Abstract
Upon mitochondrial depolarization, Parkin, a Parkinson disease-related E3 ubiquitin ligase, translocates from the cytosol to mitochondria and promotes their degradation by mitophagy, a selective type of autophagy. Here, we report that in addition to mitophagy, Parkin mediates proteasome-dependent degradation of outer membrane proteins such as Tom20, Tom40, Tom70, and Omp25 of depolarized mitochondria. By contrast, degradation of the inner membrane and matrix proteins largely depends on mitophagy. Furthermore, Parkin induces rupture of the outer membrane of depolarized mitochondria, which also depends on proteasomal activity. Upon induction of mitochondrial depolarization, proteasomes are recruited to mitochondria in the perinuclear region. Neither proteasome-dependent degradation of outer membrane proteins nor outer membrane rupture is required for mitophagy. These results suggest that Parkin regulates degradation of outer and inner mitochondrial membrane proteins differently through proteasome- and mitophagy-dependent pathways.
Keywords: Autophagy, Electron Microscopy (EM), Mitochondria, Parkinsons Disease, Proteasome, Mitophagy, Parkin
Introduction
Parkinson disease is a progressive neurodegenerative disease that is characterized by postural changes, resting tremor, muscle rigidity, and weakness (1, 2). These symptoms are mainly caused by loss of dopaminergic neurons in the substantia nigra, and mitochondrial dysfunction appears to be the primary pathogenic event. Indeed, many of the products of Parkinson disease-related genes such as α-synuclein/PARK1/4, PARKIN/PARK2, PINK1/PARK6, DJ-I/PARK7, and OMI/HTRA2 are physically and functionally linked to mitochondria (3, 4).
Parkin is a RING domain-containing E3 ubiquitin ligase, and its mutation causes autosomal recessive juvenile Parkinson disease (5). Recent studies have revealed that Parkin is important for mitochondrial quality control through degradation of damaged mitochondria. Narendra et al. (6) first demonstrated that Parkin translocates from the cytosol to depolarized mitochondria and triggers elimination of these mitochondria by autophagy, which is known as mitophagy. Targeting of Parkin to mitochondria requires PTEN-induced putative kinase 1 (Pink1),3 another Parkinson disease-associated gene product (7–14). Pink1 is an extremely unstable mitochondrial protein, but it is stabilized upon mitochondrial depolarization and subsequently recruits Parkin.
Autophagy is a membrane-mediated intracellular degradation process. A portion of cytoplasm is first enclosed by the double-membraned autophagosome, and the autophagosome then fuses with a lysosome to degrade the enclosed materials. Although autophagy has been thought to be mainly non-selective, recent studies have revealed that the autophagosomal membrane can recognize some specific proteins and organelles. Parkin-mediated autophagy of damaged mitochondria is one of the best examples of selective autophagy. However, the precise role of Parkin in the induction of mitophagy has not been fully elucidated. To date, several mitochondrial proteins, voltage-dependent anion channel 1 (VDAC1) (8), mitofusin (a mitochondrial pro-fusion factor) (11, 14, 15), Bcl-2 (16), and Drp1 (17), have been shown to be ubiquitinated by Parkin. Ubiquitination of VDAC1 may recruit the autophagy adaptor p62, which interacts with microtubule-associated protein light chain 3 (LC3) on the autophagosomal membrane (8); however, the requirement of p62 remains controversial (18–21). Ubiquitination of mitofusin may affect mitochondrial fission or fusion, which would facilitate mitophagy (11, 14, 15).
Mitochondrial degradation by autophagy has been extensively studied, whereas the involvement of the proteasome in Parkin-mediated mitochondrial degradation is less clear. As the proteasome has been found on mitochondria (22), it is possible that it has a more direct role in mitochondrial protein degradation together with Parkin.
In this study, we determined the roles of mitophagy and proteasomal degradation in Parkin-dependent degradation of depolarized mitochondria and found that proteins in the outer mitochondrial membrane (OMM) and the intermembrane space can be degraded by the proteasome, whereas those in the inner mitochondrial membrane (IMM) and the mitochondrial matrix are degraded mainly by mitophagy in cultured fibroblasts. Furthermore, we observed that Parkin induces rupture of the OMM, which is also dependent on the proteasome. These results reveal the novel Parkin-proteasome pathway and also provide new insights into maintenance of mitochondrial morphology.
EXPERIMENTAL PROCEDURES
Plasmids
HA epitope-tagged Parkin (12), enhanced green fluorescent protein (EGFP)-tagged Omp25 (23), and Su9-GFP (24) were subcloned into the pMXs-IP vector (25).
Antibodies and Reagents
Rabbit polyclonal antibodies against Tom70 (26), Tom40 (27), Tom20 (28), Tim23 (29), Tim17 (29), Tim44 (29), proteasome subunit α7 (30), and LC3 (31) have been previously described. We purchased mouse monoclonal antibodies against cytochrome c (BD Biosciences), complex III (C-III) core I (Invitrogen), and α-tubulin (DM 1A) (Sigma-Aldrich) and rabbit polyclonal antibodies against Tom20 (Santa Cruz Biotechnology). Alexa Fluor 488-conjugated anti-mouse IgG and Alexa Fluor 568-conjugated anti-rabbit IgG secondary antibodies were purchased from Invitrogen. Horseradish peroxidase-conjugated anti-mouse and anti-rabbit IgG antibodies were purchased from Jackson ImmunoResearch Laboratories.
Puromycin dehydrochloride, bafilomycin A1, actinomycin D, carbonyl cyanide m-chlorophenylhydrazone (CCCP), and etoposide were purchased from Sigma-Aldrich. Lactacystin and MG132 were purchased from the Peptide Institute Inc.
Cell Culture and Transfection
Mouse Embryonic Fibroblasts (MEFs) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, l-glutamine, and penicillin/streptomycin in an atmosphere of 5% CO2 at 37 °C. FIP200−/− (FIP200 knock-out (KO)) (32) and Atg5−/− (Atg5 KO) (33) MEFs were previously generated. DNA transfection was performed according to the manufacturer's instructions using the FuGENE 6 transfection reagent. Plasmid DNA was transfected into Plat-E packaging cells and cultured for 72 h (25). The supernatant containing retrovirus (cultured medium) was used for transfection. MEFs were incubated in the culture medium containing retrovirus for 4 h in the presence of 4–8 μm Polybrene. MEFs infected with HA-Parkin or empty virus were selected in the presence of 2 μm puromycin dehydrochloride. MEFs stably expressing HA-Parkin or empty vector were used for immunocytochemistry, immunoblot analysis, and electron microscopy. These MEFs were further infected with GFP-Omp25- or Su9-GFP-coding retroviruses and used for flow cytometry analysis.
