OH (hydroxylamine) were used as negative controls. IP, immunoprecipitate. SA, streptavidin beads. B, Brij-98 or LM lysates of J-iPRR7 cells were subjected to gel filtration on Sepharose 4B, and the separated fractions were analyzed by immunoblotting. A raft-associated transmembrane adaptor protein, LAT, was used as a control. C, Brij-98 or LM lysates of J-iPRR7 cells were subjected to density gradient ultracentrifugation, and the separated fractions were analyzed by immunoblotting. The fractions are numbered from the top. 10 represents the sediment. The lipid raft protein, LAT, was used as control.
Variable Effects of PRR7 on the Activity of Signaling Pathways in J-iPRR7
To elucidate how PRR7 affects T cell signaling pathways, we compared the phenotype of induced versus non-induced J-iPRR7 cells. Induction of PRR7 expression led to a mild spontaneous up-regulation of the activation marker CD69 in quiescent cells (Fig. 6A). Moreover, treatment of induced J-iPRR7 with PMA and ionomycin resulted in a substantial increase in IL-2 production (Fig. 6B), suggesting that these cells are in a partially activated state. In contrast, at the same time, the induced J-iPRR7 cells exhibited an attenuated calcium response (Fig. 6C) and reduced global tyrosine phosphorylation (Fig. 6D) after TCR stimulation.
Interestingly, we detected strong constitutive tyrosine phosphorylation of PRR7, which was not altered by TCR cross-linking (Fig. 6, D and E). This observation raised questions about the mechanisms mediating PRR7 phosphorylation and the role of tyrosine phosphorylation in the diverse effects of PRR7 on T cell signaling pathways.
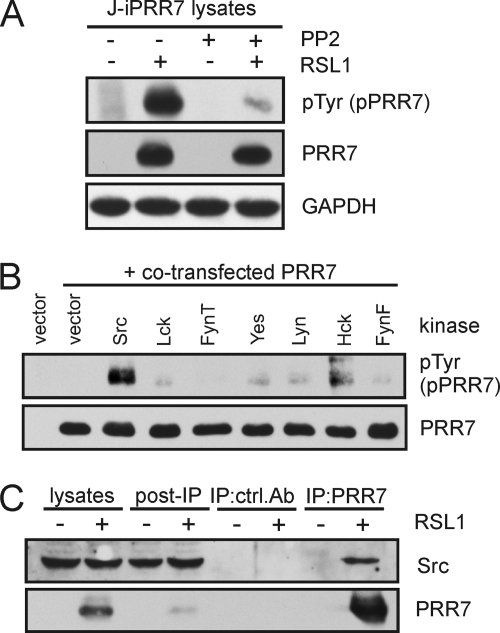
To elucidate the mechanisms of PRR7 phosphorylation, we analyzed lysates of J-iPRR7 cells treated with a specific inhibitor of Src-family kinases (SFK) PP2 by phosphotyrosine immunoblotting. As shown in Fig. 7A, PRR7 phosphorylation was almost completely inhibited in the presence of PP2, suggesting that PRR7 is constitutively phosphorylated by SFKs. To identify potential SFKs capable of PRR7 phosphorylation, constructs encoding PRR7 and various tyrosine kinases were co-transfected into COS-7 cells. The results revealed that several SFKs (most strongly Src) are able to phosphorylate PRR7 (Fig. 7B). The multiple predicted SH3 domain binding motifs in the intracellular part of PRR7 using iSPOT (29) indicated a possible association of PRR7 with SFKs (via their SH3 domains). Thus we immunoprecipitated PRR7 from the induced J-iPRR7 cells and tested the immunoprecipitates for the presence of associated SFKs by immunoblotting. Among the tested SFKs (Src, Lck, Fyn), the only identified binding partner was Src (Fig. 7C and not shown).
FIGURE 7.
