Abstract
With increasing worldwide rates of morbidity and mortality of pulmonary fibrosis, the development of effective therapeutics for this disease is of great interest. Secretoglobin (SCGB) 3A2, a novel cytokine-like molecule predominantly expressed in pulmonary airways epithelium, exhibits anti-inflammatory and growth factor activities. In the current study SCGB3A2 was found to inhibit TGFβ-induced differentiation of fibroblasts to myofibroblasts, a hallmark of the fibrogenic process, using pulmonary fibroblasts isolated from adult mice. This induction was through increased phosphorylation of STAT1 and expression of SMAD7 and decreased phosphorylation of SMAD2 and SMAD3. To demonstrate the effect of SCGB3A2 on the TGFβ signaling in vivo, a bleomycin-induced pulmonary fibrosis mouse model was used. Mice were administered bleomycin intratracheally followed by intravenous injection of recombinant SCGB3A2. Histological examination in conjunction with inflammatory cell counts in bronchoalveolar lavage fluids demonstrated that SCGB3A2 suppressed bleomycin-induced pulmonary fibrosis. Microarray analysis was carried out using RNAs from lungs of bleomycin-treated mice with or without SCGB3A2 and normal mice treated with SCGB3A2. The results demonstrated that SCGB3A2 affects TGFβ signaling and reduces the expression of genes involved in fibrosis. This study suggests the potential utility of SCGB3A2 for targeting TGFβ signaling in the treatment of pulmonary fibrosis.
Keywords: Fibroblast, Gene Expression, Interferon, Lung, Microarray, Transforming Growth Factor β (TGFβ), Animal Models, Bleomycin, Pulmonary Fibrosis, Secretory Protein
Introduction
An increasing number of people are affected by pulmonary fibrosis worldwide, with increasing morbidity and mortality rates. In the United States, the number of patients suffering from pulmonary fibrosis is about 200,000 (1, 2) (www.nhlbi.nih.gov). Recovery from pulmonary fibrosis is possible at early stages of the disease, whereas the recovery is limited once the fibrosis has progressed. A risk for developing pulmonary fibrosis increases by administration of bleomycin (BLM),2 an anticancer and antibiotic agent, used in therapy for many types of solid tumors.
Fibrosis arises from inflammation initiated by cell injury, and injured tissues are gradually replaced by collagen fibers that are produced from fibroblasts and accumulate as myofibroblasts. Damaged cells produce chemokines, which stimulate leukocytes to proliferate and produce profibrotic cytokines such as transforming growth factor β (TGFβ), a major profibrotic growth factor, and interleukin-13 (IL-13), a major profibrotic mediator. TGFβ1 induces collagen type I transcription through the SMAD signaling, whereas IL-13 stimulates macrophages to produce TGFβ (3–6). On the other hand, interferon-γ (IFNγ) inhibits collagen generation through STAT1 activation followed by sequestration of p300, which plays a pivotal role in the regulation of collagen synthesis by TGFβ (7, 8). IFNγ also induces the antagonistic SMAD7, which in turn impairs TGFβ signaling through inhibition of the SMAD3 interaction with the TGFβ receptor (9) and/or disruption of formation of the TGFβ-induced functional SMAD-DNA complex (10). Other molecules are also involved in the fibrotic process (6). These include other Th2 cytokines such as IL-4, IL-5, and IL-10, chemokines such as CCL2 and CCL3, connective tissue growth factor, and platelet-derived growth factor (3, 11). Molecules involved in various pathways leading to myofibroblast expansion are considered to be useful as therapeutic targets. To this end, a number of inhibitors and/or monoclonal antibodies against these targeted molecules have been developed and subjected to clinical trials as a means to treat fibrosis (3, 11, 12). IFNγ is one of the targeted molecules used as a new therapy for fibrosis (5, 6, 12); however, because of potentially harmful side effects, a better alternative is desirable (13).
Secretoglobin (SCGB) 3A2, previously called uteroglobin-related protein 1, is a member of the SCGB gene superfamily (14). The SCGB gene superfamily consists of 3 gene families; family 1 has 4 subfamilies, each composed of 3–11 members, family 2 consists of 2 subfamilies, each with 6–10 members, and family 3 consists of only one subfamily with 5 members (15, 16). All members of the SCGB gene superfamily are cytokine-like secreted proteins of ∼10 kDa, found only in mammals. They form homodimers or heterodimers with other members. Most functions of SCGB are still elusive, and the signaling pathways including a possible receptor(s) that transmits activities of these proteins is not known. Among the best studied member of the gene superfamily is SCGB1A1, also called uteroglobin, Clara cell secretory protein, or Clara cell 10-kDa protein that exhibits anti-inflammatory and immunomodulatory activities in lung (17–19). Studies on the mechanisms of the anti-inflammatory activity of SCGB1A1 have been carried out (20, 21). Other members such as SCGB2A2 (mammaglobin A) and SCGB1D2 (lipophilin B) are known as a cancer marker for mammary gland (22) (23).
SCGB3A2 is the second member of the SCGB family 3, subfamily A. It is predominantly expressed in lung airways. SCGB3A2 was found to play a role in suppression of lung inflammation using a mouse model for allergic airway inflammation (24) and to promote branching and maturation of mouse fetal lungs (25). MARCO (macrophage scavenger receptor with collagenous structure), expressed in alveolar macrophages in lung, was suggested as a possible receptor for SCGB3A2 (26). On the other hand, we have demonstrated the possible presence of a SCGB3A2-specific receptor on the mesenchymal cells of mouse fetal lungs (25). Despite these studies, very little is known about the biological and physiological functions of SCGB3A2 and its mechanisms of action including the receptor and the signaling pathway it provokes.
In the present study SCGB3A2 was found to inhibit the TGFβ signaling through increased STAT1 phosphorylation and expression of SMAD7 and decreased phosphorylation of SMAD2/3, thus resulting in inhibition of TGFβ-induced myofibroblast differentiation. In an in vivo mouse model, SCGB3A2 markedly suppressed BLM-induced pulmonary fibrosis, suggesting the potential use of SCGB3A2 as a novel therapeutic reagent to treat pulmonary fibrosis.