Flow Cytometry
Cells were harvested with 0.05% Trypsin-EDTA (Invitrogen) and diluted with phosphate-buffered saline (PBS). Analysis of 1 × 104 cells/sample was performed on a FACSCalibur HG (BD Biosciences). Data were analyzed using BD CellQuest Pro (BD Biosciences). Green fluorescent protein (GFP) fluorescence intensity was normalized to that of CCCP-untreated samples (set to 100%), and the relative intensity of GFP in the CCCP-treated group was calculated accordingly.
Immunocytochemistry
MEFs grown on gelatin-coated coverslips were washed twice with PBS and fixed in 4% paraformaldehyde in PBS for 10 min at room temperature. Cells were then washed twice with PBS and permeabilized with 0.1% Triton X-100 in PBS and were again washed twice with PBS. To retrieve C-III core 1 antigen, the coverslips (with cells) were put into hot ion exchange water, boiled in a microwave, and left for 5 min. After blocking with 3% bovine serum albumin in PBS for 30 min at room temperature, the cells were incubated with primary antibodies for 1 h at room temperature. Cells were washed four times with PBS followed by incubation with secondary antibodies for 1 h at room temperature. Cells were then washed five times with PBS and mounted with SlowFade Gold (Invitrogen). Samples were analyzed with a fluorescence microscope (IX81; Olympus) equipped with a charge-coupled device camera (ORCA ER; Hamamatsu Photonics). A 60× PlanAPO oil immersion lens (1.42 NA; Olympus) was used. Images were acquired using MetaMorph image analysis software version 7.1.5.0 (Molecular Devices).
Western Blotting
MEFs were washed with ice-cold PBS, harvested in cold PBS, and centrifuged at 3,000 rpm for 5 min. Cells were lysed in a lysis buffer (50 mm Tris-HCl, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitor mixture (Complete EDTA-free protease inhibitor; Roche Applied Science)). The lysate was clarified by centrifugation at 15,000 rpm for 5 min at 4 °C and was mixed with 6× sample buffer. Samples were subsequently separated by SDS-PAGE and transferred to Immobilon-P transfer membrane (Millipore). Immunoblot analysis was performed with the indicated antibodies and visualized with Immobilon Western (Millipore). The signal intensities were analyzed using an imaging analyzer (LAS-3000mini; Fujifilm) and Multi Gauge software (version 3.0; Fujifilm). Contrast and brightness adjustment was applied using Photoshop 7.0.1 (Adobe).
Electron Microscopy
MEFs were cultured on collagen-coated plastic coverslips. They were fixed in 2.5% glutaraldehyde in 0.1 m sodium phosphate buffer, pH 7.4 (phosphate buffer) for 2 h. The cells were washed in the same buffer three times and were post-fixed in 1% osmium tetroxide in 0.1 m phosphate buffer for 1 h and then dehydrated and embedded in Epon 812 according to a standard procedure (34). Ultrathin sections were stained with uranyl acetate and lead citrate and observed under a Hitachi H-7100 electron microscope.
For immunoelectron microscopy analysis of endogenous proteasome, cells were fixed with 4% paraformaldehyde in 0.1 m sodium phosphate buffer (pH 7.4) for 2 h at room temperature. The pre-embedding silver enhancement immunogold method was performed as previously described (35).
RESULTS
OMM Proteins Are Degraded Mainly in an Autophagy-independent Manner in Depolarized Mitochondria
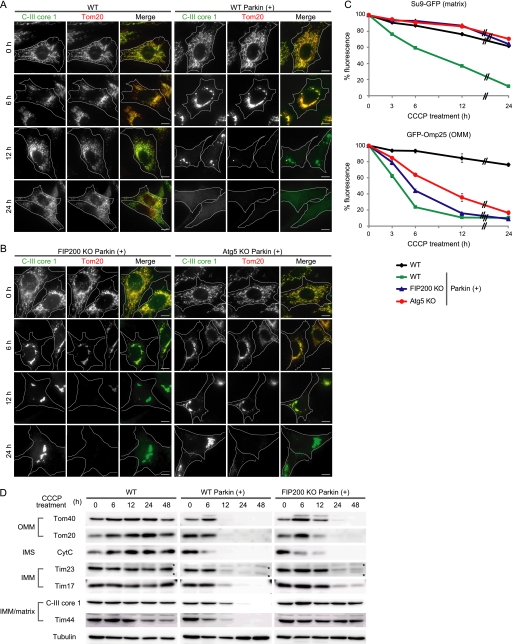
To analyze the role of Parkin in turnover of damaged mitochondria, we utilized a well established experimental system (6, 12, 19). We introduced exogenous Parkin into MEFs, in which expression of endogenous Parkin is negligible, and treated cells with the mitochondrial uncoupling reagent CCCP. In these Parkin-transfected cells, mitochondria translocated to the perinuclear region and were degraded during CCCP treatment for 24 h, whereas mitochondria remained present throughout the cytoplasm in Parkin-untransfected cells (Fig. 1A) (6, 19). In Parkin-transfected cells, we found that the signal of the OMM protein marker Tom20 disappeared more rapidly than that of the IMM/matrix protein (associating with the matrix side of the IMM) marker core 1 subunit of C-III.
FIGURE 1.