Tyrosine phosphorylation of PRR7 and association of PRR7 with Src. A, J-iPRR7 cells were induced to express PRR7 and simultaneously treated with the SFK inhibitor, PP2 (10 μm), for 18 h or left untreated. Tyrosine phosphorylation status of PRR7 was analyzed by immunoblotting with the anti-phospho-Tyr mAb (4G10). GAPDH staining served as a sample loading control. B, PRR7 was co-transfected with Src kinases into COS-7 cells. After 24 h, cells were lysed, and tyrosine phosphorylation of PRR7 was detected by immunoblotting with the anti-phospho-Tyr (4G10) mAb. C, J-iPRR7 cells were induced to express PRR7 or left uninduced for 18 h and lysed in LM buffer, and PRR7 was immunoprecipitated (IP) using the anti-PRR7-Sepharose or an isotype control mAb. Eluted proteins were analyzed by immunoblotting.
Mechanisms of PRR7-dependent Regulation of J-iPRR7 Signaling
So far, our observations suggested that PRR7 expression had a dual activating/inhibitory effect. It resulted in selective up-regulation of c-Jun and CD69, whereas at the same time it led to the inhibition of proximal TCR signaling. Among the hypothetical explanations for the inhibitory effects of PRR7 on TCR-mediated signaling was the possibility that the cells are already in the process of apoptosis advanced enough to have negative effects on TCR signal transduction. To test this, we treated J-iPRR7 cells with caspase inhibitor Z-VAD during RSL1-mediated PRR7 induction and then followed a calcium response downstream of TCR. Z-VAD treatment had a negligible effect on calcium response in PRR7-expressing J-iPRR7 cells, suggesting an apoptosis-independent mechanism of PRR7-mediated inhibition of T cell signaling (Fig. 8A).
To identify the point where PRR7 interferes with TCR signaling pathways, we treated J-iPRR7 cells with RSL1 to induce PRR7 expression, stimulated the cells by TCR cross-linking, and followed the phosphorylation of individual proteins in the cell lysates. We observed reduced phosphorylation of a number of important signaling molecules, including ZAP-70, LAT, PLCγ1, ERK1/2, and JNK (Fig. 8B) after TCR stimulation. The only remarkable exception was the up-regulation of c-Jun protein level, which was also reflected in the increased amount of the phosphorylated form of this transcription factor (Fig. 8B).
The reduction in tyrosine phosphorylation of proteins in proximal TCR signaling pathways, including ZAP-70, LAT, and PLCγ1, suggested the effects of PRR7 at the level of TCR itself. To analyze this, we followed the phosphorylation of TCRζ chain after TCR cross-linking. We observed a substantial reduction in TCRζ phosphorylation both in a basal state and after stimulation in PRR7-expressing cells (Fig. 8C). As TCRζ is a substrate SFKs, we next analyzed the activation status of these kinases by immunoblotting with antibodies specific to their activating and inhibitory tyrosines with a focus on Lck, the major Src-family member in T cells. Surprisingly, we observed reduction in the phosphorylation of both regulatory tyrosines (SFK activation loop tyrosine and Lck inhibitory tyrosine) in PRR7-expressing cells. However, we also detected a highly reproducible reduction in total Lck staining (Fig. 8D). These data showed that the decreased Lck level is probably the major consequence of PRR7 expression, which is then reflected in the reduced content of both regulatory phosphotyrosines in the samples and reduced activity of downstream signaling pathways.
Finally, to assess the role of SFK activity and tyrosine phosphorylation of PRR7 in the opposing functions of PRR7, we treated J-iPRR7 cells with SFK inhibitor PP2 during the PRR7 induction. Surprisingly, PP2 treatment had only a minor effect on c-Jun up-regulation as well as apoptosis induction (not shown), indicating that the effect of PRR7 on these pathways is independent of SFK signaling. In contrast, PP2 treatment inhibited the up-regulation of CD69 (Fig. 8E).