EXPERIMENTAL PROCEDURES
Isolation and Primary Culture of Lung Fibroblasts
Lung tissues from 7–9-week-old female mice were cut into small pieces, mounted on collagen type I-coated 60-mm plate (IWAKI, Shizuoka, Japan), and then cultured for 7 days. Fibroblasts were harvested by 0.5% trypsin and 0.53 mm EDTA in PBS, washed with DMEM supplemented with 10% FBS, plated on a 35-mm plate, and cultured for 16 h. Fibroblasts were stimulated by 10 ng/ml TGFβ (Sigma) in DMEM containing 3% FBS for 24–72 h in the presence or absence of recombinant mouse (rm) SCGB3A2 (2.5 μg/ml). The recombinant mouse SCGB3A2 (rmSCGB3A2) was purified as described (25) (detailed purification method of SCGB3A2 will be provided upon request). For simplicity “SCGB3A2” is used instead of rmSCGB3A2 throughout the manuscript. For blocking experiments of IFNγ receptor signaling, a specific antibody against IFNγ receptor (rat anti-CD119 clone GR20, BD, Tokyo, Japan) was co-cultured with SCGB3A2 or IFNγ (R & D Systems, Minneapolis, MN) for 24 h or 30 min, respectively. To determine translational regulatory mechanism, fibroblasts were incubated with SCGB3A2 (2.5 μg/ml) and cycloheximide (CHX, 1 μg/ml, WAKO, Osaka, Japan) for 3 h or pretreated with CHX 3 h before stimulation with IFNγ. To down-regulate Stat1 expression, Stat1 siRNA probes (no. 7 probe, sense strand (5′-GCAUCUUACUGAAGGUGAATT-3′) and antisense strand (5′-UUCACCUUCAGUAAGAUGCAT-3′) and no. 8 probe (sense strand 5′-GAGUUGGUUUAAUAUAUAUTT-3′) and antisense strand (5′-AUAUAUAUUAAACCAACUCAT-3′); Qiagen, Valencia, CA) were transfected into fibroblasts using Lipofectamine 2000 (Invitrogen) 48 h before addition of TGFβ and/or SCGB3A2.
Immunoblotting
Immunoblotting was performed using the following antibodies; anti-STAT1, anti-pSTAT1 (Tyr-701), anti-SMAD2, anti-pSMAD2 (Ser-465/467), anti-SMAD3 (Cell Signaling Technology, Danvers, MA), anti-pSMAD3 (Ser-423/425) (Millipore Corp. Temecula, CA), anti-SMAD7 (Santa Cruz Biotechnology, Inc. Santa Cruz, CA), anti-α-smooth muscle actin (αSMA; Sigma), and anti-β-actin (Sigma). Immunoblotting was performed as described in Kurotani et al. (25). All immunoreactive bands were visualized using ECL (GE Healthcare) or ImmunoStar LD (WAKO) with LAS-3000mini (FUJIFILM, Tokyo, Japan) and then standardized by immunoreactive band of β-actin using Multi Gauge Version 3.0 software (FUJIFILM).
Quantitative RT-PCR and RT-PCR
Total RNAs isolated using TRIzol (Invitrogen) and digested with DNase I were reverse-transcribed by Superscript II reverse transcriptase (Invitrogen). Quantitative RT-PCR (qRT-PCR) was performed with ABI Prism 7900 Sequence Detection System (Applied Biosystems, Foster City, CA) using SYBR Green master mixture. The standard curve method was used, and all data were normalized to 18 S rRNA amplified using TaqMan Ribosomal RNA Control Reagent, VIC Probe (Applied Biosystems). RT-PCR was performed using AmpliTaq Gold DNA polymerase (Applied Biosystems) with iCycler (Bio-Rad). PCR condition used was 50 °C for 2 min and 95 °C for 10 min followed by 95 °C for 15 s and 60 °C for 40 s for 40 cycles. Primers used for qRT-PCR and RT-PCR analysis are summarized in supplemental Table S1. Leukocytes isolated from normal mouse spleen were stimulated by phorbol 12-myristate 13-acetate, (10 ng/ml) (Calbiochem) and ionomycin (1 μm) (Calbiochem) for 3 h or by poly(I:C) (polyinosinic-polycytidylic acid potassium salt, 10 μg/ml; Imgenex, San Diego, CA) for 24 h. The mRNAs extracted were used as a positive control for IFNγ and IFNα (27) and for IFNβ, respectively (28).
Animals and SCGB3A2 Treatment
C57BL/6N mice (NCI-Frederick, National Institutes of Health) were maintained under a standard 12-h light/12-h dark cycle with water and chow provided ad libitum. At least ten 7–8-week-old C57BL/6N mice were prepared for each group. Eight units/kg BLM (Sigma) or PBS was directly administered once by intratracheal intubation into C57BL/6N mice using the BioLITE system (BioTex, Inc., Houston, TX). Purified SCGB3A2 (1.5 mg/kg/day) or PBS was intravenously administered to mice via the tail vein once daily for a week starting on day 14 after BLM administration. Four groups of mice were used in this study; administration of BLM followed by intravenous injection of PBS (Group 1) or SCGB3A2 (Group 2) or administration of PBS followed by intravenous injection of PBS (Group 3) or SCGB3A2 (Group 4). All animal studies were performed after approval by the National Cancer Institute Animal Care and Use Committee. SCGB3A2 used in the animal study contained endotoxin at 0.2 enzyme units/mg. The dose of SCGB3A2 was determined based on the previous study in which a total of 200 μg, but not 100 μg, exhibited growth factor activity without causing any gross abnormalities to the dam as well as the embryos (25). For microarray analysis, C57BL/6N mice (7–8 weeks old) were treated with PBS or SCGB3A2 by intravenous administration and were euthanized 12 h later.