OMM proteins are degraded mainly in an autophagy-independent manner in depolarized mitochondria. A and B, wild-type (WT) MEFs with and without stable Parkin expression (A) and FIP200 KO and Atg5 KO MEFs with Parkin expression (B) were treated with 20 μm CCCP for different time periods as indicated. Cells were immunostained for C-III core 1 (an IMM/matrix protein) and Tom20 (an OMM protein). Signal color is indicated by the colored text. Scale bars, 10 μm. C, wild-type MEFs (with and without exogenous Parkin expression) and FIP200 KO and Atg5 KO MEFs (with exogenous Parkin expression) expressing either Su9-GFP (a matrix protein) or GFP-Omp25 (an OMM protein) were treated with 20 μm CCCP for different time periods as indicated. The GFP fluorescence was quantified by flow cytometry. Data represent mean ± S.E. of three independent experiments. D, wild-type MEFs (with and without exogenous Parkin expression) and FIP200 KO MEFs (with exogenous Parkin expression) were treated with 20 μm CCCP for different time periods as indicated. The cells were analyzed by SDS-PAGE and subsequent immunoblotting with antibodies against Tom40 and Tom20 (OMM proteins), cytochrome c (CytC) (an intermembrane space (IMS) protein), Tim23 and Tim17 (IMM proteins), and C-III core 1 and Tim44 (IMM/matrix proteins), as well as α-tubulin (a loading control). Asterisks indicate nonspecific immunoreactive bands.
We next tested the involvement of autophagy in this process, which has been previously demonstrated (36, 37). The disappearance of C-III core 1 was greatly suppressed in autophagy-deficient FIP200 KO (34) and Atg5 KO MEFs (33) transfected with Parkin, and the C-III core 1 signal was easily detected even 24 h after CCCP treatment (Fig. 1B). By contrast, the Tom20 signals were affected only slightly and lost at 24 h even in these autophagy-deficient cells. Tom70, another OMM protein, showed a similar pattern (supplemental Fig. S1). These observations raised the possibility that there may be a difference in the protein degradation mechanism between the OMM and IMM.
To test this hypothesis in a quantitative manner, we established a flow cytometry-based assay. We generated MEF lines stably expressing GFP fused to the mitochondrial matrix-targeting signal of subunit 9 of F0-ATPase (Su9-GFP) (24) and GFP-Omp25, as fluorescent mitochondrial matrix and OMM markers, respectively. As expected, the Su9-GFP and GFP-Omp25 signals were largely unaffected in Parkin-untransfected MEFs but significantly decreased in Parkin-transfected wild-type MEFs in a time-dependent manner following CCCP treatment (Fig. 1C). The CCCP-induced decrease in the Su9-GFP signal was markedly inhibited in FIP200 KO and Atg5 KO MEFs. However, decrease in the GFP-Omp25 signal was ameliorated only slightly in FIP200 KO and Atg5 KO MEFs (Fig. 1C). At 24 h after CCCP treatment, most GFP-Omp25 signals were lost even in these autophagy-deficient cells. These data suggest that OMM protein degradation depends only partially on autophagy, whereas matrix protein degradation almost completely relies on autophagy.
We further confirmed these findings by immunoblot analysis with additional mitochondrial proteins. The expression levels of Tom40, Tom20, cytochrome c, Tim23, Tim17, and C-III core 1 were almost unchanged during 48-h CCCP treatment in the absence of exogenous Parkin (Fig. 1D). In Parkin-transfected wild-type MEFs, the OMM proteins Tom40 and Tom20 and the intermembrane space protein cytochrome c were almost undetectable 12 h after CCCP treatment (Fig. 1D). By contrast, small but significant amounts of the IMM proteins Tim23 and Tim17 and the IMM/matrix protein C-III core 1 and Tim44 were still detected at 12 h. This difference was observed more clearly under autophagy-deficient conditions. In FIP200 KO cells, degradation of Tom40, Tom20, and cytochrome c was retarded, but these proteins were almost undetectable at 24 h after CCCP treatment (Fig. 1D). Degradation of Tim23, Tim17, C-III core 1, and Tim44 was more markedly suppressed; these proteins were detectable even after 48-h CCCP treatment. In particular, degradation of the IMM/matrix proteins C-III core 1 and Tim44 was inhibited almost completely in FIP200 KO cells. Thus, degradation of IMM and matrix proteins seems to be more dependent on autophagy than that of OMM and intermembrane space proteins.
These data, which were collected using three different methods for several different mitochondrial proteins, suggest that mitochondrial proteins are differentially degraded in CCCP-treated cells. OMM and intermembrane space proteins appear to be degraded more rapidly, mainly through an autophagy-independent manner, than IMM and matrix proteins, which are degraded primarily by mitophagy.
CCCP Induces OMM Rupture in a Parkin-dependent and Autophagy-independent Manner
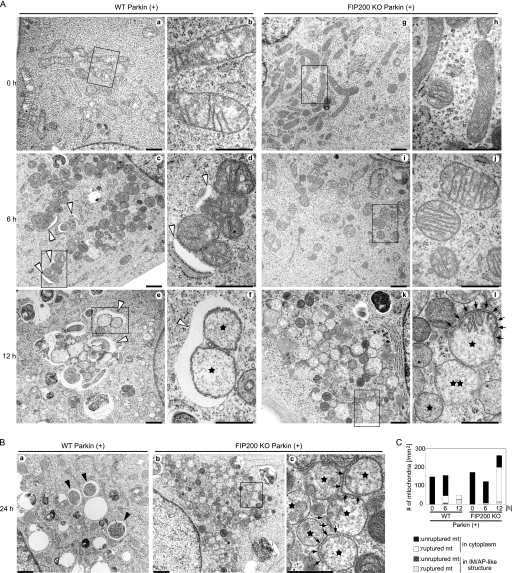
Next, using transmission electron microscopy, we examined the morphologic changes that occur during degradation of mitochondrial proteins. CCCP induced mitochondrial fragmentation irrespective of Parkin expression (Fig. 2A, and supplemental Fig. S2, A and C). At 6 h, the fragmented mitochondria clustered in the perinuclear region and became enclosed by isolation membranes in Parkin-transfected wild-type cells (Fig. 2A, panels c and d, and supplemental Fig. S2A, panels a–c). At 12 h following CCCP treatment, large numbers of mitochondria were enclosed by autophagosomes and degraded (Fig. 2A, panels e and f, and 2C). At this time point, the majority of autophagosomes contained cytoplasm without mitochondria (supplemental Fig. S2A, panel d, and 2B). At 24 h, almost no mitochondria remained, and only autophagosomes containing cytoplasm without mitochondria were observed (Fig. 2B, panel a). Most of the isolation membranes that were enclosing mitochondria were associated with the rough endoplasmic reticulum (Fig. 2A, panel d, and supplemental Fig. S2A), as frequently observed in canonical autophagy (38–40). It is, therefore, likely that the conventional isolation membrane is involved in mitophagy. Either a single mitochondrion or a cluster containing several (sometimes more than 10) mitochondria was enclosed by an autophagosome (Fig. 2A, panels c–f, and supplemental Fig. S2A). Occasionally, several distinct isolation membranes appeared to enclose a single mitochondrial cluster (supplemental Fig. S2A, panel b). In agreement with previous reports, CCCP-induced mitochondrial clustering and mitophagy were not observed in Parkin-untransfected cells (supplemental Fig. S2C). Typical mitophagy was not observed in Parkin-expressing FIP200 KO cells (Fig. 2A), although some aberrant membrane structures were generated occasionally (supplemental Fig. S2A, panels g and h, and S2B).