DISCUSSION
Thus far, PRR7 has been described in a single paper (18) as a component of the postsynaptic density (PSD) fraction of the rat forebrain, suggesting that it may play a role in modulating neural activities via interactions with the PSD-95/N-methyl-d-aspartate receptor complex or in PSD core formation. Here, we report that PRR7 is expressed in many tissues, including leukocytes, and is significantly up-regulated during T cell activation (Fig. 2, C and D). Nevertheless, even in activated T cells we had difficulties detecting PRR7 using our monoclonal antibodies on the immunoblot of whole cell lysates, which was most likely the result of both low expression level of PRR7 and suboptimal antibody reactivity. Therefore, we have used mainly Jurkat transfectants transiently or inducibly expressing recombinant PRR7. In these cells, our monoclonal antibodies could readily detect PRR7 in cell lysates and could be used in biochemical studies.
PRR7 is highly evolutionarily conserved (Table 1), suggesting that its function might be preserved across all vertebrates. The intracellular part of this protein (Fig. 1A) contains several motifs (proline-rich WW binding motif, tyrosine-based SH2 domain binding and/or internalization motifs, proline-based SH3 binding motifs, and a C-terminal PDZ-binding motif) indicating potential for interactions with other proteins.
The PDZ binding motif is responsible for the reported association with the MAGUK family scaffold protein PSD-95 in neurons (18). We assumed that in leukocytes it could be associated with the leukocyte counterpart of PSD-95, DLG1 (30). However, in our co-transfection experiments, we were not able to confirm such an association, whereas the association with PSD-95 was readily demonstrable under the same conditions (not shown). Src is another binding partner of PRR7, as identified in this work. PRR7 is most likely a substrate of Src, as we observed strong tyrosine phosphorylation of PRR7 in Jurkat cells, which was inhibited by the SFK inhibitor, PP2.
The most striking consequence of PRR7 overexpression in Jurkat T cell line was increased apoptosis. We used multiple deletion mutants to identify the region critical for apoptosis induction. Our analysis limited this region to the sequence between aa 159–171 in the WBP-1 domain of PRR7. Importantly, this region contains only a single tyrosine residue, Tyr-166, suggesting that phosphorylation of this site may be involved. However, our experiments with PP2 treatment of J-iPRR7 cells did not provide sufficient evidence for the role of SFK-mediated PRR7 tyrosine phosphorylation in apoptosis induction (not shown). We could also rule out the activation-induced cell death, as we did not observe any up-regulation of FasL in J-iPRR7 cells nor were we able to block PRR7-induced apoptosis by blocking antibodies to FasL (not shown). Thus, the mechanism of how the region around Tyr-166 mediates pro-apoptotic signaling remains unknown. One of the remaining hypotheses is phosphorylation-independent binding of a WW domain-containing protein to the tyrosine motif in the critical region. However, the identity of such a putative protein remains to be determined.
PRR7 inducibly expressed in the Jurkat cells was initially present at the plasma membrane, and later it was mainly detected in reversible, perinuclear, vesicular structures (Fig. 4, B and C). Time course analysis of PRR7 localization in the inducible system after RSL1 withdrawal suggests that the perinuclear accumulation is a continuous, active process involving removal from the plasma membrane by endocytosis (Fig. 4C).
Interestingly, the region involved in PRR7-mediated internalization partially overlaps with the section critical for PRR7-mediated apoptosis, suggesting the link between PRR7 internalization and induction of apoptosis. However, the region required for PRR7 internalization also includes the sequences outside the area critical for apoptosis. Moreover, the mutation of all PRR7 tyrosine residues that had strong inhibitory effects on apoptosis induction resulted only in a mild reduction in PRR7 internalization (not shown), suggesting that this region mediates apoptosis via other mechanisms then solely by triggering PRR7 endocytosis. On the other hand, the mutant lacking the membrane-anchoring sequences, which localized exclusively in the intracellular compartment, induced the highest degree of apoptosis. Taken together these data suggest that internalization and tyrosine phosphorylation are two independent mechanisms, which may cooperate to efficiently trigger apoptosis in J-iPRR7. An important question remains of how the region between aa 151 and 171 mediates PRR7 internalization. Tyrosines 153 and 166 within this critical region are part of potential tyrosine-based internalization sequences fitting the consensus YXXΦ (31). Tyrosine to phenylalanine mutations within similar motifs can substantially reduce the rate of endocytosis (32). Mutations of PRR7 tyrosines had only a mild effect on PRR7 internalization, suggesting that these motifs are not genuine internalization sequences.