Bronchoalveolar Lavage
On day 21 after administration of BLM, mice were euthanized and subjected to bronchoalveolar lavage. Bronchoalveolar lavage fluid (BALF) was obtained by intratracheal instillation of 1 ml of PBS into the lung while it was kept in the thoracic cavity. Cells in the BALF were centrifuged at 500 × g and subjected to Diff-Quick (Baxter Healthcare, Miami, FL) staining. Cells were identified and counted using 2000–2500 cells from 10 fields randomly chosen. Tissues were fixed in 4% paraformaldehyde or stored at −80 °C for later RNA preparation.
Pathology
The whole lung was inflated and fixed with 4% paraformaldehyde. Lung tissues were embedded in paraffin, and 4-μm whole lung sections were prepared. Hematoxylin and eosin staining was carried out for assessment of BLM-induced fibrosis. Masson's Trichrome staining (Sigma) was used to detect collagen fibers. Immunohistochemical staining for SCGB3A2, pSMAD2, pSMAD3, and SMAD2/3 was performed in BLM-treated lungs using anti-mouse SCGB3A2 antibody (produced in our laboratory) (14), anti-pSMAD2(ser465/467) (Cell Signaling Technology), anti-pSMAD3 (Epitomics, Burlingame, CA), and anti-SMAD2/3 (BD Biosciences), respectively. For antigen-retrieval, the sections were incubated in Tris-EDTA (pH 6.0) at a temperature over 95 °C for 10 min before protein blocking. The immunoreactivities were enhanced by ABC method (Vector Laboratories, Burlingame, CA) followed by visualization using diaminobenzidine (DakoCytomation, Carpinteria, CA) and counterstained with hematoxylin. Immunocytochemical staining for αSMA was performed as described (29, 30). Briefly, fibroblasts cultured on 8-well Lab Tek Chamber Glass Slide (Nalge Nunc International, Naperville, IL) with and without TGFβ and/or SCGB3A2 were incubated at room temperature in PBS containing 0.1% Triton X 100 for 15 min followed by fixing with 4% paraformaldehyde for 15 min at room temperature. Cells were incubated with anti-αSMA antibody (Sigma, 1:400) overnight at 4 °C after blocking with 3% BSA-PBS for 1 h at room temperature. Cells were incubated with Alexa Fluor 488 goat anti-mouse IgG (Invitrogen) for 30 min at room temperature in dark. Nuclei were stained by 1 mg/ml DAPI (4′,6-diamino-2-phenylindole) (47). The αSMA signal was observed using Nikon EC (Nikon, Tokyo, Japan).
Fibrosis Assessment (Grading)
The grade of BLM-induced pulmonary fibrosis was determined based on the percentage of the fibrotic area in the whole lung section as follows; the presence of fibrosis in 0–25% of the lung for Grade 1, 26–50% of the lung for Grade 2, 51–75% of the lung for Grade 3, and 76–100% of the lung for Grade 4. No fibrosis, but with inflammation including a few infiltrating foci of lymphocytes or a very small granulomas, was considered Grade 0.
DNA Microarray
Total RNAs were purified using TRIzol and RNAeasy (Qiagen, Valencia, CA) from lungs of four groups (Group 1, 2, 3, and 4) of mice as described above or mice 12 h after SCGB3A2 treatment. Ten (group mice analysis) or 20 μg (SCGB3A2 treatment) of purified total RNA were reverse-transcribed to label with Cy3 and Cy5 (GE Healthcare) using the FairPlay Microarray Labeling kit (Stratagene, La Jolla, CA) or SuperScriptTM Indirect cDNA Labeling Core kit (Invitrogen), respectively. Microarray analysis was carried out using individually isolated RNA from at least 5 mice in each group and mouse array chips (45K) obtained from the NCI Microarray Facility. Experiments and analysis were performed according to the manufacturer's instruction and the instruction of the Center for Cancer Research, NCI (nciarray.nci.nih.gov). Gene Ontology (GO) analysis (31) and pathway analysis were performed using a mouse gene data (Mm-Std_20060628.gdb) and a mouse pathway data (Mm_Contributed_20070917) from MAPPFinder (32). Up-regulated genes were sorted based on scores (ratio of Cy5 per Cy3) more than 2.0 (average) between Group 1 and Group 3, more than 1.5 (average) between Group1 and Group2, and those up-regulated in all reactions.
Statistical Analysis
Data are shown as the mean ± S.D. from indicated numbers of independent experiments. Statistical analysis was performed one-way ANOVA followed by Bonferroni multiple comparison test.
RESULTS
Effect of SCGB3A2 on Differentiation of Fibroblasts into Myofibroblasts
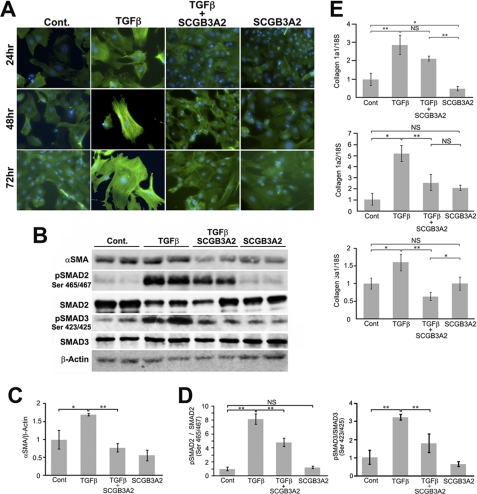