FIGURE 2.
CCCP induces OMM rupture in a Parkin-dependent and autophagy-independent manner. A, WT (panels a–f) and FIP200 KO (panels g–l) MEFs stably expressing Parkin were incubated for 0 h (panels a, b, g, and h), 6 h (panels c, d, i, and j), or 12 h (panels e, f, k, and l) with 20 μm CCCP. These cells were fixed and subjected to conventional electron microscopy analysis. Rectangles show enlarged areas in panels b, d, f, h, j, and l. Stars indicate mitochondria with ruptured OMM. Double stars indicate mitochondria with ruptured OMM and IMM. Mitochondrial clusters are enclosed by isolation membranes (white arrowheads) (panels c, d, e, and f). Two ruptured mitochondria are enclosed by a single isolation membrane (panel f). Arrows indicate residual OMM (panel l). Scale bars, 1 μm (panels a, c, e, g, i, and k) and 500 nm (panels b, d, f, h, j, and l). B, wild-type (panel a) and FIP200 KO (panels b and c) MEFs stably expressing Parkin were incubated for 24 h with 20 μm CCCP and subjected to electron microscopy. Black arrowheads indicate autophagosomes without mitochondria. After 24 h of CCCP treatment, almost all mitochondria disappeared in wild-type cells, but ruptured mitochondria (indicated by stars) accumulated in FIP200 KO cells. Arrows indicate residual OMM (panel c). Scale bars, 1 μm (panels a and b) and 500 nm (panel c). C, wild-type MEFs stably expressing Parkin were incubated with 20 μm CCCP for different time periods as indicated. The number of ruptured and unruptured mitochondria (mt) in the cytoplasm or inside autophagosomes was counted (per mm2) from at least eight randomly selected cells. IM, isolation membranes; AP, autophagosomes.
Besides these mitophagy-related membrane organizations, we observed morphological changes in mitochondria themselves. The CCCP treatment caused mitochondrial swelling, rupture of the OMM, and loss of cristae structure. These changes could be observed at 6 h and became prominent at 12 h (Fig. 2A, panels e and f, and 2C). Rupture of the OMM was Parkin-dependent because it was not observed in Parkin-untransfected wild-type and FIP200 KO MEFs even 24 h after CCCP treatment (supplemental Fig. S2C). The swollen mitochondria with ruptured OMM accumulated in Parkin-transfected FIP200 KO cells, probably because these mitochondria were not eliminated by mitophagy (Fig. 2A, panels k and l, 2B, panels b and c, and 2C). Rupture of the OMM was often partial; residual OMM fragments were frequently observed on ruptured mitochondria. Portions of mitochondria with OMM retained cristae inside (Fig. 2A, panel l, arrows). By contrast, portions that lost the OMM contained destructed cristae (Fig. 2A, panel l). This suggests that the OMM is important for maintenance of the cristae structure. On these ruptured mitochondria, isolation membranes seemed to appear on the mitochondrial surface that had residual OMM (supplemental Fig. S2A, panel f, arrows), suggesting that some adaptor protein(s) on the OMM such as p62 may be involved in mitochondrial recognition (8, 18, 19). Occasionally, even the IMM appeared to be ruptured (Fig. 2A, panel l, double star). Both unruptured and ruptured mitochondria could be found in autophagosomes, suggesting that OMM rupture is not a prerequisite for mitophagy (Fig. 2, A and C). All these data suggest that CCCP induces OMM rupture and probably the subsequent destruction of cristae in a Parkin-dependent, but autophagy-independent manner.
OMM Protein Degradation Is Inhibited by Proteasome Inhibitors
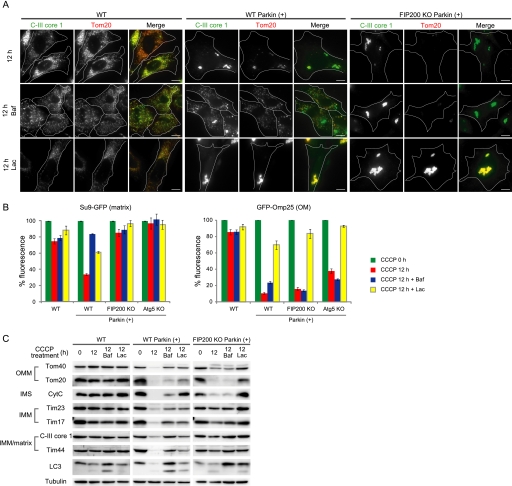
We next determined the mechanism underlying OMM protein degradation. Immunocytochemical analysis showed that bafilomycin A1, a specific inhibitor of vacuolar-type H+-ATPase, had only a weak inhibitory effect on CCCP-induced loss of Tom20 and Tom70 signals in Parkin-transfected cells (bafilomycin A1 treatment causes redistribution of mitochondria throughout cells, suggesting that mitochondria are not subjected to aggregation once they are enclosed by autophagosomes) (Fig. 3A and supplemental Fig. S3A). By contrast, Tom20 and Tom70 signals in both wild-type and FIP200 KO cells markedly recovered following treatment with the proteasomal inhibitor lactacystin, although mitochondrial clustering was not inhibited (Fig. 3A and supplemental Fig. S3A). MG132, another proteasomal inhibitor, similarly affected the level of Tom20 and Tom70 expression (supplemental Fig. S3B, and data not shown). Lactacystin and MG132 had no effect on degradation of these proteins in Parkin-untransfected cells in this experiment (Fig. 3A and supplemental Fig. S3).
FIGURE 3.