Other phenotypic features of the induced J-iPRR7 cells were also quite striking. They exhibited some traits of partially activated or “primed” T cells, including a spontaneous increase in the expression of the surface activation marker, CD69, and strongly enhanced IL-2 production after PMA and ionomycin stimulation (Fig. 6, A and B). On the other hand, proximal elements of immunoreceptor signaling cascades (tyrosine phosphorylation of cytoplasmic substrates and cytoplasmic calcium elevation upon TCR cross-linking) were clearly diminished or down-regulated (Fig. 6, C and D, and 8, B–D). A likely explanation is that PRR7 by a so-far unknown mechanism down-regulates protein levels of a critical SFK Lck, which in turn results in the reduction of the activity of all the downstream pathways. On the other hand, a concomitant positive regulatory function of PRR7 was evidenced by a strong up-regulation of AP-1 transcription factor component c-Jun accompanied by a mild induction of the activation marker CD69 in the cells where PRR7 expression was induced (Figs. 6A and 8B). Intriguingly, c-Jun up-regulation was not affected by PP2 treatment (not shown), suggesting that this phenomenon is independent of Src or other SFKs. Moreover, we did not see any up-regulation of upstream pathways such as JNK or Erk that could lead to c-Jun up-regulation. c-Jun turnover is regulated via ubiquitin-dependent degradation, and thus a plausible hypothesis is that PRR7 somehow interferes with this process. This could lead to up-regulation of c-Jun protein levels independently of the activity of upstream MAP kinase pathways. Interestingly, one of the proteins involved in the regulation of c-Jun turnover is Itch, an E3 ubiquitin ligase of NEDD4 family (33) that also possesses multiple WW domains and could potentially interact with PRR7. However, we could not detect any Itch protein in PRR7 immunoprecipitates (not shown), although it remains possible that the interaction is labile or that some other member of the NEDD4 family is involved in such an interaction.
It can be speculated that increased c-Jun/AP-1 activity is responsible for the primed state of PRR7 expressing Jurkat, resulting in a high level of IL-2 production after stimulation. Although AP-1 sites are important components of the IL-2 promoter (34, 35), additional factors such as NF-AT and NF-κB are required for the full activity. If these are provided, e.g. after stimulation with PMA and ionomycin, a strong response can result. Regulation of CD69 expression also involves AP-1, but in contrast to c-Jun, CD69 up-regulation could be inhibited by PP2. A somewhat speculative explanation can be that tonic signaling by TCR (which is dependent on SFK) is required for CD69 expression in addition to c-Jun activity. In Jurkat cells, basal TCR signaling results in low level of CD69 expression (36). It is likely that it can be further potentiated by c-Jun overexpression while still being dependent on SFK-driven basal signaling. In line with this explanation is the observation that PRR7-mediated CD69 up-regulation could be partially blocked by inhibitors of several pathways (NF-AT, NF-κB, and p38 MAPK, SAPK/JNK) known to be initiated by TCR/SFK mediated signaling (not shown).
Although our present results show that PRR7 is able to influence T cell physiology rather dramatically, it remains to be determined whether it plays a regulatory role under more physiological conditions, e.g. in the natural process of T cell activation (37). Such roles may be indicated by the evidently tightly controlled expression of PRR7 in resting T cells and its marked up-regulation upon activation (Fig. 2). In contrast, cell types other than T lymphocytes, e.g. fibroblasts, may be much less sensitive to increased levels of PRR7 expression; in this context it should be noted that overexpression of full-length PRR7 was well tolerated by COS-7 or HEK293FT cells. Clearly, preparation of conditional Prr7 gene knock-out mice may provide more definitive answers concerning the roles of PRR7 in various types of cells and tissues.