It was previously demonstrated that SCGB3A2 has at least two biological functions; an anti-inflammatory function was revealed using an ovalbumin-induced allergic airway inflammation model (24), and growth factor activity was studied using ex vivo fetal lung organ cultures and in vivo injection of SCGB3A2 to pregnant mice (25). To obtain insight into whether SCGB3A2 plays any additional roles in lung physiology and/or diseases, primary fibroblasts obtained from adult mouse lungs were used as an in vitro model of fibrosis in which the effect of SCGB3A2 on TGFβ-induced differentiation of fibroblasts into myofibroblasts was studied (33). Inhibition of this transformation would indicate a role for SCGB3A2 in controlling fibrosis. As expected, TGFβ treatment resulted in differentiation of pulmonary fibroblasts to myofibroblasts within 24 h as clearly seen by the characteristic myofibroblast morphology and the robust expression of αSMA (Fig. 1A, TGFβ, αSMA is seen in green). In contrast, treatment of fibroblasts with both TGFβ and SCGB3A2 together (TGFβ+SCGB3A2) exhibited morphology and the level of αSMA expression similar to those of normal fibroblasts (Fig. 1A, Cont. versus TGFβ+SCGB3A2). SCGB3A2 alone did not have any effect on the morphology of fibroblasts (Fig. 1A, SCGB3A2). Note that the number of cells did not significantly differ among different culture groups for at least up to 72 h (data not shown). The level of αSMA protein was increased by TGFβ treatment, which returned to control levels in cells treated with SCGB3A2 as determined by Western blotting performed after 24 h of stimulation with TGFβ (Fig. 1, B and C). SCGB3A2 alone did not have a significant effect on the level of αSMA protein as compared with control. When SMAD2 and SMAD3, the major molecules in the TGFβ signaling pathway, were examined, phosphorylated SMAD2 (pSMAD2) and SMAD3 (pSMAD3) were increased by TGFβ treatment as expected, and combined treatment with TGFβ and SCGB3A2 partially reversed the increase (Fig. 1, B and D). Again, SCGB3A2 alone did not have any effect on SMAD2 and SMAD3 phosphorylation. Furthermore, qRT-PCR analysis revealed that the expression of collagen 1a2 and collagen 3a1 was decreased in fibroblasts stimulated by TGFβ and SCGB3A2 together as compared with those treated with TGFβ alone (Fig. 1E). Because production of a complete collagen fiber requires three collagens (34), the reduced expression of two collagen genes is likely to account for the reduced Type I and Type III collagen fibers. The effect of SCGB3A2 on the reduced expression of collagen genes was abolished when SCGB3A2 was boiled before the addition to the media, suggesting that the effect of SCGB3A2 was not due to any contaminants in SCGB3A2 preparation (supplemental Fig. S1). These results demonstrated that SCGB3A2 inhibited TGFβ-induced differentiation of primary lung fibroblasts to myofibroblasts.
FIGURE 1.
Inhibition of differentiation from fibroblast to myofibroblast by SCGB3A2. A, representative immunocytochemistry for αSMA (n = 4) is shown. αSMA was visualized as a green signal. Fibroblasts isolated from adult mouse lungs were cultured in the absence (Cont.; normal fibroblast) or the presence of SCGB3A2, TGFβ, or TGFβ and SCGB3A2 together for 24–72 h. All the images were taken at ×600 (original magnification). B, representative immunoblotting results for αSMA, phosphorylated SMAD2 (pSMAD2), total SMAD2, phosphorylated SMAD3 (pSMAD3), total SMAD3, and β-actin using cells harvested after 24 h stimulation are shown. C, and D, densitometric analysis of the immunoblot signals in B is shown as the mean ± S.D. (n = 4) for αSMA (C) and pSMAD2/SMAD2 and pSMAD3/SMAD3 ratios (D). E, qRT-PCR for collagen 1a1, collagen 1a2, and collagen 3a1 is shown. Fibroblasts were harvested 24 h after stimulation and were subjected to qRT-PCR analysis to determine the level of collagen genes expression. Cont, normal fibroblast as control; TGFβ, stimulated by TGFβ; TGFβ+SCGB3A2, administration of both TGFβ and SCGB3A2; SCGB3A2, fibroblast stimulated by SCGB3A2 only. The graph shows the mean ± S.D. from 4–8 lungs per group, each in triplicate. *, p < 0.05; **, p < 0.01; NS, not significant.
Induction of STAT1 Phosphorylation and SMAD7 Expression by SCGB3A2
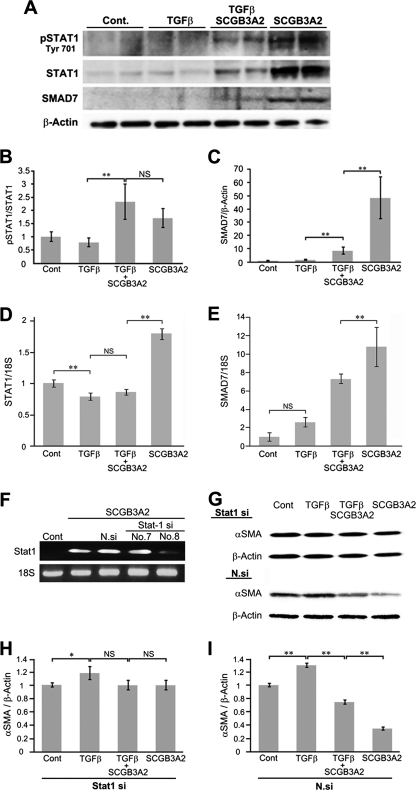
Immunoblotting was performed to examine levels of SMAD7 protein and the status of STAT1 phosphorylation in the presence or absence of TGFβ and/or SCGB3A2 (Fig. 2). It is well documented that IFNγ treatment results in phosphorylation of STAT1 and induces SMAD7 expression, which interferes with the TGFβ signaling (9, 35). In mouse lung primary fibroblasts, the expression of SMAD7, STAT1, and phosphorylated STAT1 was hardly detectable in control and TGFβ-treated cells (Fig. 2A). In contrast, the level of STAT1, phosphorylated STAT1 (pSTAT1), and SMAD7 was markedly increased when fibroblasts were treated with SCGB3A2 (Fig. 2, A–C). TGFβ dramatically inhibited the induction of STAT1 and SMAD7 by SCGB3A2 (see TGFβ+SCGB3A2 versus SCGB3A2), whereas the pSTAT1/STAT1 ratio stayed at similar levels with TGFβ+SCGB3A2 and SCGB3A2-only treatments (Fig. 2B). The SCGB3A2-induced increase of STAT1 and SMAD7 expression was due to an mRNA increase as demonstrated by qRT-PCR (Fig. 2, D and E, respectively). When STAT1 siRNA was transfected to mouse lung primary fibroblasts followed by treatment with TGFβ and/or SCGB3A2, TGFβ-induced αSMA expression levels stayed the same with and without SCGB3A2 (Fig. 2, G and H). With control nonspecific negative siRNA, TGFβ-induced αSMA expression was reduced by SCGB3A2 as expected (Fig. 2I), confirming that the effect of SCGB3A2 is through STAT1. These data demonstrated that SCGB3A2 enhanced expression of SMAD7 and STAT1 and phosphorylation of STAT1, resulting in inhibition of the TGFβ signaling pathway.