OMM protein degradation is inhibited by proteasome inhibition. A, WT and FIP200 KO MEFs (with and without exogenous Parkin expression) were treated with 20 μm CCCP for 12 h in the presence or absence of 0.2 μm bafilomycin A1 (Baf) or 5 μm lactacystin (Lac). Cells were immunostained for C-III core 1 and Tom20. Signal color is indicated by the colored text. Scale bars, 10 μm. B, degradation of GFP-Omp25 and Su9-GFP was analyzed by flow cytometry as in Fig. 1C. Wild-type and autophagy-deficient (FIP200 KO and Atg5 KO) MEFs were treated with 20 μm CCCP for 12 h in the presence or absence of 0.2 μm bafilomycin A1 or 5 μm lactacystin. Data represent mean ± S.E. of three independent experiments. C, wild-type and FIP200 KO MEFs (with and without exogenous Parkin expression) were treated with 20 μm CCCP in the presence or absence of 0.2 μm bafilomycin A1 or 5 μm lactacystin for 12 h. The cells were analyzed by immunoblotting using different antibodies as indicated. IMS, intermembrane space; CytC, cytochrome c.
This unexpected effect of lactacystin was also observed in the flow cytometry-based degradation assay. CCCP-induced GFP-Omp25 degradation was significantly inhibited by treatment with lactacystin (Fig. 3B) or MG132 (data not shown) and almost fully restored in autophagy-deficient cells.
Finally, we confirmed the effect of lactacystin by immunoblot analysis. CCCP-induced degradation of Tom40, Tom20, and cytochrome c was partially suppressed by bafilomycin A1 in wild-type cells, but not in autophagy-defective FIP200 KO cells (Fig. 3C). The lysosome-inhibitory effect of bafilomycin A1 was confirmed; bafilomycin A1 increased the levels of the autophagosome marker LC3-II by suppression of its degradation in wild-type cells, but not in FIP200 KO cells. As expected, lactacystin showed inhibitory effects on degradation of Tom40, Tom20, and cytochrome c. In FIP200 KO cells, the effect of lactacystin was prominent; degradation of Tom20 and cytochrome c was almost completely suppressed (Fig. 3C). Although less efficient, degradation of Tom40 was also suppressed by lactacystin. These data suggest that the proteasome is involved in Parkin-dependent degradation of OMM proteins induced by CCCP.
Proteasome Inhibition Suppresses OMM Rupture but Not Mitophagy
In agreement with our results using autophagy-deficient cells, degradation of IMM and matrix proteins was inhibited by treatment with bafilomycin A1 in wild-type cells (Fig. 3, B and C). However, we found that lactacystin also mildly inhibited loss of the matrix model protein Su9-GFP in the flow cytometry-based degradation assay (Fig. 3B). This effect was confirmed by immunoblot analysis of the IMM and IMM/matrix proteins; lactacystin inhibited degradation of Tim23, Tim17, C-III core 1, and Tim44 in Parkin-transfected wild-type cells (Fig. 3C). There are two possible explanations for these findings; proteasome inhibition may suppress mitophagy, or it may inhibit mitophagy-independent degradation of IMM and matrix proteins. We assumed that the latter was more likely because the inhibitory effect of lactacystin on IMM and matrix protein degradation was still observed in FIP200 KO MEFs (Fig. 3, B and C).
We thus determined whether or not mitophagy itself was inhibited by lactacystin using electron microscopy. We observed similar or slightly increased numbers of autophagosomes with and without mitochondria in lactacystin-treated cells as compared with untreated cells (Fig. 4, A and C). Enclosed mitochondria were indeed degraded in autolysosomes (Fig. 4A, panel g, double stars). Thus, lactacystin does not inhibit mitophagy. In this experiment, we also found that lactacystin restored the integrity of the OMM. Although 12-h CCCP treatment caused OMM rupture in almost all mitochondria, lactacystin suppressed the rupture almost completely both in wild-type MEFs (Fig. 4A, panels c–f, and 4B) and in FIP200 KO MEFs (Fig. 4A panels j and k, and 4B). The structure of the cristae was restored accordingly. Disruption of the IMM, which was observed in some CCCP-treated mitochondria (Fig. 2A, panel l), was not detected in lactacystin-treated cells. Lactacystin did not inhibit mitochondrial fragmentation and clustering (Fig. 4A). These data suggest that proteasomal activity is critical for OMM rupture, which would eventually cause mitophagy-independent protein degradation of the intermembrane space, IMM, and matrix or whole mitochondrial loss.
FIGURE 4.
Proteasome inhibition suppresses OMM rupture but not mitophagy. A, wild-type (panels a–g) and FIP200 KO (panels h–k) MEFs stably expressing Parkin were incubated with 20 μm CCCP for 12 h with (panels c–g, j, and k) or without (panels a, b, h, and i) 5 μm lactacystin (Lac) and then subjected to electron microscopy. Rectangles show enlarged areas in panels b, d, i, and k. Autophagosomes without mitochondria (black arrowheads) and with mitochondria (white arrowheads) are shown. Single stars indicate mitochondria with ruptured OMM (panels b and i). In panel g, double stars indicate a degrading mitochondrion inside an autophagic vacuole. In panels b and i, arrows indicate residual OMM. Scale bars, 1 μm (panels a, c, h, and j) and 500 nm (panels b, d, e–g, i, and k). B, the ratio of ruptured and unruptured mitochondria (mt) in isolation membrane (IM)/autophagosome (AP)-like structures or in the cytoplasm was calculated from at least eight randomly selected cells. Lac, lactacystin. C, the number of IM/AP-like structures with (white columns) and without (black columns) mitochondria was counted from at least eight randomly selected cells. For bafilomycin A1 (Baf) treatment, cells were incubated with 20 μm CCCP and 0.2 μm bafilomycin A1 for 12 h. Selected images of wild-type cells are shown in supplemental Fig. S4.
Bafilomycin A1 treatment caused accumulation of large autophagic vacuoles containing mitochondria in wild-type cells, but not in FIP200 KO cells (Fig. 4C and supplemental Fig. S4). Consistent with the results obtained by fluorescence microscopy (Fig. 3A), the bafilomycin A1 treatment impaired mitochondrial aggregation (supplemental Fig. S4e). Bafilomycin A1 treatment did not inhibit OMM rupture of mitochondria in the cytoplasm (supplemental Fig. S4).