Acknowledgments
We thank all colleagues who provided the cells, vectors, and antibodies as indicated under “Experimental Procedures.”
This work was supported in part by Academy of Sciences of the Czech Republic Project AV0Z50520514, GACR (Project MEM/09/E011), and by the Center of Molecular and Cellular Immunology (Project 1M0506, Ministry of Education, Youth, and Sports of the Czech Republic).
- TCR
- T cell receptor
- aa
- amino acid
- Brij-98
- polyoxyethylene 20 oleyl ether
- J-iPRR7
- Jurkat clones with PRR7-inducible expression
- LM
- lauryl maltoside (n-dodecyl-β-d-maltoside)
- PBL
- peripheral blood lymphocytes
- PSD
- postsynaptic density
- RSL1
- RheoSwitch Ligand 1, diacylhydrazine [(N-(2-ethyl-3-methoxybenzoyl)-N′-(3,5-dimethylbenzoyl)-N′-tert-butylhydrazine]
- SFK
- Src family kinase
- TRAP
- transmembrane adaptor protein
- PMA
- phorbol 12-myristate 13-acetate
- PHA
- phytohemagglutinin
- qPCR
- quantitative PCR
- Z-VAD-FMK
- carbobenzoxy-valyl-alanyl-aspartyl-[O-methyl]-fluoromethylketone
- PLC
- phospholipase C.
REFERENCES
- 1. Simeoni L., Kliche S., Lindquist J., Schraven B. (2004) Curr. Opin. Immunol. 16, 304–313 [DOI] [PubMed] [Google Scholar]
- 2. Zhang W., Trible R. P., Samelson L. E. (1998) Immunity 9, 239–246 [DOI] [PubMed] [Google Scholar]
- 3. Brdiĉka T., Cerný J., Horejŝí V. (1998) Biochem. Biophys. Res. Commun. 248, 356–360 [DOI] [PubMed] [Google Scholar]
- 4. Brdicka T., Pavlistová D., Leo A., Bruyns E., Korínek V., Angelisová P., Scherer J., Shevchenko A., Hilgert I., Cerný J., Drbal K., Kuramitsu Y., Kornacker B., Horejsí V., Schraven B. (2000) J. Exp. Med. 191, 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kawabuchi M., Satomi Y., Takao T., Shimonishi Y., Nada S., Nagai K., Tarakhovsky A., Okada M. (2000) Nature 404, 999–1003 [DOI] [PubMed] [Google Scholar]
- 6. Brdicka T., Imrich M., Angelisová P., Brdicková N., Horváth O., Spicka J., Hilgert I., Lusková P., Dráber P., Novák P., Engels N., Wienands J., Simeoni L., Osterreicher J., Aguado E., Malissen M., Schraven B., Horejsí V. (2002) J. Exp. Med. 196, 1617–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Janssen E., Zhu M., Zhang W., Koonpaew S., Zhang W. (2003) Nat. Immunol. 4, 117–123 [DOI] [PubMed] [Google Scholar]
- 8. Brdicková N., Brdicka T., Angelisová P., Horváth O., Spicka J., Hilgert I., Paces J., Simeoni L., Kliche S., Merten C., Schraven B., Horejsí V. (2003) J. Exp. Med. 198, 1453–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hur E. M., Son M., Lee O. H., Choi Y. B., Park C., Lee H., Yun Y. (2003) J. Exp. Med. 198, 1463–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhu M., Janssen E., Leung K., Zhang W. (2002) J. Biol. Chem. 277, 46151–46158 [DOI] [PubMed] [Google Scholar]
- 11. Marie-Cardine A., Kirchgessner H., Bruyns E., Shevchenko A., Mann M., Autschbach F., Ratnofsky S., Meuer S., Schraven B. (1999) J. Exp. Med. 189, 1181–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bruyns E., Marie-Cardine A., Kirchgessner H., Sagolla K., Shevchenko A., Mann M., Autschbach F., Bensussan A., Meuer S., Schraven B. (1998) J. Exp. Med. 188, 561–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu Y., Zhang W. (2008) J. Leukoc. Biol. 84, 842–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Horejsí V. (2004) Immunol. Lett. 92, 43–49 [DOI] [PubMed] [Google Scholar]