FIGURE 2.
Phosphorylation of STAT1 and SMAD7 expression by SCGB3A2. A, immunoblotting for phosphorylated STAT1 (pSTAT1), total STAT1, SMAD7, and β-actin as control is shown. B and C, shown is a graph for densitometric analysis of the immunoblot signals shown in A for pSTAT1/STAT1 (B) and SMAD7/β-actin (C). The mean ± S.D. (n = 4) is shown. D and E, qRT-PCR for STAT1 (D) and SMAD7 (E) using 18 S as an internal control is shown. NS, not significant. F–I, effect of STAT1 siRNA or nonspecific control siRNA on the level of αSMA is shown. F, qRT-PCR to confirm knockdown of STAT1 mRNA with STAT1 siRNA probes is shown. G, immunoblotting for αSMA and β-actin using cells transfected with no. 8-STAT1 siRNA probe (STAT1 si) or nonspecific negative siRNA probe (N.si). H and I, shown is a graph for densitometric analysis of the immunoblot signals shown in G for αSMA/β-actin with a STAT1 siRNA probe (H) and nonspecific negative siRNA probe (I). The mean ± S.D. (n = 4) is shown. Fibroblasts were harvested 24 h after stimulation. Cont, normal fibroblast as control; TGFβ, stimulated by TGFβ; TGFβ+SCGB3A2, administration of both TGFβ and SCGB3A2; SCGB3A2, fibroblast stimulated by SCGB3A2 only. **, p < 0.01, NS, not significant. Representative immunoblotting results are shown.
Relationship between SCGB3A2 and Interferon γ Receptor
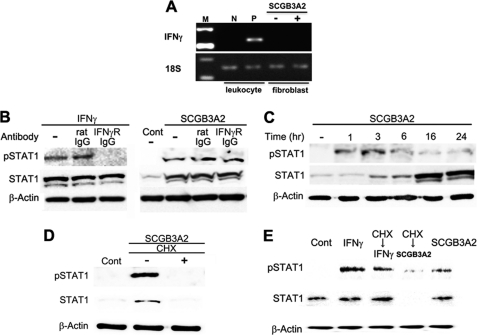
Several cytokines are known to activate STAT1. Among them, the key cytokine that activates STAT1 is IFNγ (36). IFNγ is known to improve fibrosis through phosphorylation of STAT1 (5, 6, 12), and its clinical effect on pulmonary fibrosis is well documented (37). IFNα and IFNβ also activate STAT1 and inhibit fibrosis (38–40). RT-PCR analysis using lung primary fibroblasts demonstrated that IFNγ was not expressed in lung primary fibroblasts regardless of SCGB3A2 treatment (Fig. 3A). Similarly, neither IFNα nor IFNβ was expressed in lung primary fibroblasts stimulated by SCGB3A2 (supplemental Fig. S2). In the following studies, we focused on the relationship between SCGB3A2 and IFNγ and its receptor. When IFNγ receptor-specific neutralizing antibody was added to the culture, IFNγ-induced phosphorylation of STAT1 was inhibited, whereas it did not inhibit phosphorylation of STAT1 induced by SCGB3A2 (Fig. 3B). Interestingly, it took ∼3 h for the SCGB3A2-induced phosphorylation of STAT1 to reach maximum levels (Fig. 3C). This was unusually long as compared with the IFNγ-induced STAT1 phosphorylation, which reached maximal levels within 30 min of stimulation (41, 42) (supplemental Fig. S3). Furthermore, SCGB3A2-stimulated STAT1 phosphorylation was suppressed in the presence of CHX (Fig. 3D). In contrast, IFNγ-stimulated STAT1 phosphorylation was unchanged with and without CHX treatment (Fig. 3E). These data indicated that SCGB3A2 promoted phosphorylation of STAT1 in a manner independent of IFNγ receptor and through a CHX-sensitive intermediate molecule. These data further suggested that the SCGB3A2-STAT1 signaling may have been through a SCGB3A2-specific receptor.
FIGURE 3.
Relationship between SCGB3A2 and IFNγ receptor. A, representative RT-PCR for IFNγ in fibroblasts in the presence or absence of SCGB3A2 (n = 3) is shown. Leukocytes stimulated by phorbol 12-myristate 13-acetate (10 ng/ml) and ionomycin (1 μm) for 3 h were used as a positive control for IFNγ. B, shown are blocking experiments for IFNγ receptor signaling. A representative result is shown (n = 3). pSTAT1 was detected in the presence of anti-IFNγ receptor antibody and SCGB3A2 (right panel). C, a representative time-course result of STAT1 phosphorylation by SCGB3A2 (n = 3) is shown. D and E, protein synthesis blocking experiments are shown. Fibroblasts were incubated with both SCGB3A2 and CHX (1 μg/ml) for 3 h (D and E for CHX→SCGB3A2) or were preincubated with CHX (1 μg/ml) for 3 h before stimulation with IFNγ for 15 min (E). A representative result is shown (n = 3).