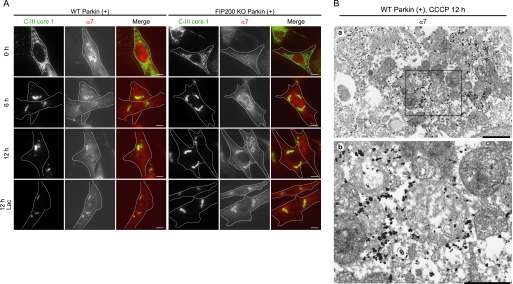
Proteasomes Are Recruited to Depolarized Mitochondria
OMM protein degradation occurs in two possible locations. First, upon CCCP-induced Parkin translocation, the OMM proteins may be extracted from the mitochondrial membrane and delivered to the proteasomes, or second, the proteasomes may be recruited to damaged mitochondria. Our observation that proteasome inhibitor treatments caused accumulation of Tom20 and Tom70 on the mitochondrial membrane in CCCP-treated cells (Fig. 3A, and supplemental Fig. S3) suggests that the latter possibility is more likely than the former. We tested this hypothesis by co-staining C-III core 1 and the proteasome α7 subunit. Under normal conditions, the proteasomes were diffusely present in both the nucleus and the cytoplasm (Fig. 5A). However, following CCCP treatment, proteasomes translocated to the perinuclear regions and co-localized with clustered mitochondria in Parkin-transfected wild-type and FIP200 KO MEFs (Fig. 5A). Translocation was also observed in lactacystin-treated cells (Fig. 5A). Immunoelectron microscopy also revealed the presence of the proteasome on mitochondria, most of which are morphologically altered (Fig. 5B). These data suggest that the proteasomes are recruited to depolarized mitochondria and degrade OMM proteins.
FIGURE 5.
Proteasomes are recruited to depolarized mitochondria. A, wild-type and FIP200 KO MEFs stably expressing Parkin were treated with 20 μm CCCP for different periods of time as indicated in the presence or absence of 5 μm lactacystin (Lac). The cells were immunostained for C-III core 1 and α7 subunit of the proteasome. Signal color is indicated by the colored text. Scale bars, 10 μm. B, wild-type MEFs stably expressing Parkin were treated with 20 μm CCCP for 12 h and subjected to immunoelectron microscopy using anti-proteasomal α7 subunit antibody. Scale bars, 1 μm (panel a) and 500 nm (panel b).
DISCUSSION
We have shown that Parkin mediates OMM protein degradation and rupture, which depend on proteasomal activity in depolarized mitochondria; Fig. 6 shows a model of the proposed mechanism. Parkin translocates to depolarized mitochondria and induces degradation of OMM proteins in a proteasome-dependent manner. The proteasome is also recruited to these depolarized mitochondria. This is consistent with recent studies that p97 (also known as valosin-containing protein, VCP) can be recruited to mitochondria and is involved in OMM protein degradation (41, 42). Massive degradation or specific degradation of some critical protein(s) causes OMM rupture, which would trigger degradation of intermembrane space proteins. OMM rupture exposes the IMM to the cytoplasmic environment and also causes vigorous morphological changes of the IMM structure, which may eventually lead to secondary degradation of IMM and matrix proteins. This model could explain why Tim23 and Tim17 are relatively more sensitive to degradation than C-III core 1 and Tim44, as Tim23 and Tim17, but not C-III core 1 and Tim44, are exposed to the intermembrane space (43, 44). These novel roles of Parkin are neither dependent on nor required for its well known role in mitophagy induction; we observed that whole mitochondria with either ruptured or unruptured mitochondria were degraded by mitophagy and that lactacystin did not inhibit mitochondrial sequestration by autophagosomes. Therefore, Parkin has three distinct functions: clustering of depolarized mitochondria, mitophagy induction, and OMM protein degradation and rupture.
FIGURE 6.
Proposed model of mitochondrial protein degradation and OMM rupture. Reduction in mitochondrial membrane potential (ΔΨm) causes fragmentation of mitochondria. Parkin is recruited to the OMM, resulting in clustering of mitochondria at the perinuclear region. Some OMM proteins (X) are ubiquitinated (Ub) directly or indirectly by Parkin and then degraded by the proteasome, which may eventually cause OMM rupture. Both unruptured and ruptured mitochondria are enclosed by autophagosomes and degraded by mitophagy. The step blocked by lactacystin is indicated.
During revision of this study, three independent groups reported that OMM proteins including mitofusin 1/2, Tom20, Tom70, and VDAC can be degraded in a Parkin- and proteasome-dependent manner (17, 41, 45). Two of them suggest that degradation of mitofusin or other OMM proteins could promote mitophagy (41, 45). These observations are different from that of our ultrastructural analysis suggesting that proteasomal degradation is not essential for mitophagy (Fig. 4). The exact reason behind this discrepancy is currently unknown, but this may be due to difference in experimental systems. Alternatively, it is still possible that although proteasomal degradation is not essential, it may be important for efficient mitophagy.
Our data and the data of other groups have shown that many OMM proteins could be degraded in a Parkin-dependent manner (8, 11, 14–17, 41, 45). It is unknown whether all these proteins are directly recognized and ubiquitinated by Parkin. It is known that the proteasome can degrade non-ubiquitinated proteins in trans, if these proteins are recruited to the proteasome by other mediator proteins (46–48). Because proteasome (Fig. 5) and p97 can be recruited to depolarized mitochondria (41), such an indirect mechanism may be involved. Alternatively, rupture of the OMM may induce degradation of other OMM proteins as secondary events.
Our results also emphasize that it is critically important to use IMM or matrix proteins as indicators of mitophagy; analysis of OMM proteins alone may merely represent proteasomal degradation instead of mitophagy. Quantification of the mitochondrial DNA copy number would be another ideal method for monitoring mitophagy (20).
Although the biological role of Parkin in OMM degradation and rupture is intriguing, its pathophysiological relevance is not clear. Similar OMM rupture has been observed in apoptotic cells (49–51), but we detected neither activation of caspase-3 nor nuclear fragmentation in these cells (data not shown). The involvement of the ubiquitin-proteasome system including other E3 ligases in OMM protein degradation and quality control has been reported (52–57). Parkin may also have a role in quality control of mitochondria through turnover of OMM proteins under pathophysiological conditions. If mitochondrial damage is massive, Parkin may trigger whole mitochondrial degradation by mitophagy. Such cooperation between the ubiquitin-proteasome system and mitophagy has also been observed in fission yeast; proteasome dysfunction causes mitophagy in G0 phase to reduce the level of reactive oxygen species (58). Although more studies will be required, particularly using primary neurons, Parkin could be a key factor that regulates cellular homeostasis through mitochondrial turnover in both an autophagy-dependent and a proteasome-dependent manner.