- 15. Horejsí V., Otáhal P., Brdicka T. (2010) FEBS J. 277, 4383–4397 [DOI] [PubMed] [Google Scholar]
- 16. Fuller D. M., Zhang W. (2009) Immunol. Rev. 232, 72–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Koelsch U., Schraven B., Simeoni L. (2008) J. Immunol. 181, 5930–5939 [DOI] [PubMed] [Google Scholar]
- 18. Murata Y., Doi T., Taniguchi H., Fujiyoshi Y. (2005) Biochem. Biophys. Res. Commun. 327, 183–191 [DOI] [PubMed] [Google Scholar]
- 19. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001) J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 20. Hobbs S., Jitrapakdee S., Wallace J. C. (1998) Biochem. Biophys. Res. Commun. 252, 368–372 [DOI] [PubMed] [Google Scholar]
- 21. Lessard J., Aicha S. B., Fournier A., Calvo E., Lavergne E., Pelletier M., Labrie C. (2007) Prostate 67, 808–819 [DOI] [PubMed] [Google Scholar]
- 22. Qiao L., Schürmann G., Betzler M., Meuer S. C. (1991) Gastroenterology 101, 1529–1536 [DOI] [PubMed] [Google Scholar]
- 23. Fraser J. D., Goldsmith M. A., Weiss A. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 7133–7137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Otáhal P., Angelisová P., Hrdinka M., Brdicka T., Novák P., Drbal K., Horejsí V. (2010) J. Immunol. 184, 3689–3696 [DOI] [PubMed] [Google Scholar]
- 25. Wan J., Roth A. F., Bailey A. O., Davis N. G. (2007) Nat. Protoc. 2, 1573–1584 [DOI] [PubMed] [Google Scholar]
- 26. Finn R. D., Mistry J., Tate J., Coggill P., Heger A., Pollington J. E., Gavin O. L., Gunasekaran P., Ceric G., Forslund K., Holm L., Sonnhammer E. L., Eddy S. R., Bateman A. (2010) Nucleic Acids Res. 38, D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen H. I., Sudol M. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 7819–7823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Saras J., Heldin C. H. (1996) Trends Biochem. Sci. 21, 455–458 [DOI] [PubMed] [Google Scholar]
- 29. Brannetti B., Helmer-Citterich M. (2003) Nucleic Acids Res. 31, 3709–3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Round J. L., Tomassian T., Zhang M., Patel V., Schoenberger S. P., Miceli M. C. (2005) J. Exp. Med. 201, 419–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bonifacino J. S., Traub L. M. (2003) Annu. Rev. Biochem. 72, 395–447 [DOI] [PubMed] [Google Scholar]
- 32. Jadot M., Canfield W. M., Gregory W., Kornfeld S. (1992) J. Biol. Chem. 267, 11069–11077 [PubMed] [Google Scholar]
- 33. Gao M., Labuda T., Xia Y., Gallagher E., Fang D., Liu Y. C., Karin M. (2004) Science 306, 271–275 [DOI] [PubMed] [Google Scholar]
- 34. Jain J., Valge-Archer V. E., Rao A. (1992) J. Immunol. 148, 1240–1250 [PubMed] [Google Scholar]
- 35. Castellanos M. C., Muñoz C., Montoya M. C., Lara-Pezzi E., López-Cabrera M., de Landázuri M. O. (1997) J. Immunol. 159, 5463–5473 [PubMed] [Google Scholar]
- 36. Roose J. P., Mollenauer M., Ho M., Kurosaki T., Weiss A. (2007) Mol. Cell. Biol. 27, 2732–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Krammer P. H., Arnold R., Lavrik I. N. (2007) Nat. Rev. Immunol. 7, 532–542 [DOI] [PubMed] [Google Scholar]