Inhibition of BLM-induced Lung Fibrosis by SCGB3A2
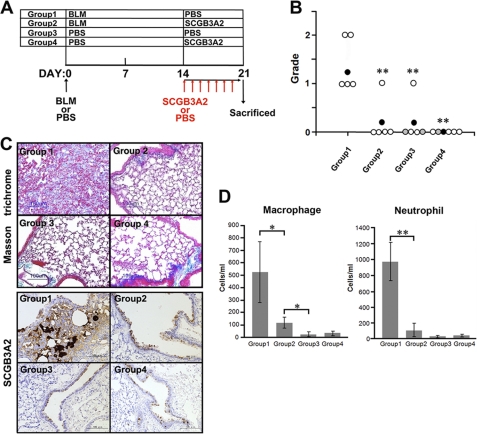
To validate these in vitro data that SCGB3A2 suppresses the TGFβ signaling pathway resulting in reduction of fibrosis, mice were subjected to a pulmonary fibrosis model. Pulmonary fibrosis was induced by direct administration of BLM or PBS as control to mice by intratracheal intubation. The histopathology of the lungs on day 14 (2 weeks) after BLM treatment did not reveal any fibrosis regardless of treatment regimens (data not shown). At day 14, mice received the first of 7 daily consecutive intravenous injections of SCGB3A2 or PBS through the tail vein (Fig. 4A). On day 21 after BLM administration, pulmonary fibrosis was observed in the lungs of BLM-treated group of mice (Group 1). The extent of fibrosis was rated as grade 1 to grade 2 (Fig. 4B). In contrast, mice in Group 2 that received BLM and SCGB3A2 did not develop any pulmonary fibrosis except one mouse of five tested. In Group 3, one mouse exhibited small fibrotic lesions, whereas no fibrosis was observed in Group 4 mice. Collagen fibers were found to focally occupy the alveolar space of mice in Group 1 but not in other groups of lungs as determined by Masson's Trichrome staining, which detects collagen fibers (Fig. 4C, upper panel). Excessive SCGB3A2 expression was focally found in a part of airway epithelial cells and the foci of fibrosis of Group 1 mouse lungs (Fig. 4C, lower panel). In contrast, other groups of lungs (Groups 2–4) expressed SCGB3A2 at similar levels in airway epithelial cells as expected, although Group 2 lungs appeared to have slightly higher expression than Group 3 and 4 lungs. When the expression of total SAMD2/3 as well as pSMAD2 and pSMAD3 was examined by immunohistochemistry, all were highly up-regulated in most of airway epithelial cells as well as the foci of fibrosis in Group 1 mouse lungs, whereas expression stayed at similar levels and patterns in other groups of lungs including those of BLM+SCGB3A2 administered mouse lungs (Group 2) (supplemental Fig. S4). The latter results suggested that SCGB3A2 appeared to have suppressed expression of SMAD2/3 and/or pSMAD2 and pSMAD3 in epithelial as well as parenchymal cells. The numbers of macrophages and neutrophils in BALF were enhanced by BLM treatment, whereas they were markedly reduced to levels close to that of controls in the SCGB3A2-treated group (Fig. 4D). The number of lymphocytes was not different with statistical significance among the four groups (data not shown). These data indicated that development of BLM-induced fibrosis was suppressed by the daily administration of SCGB3A2 on the third week of BLM treatment.
FIGURE 4.
Inhibition of BLM-induced lung fibrosis by SCGB3A2. A, shown is a scheme to prepare BLM-induced fibrosis model mice. BLM or PBS was administered to the lungs of C57BL/6N mice by intratracheal intubation at day 0. SCGB3A2 or PBS was administered to mice by intravenous injection once daily for a week starting at day 14 followed by euthanasia at day 21. The four groups of mice were subjected to the studies as shown. PBS was administered as a control in Groups 3 and 4. Whole experiments were carried out at least twice. B, grading of fibrosis carried out as described under “Experimental Procedures.” BLM administration resulted in production of fibrous tissue, focally or diffusely in lungs of Group 1, whereas no fibrous tissue formation was observed in four of five lungs exposed to BLM followed by SCGB3A2 treatment in Group 2. White circle, no lesions; black circle, mean (n = 5); gray, presence of a few infiltrating foci of lymphocytes or very small granulomas. **, p < 0.01, Group 1 versus Groups 2, 3, or 4. C, Masson's Trichrome staining and immunohistochemistry for SCGB3A2 are shown. Collagen fibers were focally detected as a blue color by Masson trichrome staining in the parenchyma of the lungs of mice in Group 1 but not the lungs of other groups. Excessive SCGB3A2 expression was found in a part of the epithelial cells and in the foci of fibrosis as a brown color by immunohistochemistry in Group 1. Representative staining is shown (n = 5). D, shown are the number of macrophages and neutrophils in BALF. The number of macrophages and neutrophils increased by BLM (Group 1) was significantly decreased by administration of SCGB3A2 (Group 2), especially neutrophils which reduced to the levels of control (Group 3 and Group 4). The graph shows the mean ± S.D. from 5 lungs per group (n = 5). *, p < 0.05; **, p < 0.01.
Alteration of Gene Expression by BLM and/or SCGB3A2
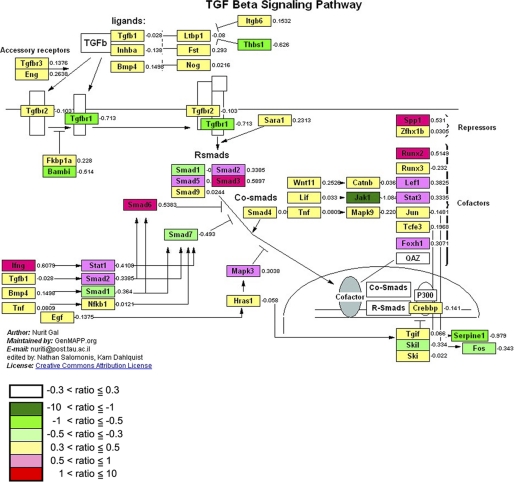
To determine the genes whose expression was altered in the BLM-induced lung fibrosis and/or SCGB3A2-induced reduction of fibrosis, microarray analysis was carried out using mRNAs obtained from lungs of mice between Group 1 (BLM-treated) and Group 3 (PBS control) and Group 1 (BLM-treated) and Group 2 (BLM- and SCGB3A2-treated). With a cutoff of >1.5-fold change in expression, 1646 and 1275 genes were, respectively, up- and down-regulated by BLM as compared with control PBS (between Group 1 and Group 3), and 346 and 919 genes were, respectively, up- and down-regulated by SCGB3A2 treatment as compared with no SCGB3A2 treatment in BLM treated mice (between Group 1 and Group 2). Using these genes, GO (gene ontology) analysis was performed for the changes caused by BLM and for the effect of SCGB3A2 in BLM-treated mice (heat map results are shown in supplemental Fig. S5). The results revealed that “Inflammatory response” and “Response to wounding” under “Biological Process” and “Extracellular region part,” “Extracellular region,” and “Extracellular space” under “Cellular Component” were overexpressed by BLM injury (Table 1) and suppressed by administration of SCGB3A2 (Table 2). Additionally, to demonstrate genes that are altered by SCGB3A2, microarray analysis was carried out using lung RNAs of normal mice that were intravenously administered SCGB3A2 and were euthanized 12 h later. GO analysis revealed that “Signal transducer activity” and “Molecular transducer activity” categorized by only one term “Molecular Function” were up-regulated by short-term treatment of SCGB3A2 (Table 3). The TGFβ signaling was among the top pathways identified by the Pathway Mapping program (Fig. 5), further supporting our conclusion that SCGB3A2 affects the TGFβ signaling pathway.