Supplementary Material
Acknowledgments
We thank Drs. N. Matsuda and K. Tanaka (Tokyo Metropolitan Institute of Medical Science) for the HA-Parkin plasmid, Dr. K. Mihara (Kyushu University) for the EGFP-Omp25 plasmid and Tom20, Tom40, and Tom70 antibodies, Dr. S. Murata (The University of Tokyo) for the proteasome α7 antibody, Dr. T. Kitamura (The University of Tokyo) for the retroviral vectors and Plat-E cells, and Dr. Jun-Lin Guan (University of Michigan) for FIP200 KO MEFs.
This work was supported in part by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology, Japan (to N. I. and N. M.), the Takeda Science Foundation (to N. M.), and the Funding Program for Next Generation World-Leading Researchers (to N. I. and N. M.).
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- Pink1
- PTEN-induced putative kinase 1
- PTEN
- phosphatase and tensin homolog
- Atg
- autophagy-related
- CCCP
- carbonyl cyanide m-chlorophenylhydrazone
- C-III
- complex III
- FIP200
- focal adhesion kinase family interacting protein of 200 kDa
- IMM
- inner mitochondrial membrane
- OMM
- outer mitochondrial membrane
- LC3
- microtubule-associated protein light chain 3
- MEF
- mouse embryonic fibroblast
- Su9
- subunit 9 of F0-ATPase
- Tim
- translocase of inner membrane
- Tom
- translocase of outer membrane
- VDAC1
- voltage-dependent anion channel 1.
REFERENCES
- 1. Dickson D. W., Braak H., Duda J. E., Duyckaerts C., Gasser T., Halliday G. M., Hardy J., Leverenz J. B., Del Tredici K., Wszolek Z. K., Litvan I. (2009) Lancet Neurol. 8, 1150–1157 [DOI] [PubMed] [Google Scholar]
- 2. Obeso J. A., Rodriguez-Oroz M. C., Goetz C. G., Marin C., Kordower J. H., Rodriguez M., Hirsch E. C., Farrer M., Schapira A. H., Halliday G. (2010) Nat. Med. 16, 653–661 [DOI] [PubMed] [Google Scholar]
- 3. Abou-Sleiman P. M., Muqit M. M., Wood N. W. (2006) Nat. Rev. Neurosci. 7, 207–219 [DOI] [PubMed] [Google Scholar]
- 4. Zhu J., Chu C. T. (2010) J. Alzheimers Dis. 20, Suppl. 2, S325–S334 [DOI] [PubMed] [Google Scholar]
- 5. Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., Yokochi M., Mizuno Y., Shimizu N. (1998) Nature 392, 605–608 [DOI] [PubMed] [Google Scholar]
- 6. Narendra D., Tanaka A., Suen D. F., Youle R. J. (2008) J. Cell Biol. 183, 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R. L., Kim J., May J., Tocilescu M. A., Liu W., Ko H. S., Magrané J., Moore D. J., Dawson V. L., Grailhe R., Dawson T. M., Li C., Tieu K., Przedborski S. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 378–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Geisler S., Holmström K. M., Skujat D., Fiesel F. C., Rothfuss O. C., Kahle P. J., Springer W. (2010) Nat. Cell Biol. 12, 119–131 [DOI] [PubMed] [Google Scholar]
- 9. Narendra D. P., Jin S. M., Tanaka A., Suen D. F., Gautier C. A., Shen J., Cookson M. R., Youle R. J. (2010) PLoS Biol. 8, e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kawajiri S., Saiki S., Sato S., Sato F., Hatano T., Eguchi H., Hattori N. (2010) FEBS Lett. 584, 1073–1079 [DOI] [PubMed] [Google Scholar]
- 11. Ziviani E., Tao R. N., Whitworth A. J. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 5018–5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C. A., Sou Y. S., Saiki S., Kawajiri S., Sato F., Kimura M., Komatsu M., Hattori N., Tanaka K. (2010) J. Cell Biol. 189, 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rakovic A., Grünewald A., Seibler P., Ramirez A., Kock N., Orolicki S., Lohmann K., Klein C. (2010) Hum. Mol. Genet. 19, 3124–3137 [DOI] [PubMed] [Google Scholar]
- 14. Gegg M. E., Cooper J. M., Chau K. Y., Rojo M., Schapira A. H., Taanman J. W. (2010) Hum. Mol. Genet. 19, 4861–4870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Poole A. C., Thomas R. E., Yu S., Vincow E. S., Pallanck L. (2010) PLoS One 5, e10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen D., Gao F., Li B., Wang H., Xu Y., Zhu C., Wang G. (2010) J. Biol. Chem. 285, 38214–38223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang H., Song P., Du L., Tian W., Yue W., Liu M., Li D., Wang B., Zhu Y., Cao C., Zhou J., Chen Q. (2011) J. Biol. Chem. 286, 11649–11658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ding W. X., Ni H. M., Li M., Liao Y., Chen X., Stolz D. B., Dorn G. W., 2nd, Yin X. M. (2010) J. Biol. Chem. 285, 27879–27890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee J. Y., Nagano Y., Taylor J. P., Lim K. L., Yao T. P. (2010) J. Cell Biol. 189, 671–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Okatsu K., Saisho K., Shimanuki M., Nakada K., Shitara H., Sou Y. S., Kimura M., Sato S., Hattori N., Komatsu M., Tanaka K., Matsuda N. (2010) Genes Cells 15, 887–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Narendra D., Kane L. A., Hauser D. N., Fearnley I. M., Youle R. J. (2010) Autophagy 6, 1090–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nakagawa T., Shirane M., Iemura S., Natsume T., Nakayama K. I. (2007) Genes Cells 12, 709–719 [DOI] [PubMed] [Google Scholar]
- 23. Horie C., Suzuki H., Sakaguchi M., Mihara K. (2002) Mol. Biol. Cell 13, 1615–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Eura Y., Ishihara N., Yokota S., Mihara K. (2003) J. Biochem. 134, 333–344 [DOI] [PubMed] [Google Scholar]