TABLE 1.
GO terms for overexpressed genes in BLM-injured lung
| Gene ontology term | Cluster frequencya | Total frequencyb | Corrected p valuec |
|---|---|---|---|
| % | % | ||
| Biological Process, cell adhesion | 6.40 | 3.50 | 2.92 × 10−5 |
| Biological adhesion | 6.40 | 3.50 | 2.92 × 10−5 |
| Antigen processing and presentation of exogenous peptide antigen | 0.70 | 0.10 | 0.00654 |
| Immune system process | 6.30 | 3.90 | 0.01711 |
| Inflammatory response | 2.70 | 1.30 | 0.01977 |
| Antigen processing and presentation of peptide antigen via MHC class II | 0.60 | 0.10 | 0.02221 |
| Antigen processing and presentation of exogenous peptide antigen via MHC class II | 0.60 | 0.10 | 0.02221 |
| Antigen processing and presentation of peptide of polysaccharide antigen via MHC class II | 0.60 | 0.10 | 0.04108 |
| Response to wounding | 3.30 | 1.80 | 0.04663 |
| Cellular Component | |||
| Extracellular region part | 18.00 | 12.70 | 5.62 × 10−7 |
| Extracellular matrix | 4.00 | 1.70 | 1.05 × 10−6 |
| Proteinaceous extracellular matrix | 3.39 | 1.70 | 1.68 × 10−6 |
| Extracellular region | 20.40 | 15.10 | 5.57 × 10−6 |
| Extracellular space | 16.80 | 12.00 | 7.62 × 10−6 |
| Molecular function | |||
| Carbohydrate binding | 3.00 | 1.60 | 0.04087 |
a Frequency of Entrez Gene IDs appeared in a given GO term based on those having over 0.585 in log ratio.
b Frequency of Entrez Gene IDs appeared in a given GO term based on all genes.
c p value corrected using the Bonferroni method.
TABLE 2.
GO terms for suppressed genes by SCGB3A2 in BLM-injured lung
| Gene ontology term | Cluster frequencya | Total frequencyb | Corrected p valuec |
|---|---|---|---|
| % | % | ||
| Biological process | |||
| Defense response | 6.10 | 2.50 | 6.78 × 10−5 |
| Response to external stimulus | 6.40 | 2.70 | 7.91 × 10−5 |
| Response to wounding | 4.50 | 1.80 | 0.00172 |
| Response to stimulus | 15.80 | 10.60 | 0.00896 |
| Inflammatory response | 3.20 | 1.30 | 0.03692 |
| Cellular component | |||
| Extracellular region | 24.50 | 15.10 | 1.82 × 10−9 |
| Extracellular region part | 20.40 | 12.70 | 2.26 × 10−7 |
| Extracellular space | 19.50 | 12.00 | 3.22 × 10−7 |
a Frequency of Entrez Gene IDs appeared in a given GO term based on those having over 0.585 in log ratio.
b Frequency of Entrez Gene IDs appeared in a given GO term based on all genes.
c p value corrected using the Bonferroni method.
TABLE 3.
GO terms overexpressed by SCGB3A2 in normal lung
| Gene ontology term, molecular function | Cluster frequencya | Total frequencyb | Corrected p valuec |
|---|---|---|---|
| % | % | ||
| Signal transducer activity | 20.7 | 15.9 | 0.0378 |
| Molecular transducer activity | 20.7 | 15.9 | 0.0378 |
a Frequency of Entrez Gene IDs appeared in a given GO term based on those having over 0.585 in log ratio.
b Frequency of Entrez Gene IDs appeared in a given GO term based on all genes.
c p value corrected using the Bonferroni method.
FIGURE 5.
Effect of SCGB3A2 on TGFβ signaling pathway. Pathway analysis of microarray data showed that the TGFβ signaling pathway was affected by administration of SCGB3A2. Median -fold differences from seven separate samples for each gene on which microarray analysis were run are color-coded based on the scale shown at the bottom left.
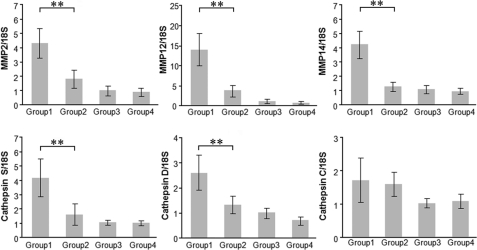
Further analysis revealed that the expression of matrix metalloproteinases (MMPs) and other extracellular matrix (ECM)-degrading enzymes that degrade damaged tissues as well as enzymes synthesizing collagen I, fibronectin, hyaluronic acid, and other components of wound provisional ECM was altered after BLM and/or SCGB3A2 treatment (supplemental Table S2). qRT-PCR was performed to confirm the alterations of gene expression in BLM-treated lungs with (Group 2) and without (Group 1) SCGB3A2 (Fig. 6). High levels of MMP2, MMP12, and MMP14 expression in BLM-treated lungs (Group 1) were significantly decreased by the administration of SCGB3A2 (Group 2); in particular, MMP14 returned to the levels of the “no BLM treatment” groups (Group 3 and 4). Cathepsin S and D that belong to a group of proteinases were also highly expressed in BLM-treated group (Group 1), and decreased to a level similar to control by SCGB3A2 (Group 2). Cathepsin C levels did not show any statistically significant difference among the four groups of mice. These data further indicated a role for SCGB3A2 in suppression of lung fibrosis induced by BLM.