- 25. Kitamura T., Koshino Y., Shibata F., Oki T., Nakajima H., Nosaka T., Kumagai H. (2003) Exp. Hematol. 31, 1007–1014 [PubMed] [Google Scholar]
- 26. Suzuki H., Maeda M., Mihara K. (2002) J. Cell Sci. 115, 1895–1905 [DOI] [PubMed] [Google Scholar]
- 27. Suzuki H., Okazawa Y., Komiya T., Saeki K., Mekada E., Kitada S., Ito A., Mihara K. (2000) J. Biol. Chem. 275, 37930–37936 [DOI] [PubMed] [Google Scholar]
- 28. Iwahashi J., Yamazaki S., Komiya T., Nomura N., Nishikawa S., Endo T., Mihara K. (1997) J. Biol. Chem. 272, 18467–18472 [DOI] [PubMed] [Google Scholar]
- 29. Ishihara N., Mihara K. (1998) J. Biochem. 123, 722–732 [DOI] [PubMed] [Google Scholar]
- 30. Hamazaki J., Sasaki K., Kawahara H., Hisanaga S., Tanaka K., Murata S. (2007) Mol. Cell. Biol. 27, 6629–6638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hosokawa N., Hara Y., Mizushima N. (2006) FEBS Lett. 580, 2623–2629 [DOI] [PubMed] [Google Scholar]
- 32. Gan B., Peng X., Nagy T., Alcaraz A., Gu H., Guan J. L. (2006) J. Cell Biol. 175, 121–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. (2004) Nature 432, 1032–1036 [DOI] [PubMed] [Google Scholar]
- 34. Hara T., Takamura A., Kishi C., Iemura S., Natsume T., Guan J. L., Mizushima N. (2008) J. Cell Biol. 181, 497–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yoshimori T., Yamagata F., Yamamoto A., Mizushima N., Kabeya Y., Nara A., Miwako I., Ohashi M., Ohsumi M., Ohsumi Y. (2000) Mol. Biol. Cell 11, 747–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chu C. T. (2010) Hum. Mol. Genet. 19, R28–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tanaka A. (2010) FEBS Lett. 584, 1386–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Eskelinen E. L. (2005) Autophagy 1, 1–10 [DOI] [PubMed] [Google Scholar]
- 39. Ylä-Anttila P., Vihinen H., Jokitalo E., Eskelinen E. L. (2009) Autophagy 5, 1180–1185 [DOI] [PubMed] [Google Scholar]
- 40. Hayashi-Nishino M., Fujita N., Noda T., Yamaguchi A., Yoshimori T., Yamamoto A. (2009) Nat. Cell Biol. 11, 1433–1437 [DOI] [PubMed] [Google Scholar]
- 41. Tanaka A., Cleland M. M., Xu S., Narendra D. P., Suen D. F., Karbowski M., Youle R. J. (2010) J. Cell Biol. 191, 1367–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xu S., Peng G., Wang Y., Fang S., Karbowski M. (2011) Mol. Biol. Cell 22, 291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang Z., Huang L., Shulmeister V. M., Chi Y. I., Kim K. K., Hung L. W., Crofts A. R., Berry E. A., Kim S. H. (1998) Nature 392, 677–684 [DOI] [PubMed] [Google Scholar]
- 44. Rehling P., Brandner K., Pfanner N. (2004) Nat. Rev. Mol. Cell Biol. 5, 519–530 [DOI] [PubMed] [Google Scholar]
- 45. Chan N. C., Salazar A. M., Pham A. H., Sweredoski M. J., Kolawa N. J., Graham R. L., Hess S., Chan D. C. (2011) Hum. Mol. Genet., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Murakami Y., Matsufuji S., Kameji T., Hayashi S., Igarashi K., Tamura T., Tanaka K., Ichihara A. (1992) Nature 360, 597–599 [DOI] [PubMed] [Google Scholar]
- 47. Newman R. M., Mobascher A., Mangold U., Koike C., Diah S., Schmidt M., Finley D., Zetter B. R. (2004) J. Biol. Chem. 279, 41504–41511 [DOI] [PubMed] [Google Scholar]
- 48. Prakash S., Inobe T., Hatch A. J., Matouschek A. (2009) Nat. Chem. Biol. 5, 29–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Angermüller S., Künstle G., Tiegs G. (1998) J. Histochem. Cytochem. 46, 1175–1183 [DOI] [PubMed] [Google Scholar]
- 50. Feldmann G., Haouzi D., Moreau A., Durand-Schneider A. M., Bringuier A., Berson A., Mansouri A., Fau D., Pessayre D. (2000) Hepatology 31, 674–683 [DOI] [PubMed] [Google Scholar]
- 51. Zischka H., Larochette N., Hoffmann F., Hamöller D., Jägemann N., Lichtmannegger J., Jennen L., Müller-Höcker J., Roggel F., Göttlicher M., Vollmar A. M., Kroemer G. (2008) Anal. Chem. 80, 5051–5058 [DOI] [PubMed] [Google Scholar]
- 52. Yonashiro R., Ishido S., Kyo S., Fukuda T., Goto E., Matsuki Y., Ohmura-Hoshino M., Sada K., Hotta H., Yamamura H., Inatome R., Yanagi S. (2006) EMBO J. 25, 3618–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Karbowski M., Neutzner A., Youle R. J. (2007) J. Cell Biol. 178, 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Neutzner A., Benard G., Youle R. J., Karbowski M. (2008) Ann. N.Y. Acad. Sci. 1147, 242–253 [DOI] [PubMed] [Google Scholar]
- 55. Cohen M. M., Leboucher G. P., Livnat-Levanon N., Glickman M. H., Weissman A. M. (2008) Mol. Biol. Cell 19, 2457–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yonashiro R., Sugiura A., Miyachi M., Fukuda T., Matsushita N., Inatome R., Ogata Y., Suzuki T., Dohmae N., Yanagi S. (2009) Mol. Biol. Cell 20, 4524–4530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Livnat-Levanon N., Glickman M. H. (2011) Biochim. Biophys. Acta 1809, 80–87 [DOI] [PubMed] [Google Scholar]
- 58. Takeda K., Yoshida T., Kikuchi S., Nagao K., Kokubu A., Pluskal T., Villar-Briones A., Nakamura T., Yanagida M. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 3540–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.