FIGURE 6.
Confirmation of microarray data by qRT-PCR. Expression levels of MMP2, MMP12, MMP14, cathespin S, cathepsin D, and cathepsin C mRNAs relative to 18 S were determined by qRT-PCR using mRNAs prepared from mouse lungs of Groups 1–4 as described in Fig. 4. The graph shows the mean ± S.D. from 4–9 lungs per group, each in triplicate. **, p < 0.01.
DISCUSSION
This study revealed that SCGB3A2 possesses anti-fibrotic activity as revealed by in vitro cell culture studies with primary lung fibroblasts and in vivo studies using a BLM-induced pulmonary fibrosis model mouse. SCGB3A2, a member of the SCGB gene superfamily composed of secretory proteins of small molecular weight, is predominantly expressed in lung airways (14, 43). The most studied member of the SCGB gene superfamily, namely SCGB1A1, also called uteroglobin, Clara cell 10-kDa protein, or Clara cell secretory protein, is a multifunctional protein with anti-inflammatory/immunomodulatory properties with manifestation of antichemotactic, antiallergic, antitumorigenic, and embryonic growth-stimulatory activities (19). The current study revealed that the related protein SCGB3A2 at therapeutic levels has novel biological activity toward induced fibrosis in addition to its known anti-inflammatory (24) and growth factor activities (25). SCGB1A1 was previously suggested to be a novel cytokine (44). The current results support the notion that the SCGB gene superfamily may be a novel cytokine family. The anti-inflammatory, growth factor and anti-fibrotic activities that SCGB3A2 possesses may suggest a potential use for this protein in the treatment of many lung diseases, including lung fibrosis as demonstrated in this study.
BLM administered by intratracheal intubation induced pulmonary fibrosis, which focally occupied the pulmonary parenchyma by 3 weeks after BLM administration. Interestingly, BLM-induced fibrosis was almost completely suppressed by SCGB3A2 treatment, which interfered with the infiltration of neutrophils and macrophages into lung and the expression of fibrosis related-genes such as collagens, fibronectin, elastin, cathepsins, and MMPs, all of which were up-regulated by BLM. The development of fibrosis was hardly detectable by the end of 2 weeks after BLM administration, suggesting that SCGB3A2 might inhibit development of fibrosis when given at its early stages.
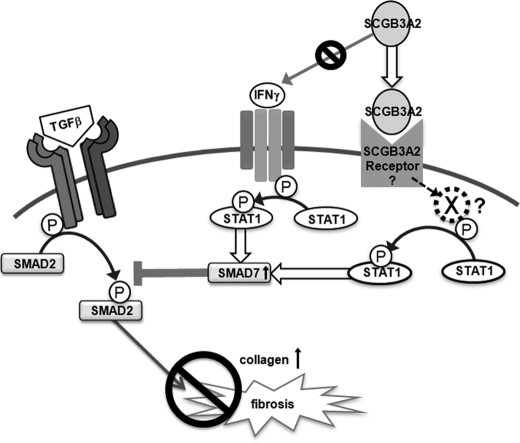
The anti-fibrotic activity of SCGB3A2 appears to be exerted through STAT1 phosphorylation, induction of SMAD7, and inhibition of SMAD2/3 phosphorylation, which results in suppression of the TGFβ signaling, ultimately leading to the inhibition of myofibroblasts formation. Interestingly, this is the exact pathway that IFNγ exhibits its anti-fibrotic activity (9, 35). We initially hypothesized that SCGB3A2 may induce the expression of IFNγ, thereby suppressing TGFβ signaling. However, the SCGB3A2 pathway is likely to be distinct from the IFNγ pathway based on the following reasons; 1) IFNγ-induced phosphorylation of STAT1 and increased expression of SMAD7 usually occurs within 10–30 min after IFNγ stimulation (supplemental Fig. S3) (9, 41, 42, 45), whereas it took ∼3 h for SCGB3A2 to induce maximum levels of STAT1 phosphorylation, 2) SCGB3A2 did not induce IFNγ mRNA expression as determined by RT-PCR, 3) IFNγ receptor neutralizing antibody (46) did not block SCGB3A2-induced STAT1 phosphorylation, and 4) CHX ablated the expression of pSTAT1 in fibroblasts treated with SCGB3A2 but not IFNγ. These results suggest the involvement of a newly synthesized protein upon SCGB3A2 stimulation other than IFNγ in the SCGB3A2-pSTAT1 pathway. In this regard it is interesting to note that STAT1 was induced by SCGB3A2 in a CHX-sensitive fashion. SMAD7 was also induced by SCGB3A2. The SCGB3A2-induced increase of STAT1 and SMAD7 expression was dramatically inhibited by TGFβ at the mRNA levels. However the pSTAT1/STAT1 ratio stayed the same. The involvement of STAT1 in the SCGB3A2 pathway was confirmed by STAT1 siRNA experiments, in which no decrease of αSMA was observed in TGFβ+SCGB3A2-treated cells as compared with TGFβ only treatment. These results together suggest that the SCGB3A2-pSTAT1-SMAD7 signaling pathway may be through a SCGB3A2-specific receptor that is distinct from the IFNγ receptor (Fig. 7). We previously suggested the presence of a SCGB3A2-specific receptor-like molecule on the surface of pulmonary mesenchymal cells (25). The nature of the CHX-sensitive intermediate molecule(s), a SCGB3A2-specific receptor, and the mechanism(s) for the inhibitory effect of TGFβ on the induction of STAT1 and SMAD7 by SCGB3A2 are currently not known. Further experiments are required to address these questions. Other cytokines/molecules such as IFNα, IFNβ, epidermal growth factor, growth hormone, and estrogen are known to activate STAT1 (47). Among them, neither IFNα nor IFNβ was induced by SCGB3A2 in mouse lung primary fibroblasts, suggesting that they are not likely involved in the SCGB3A2-pSTAT1-SMAD7 pathway. The involvement of the other cytokines/molecules in the SCGB3A2-pSTAT1 pathway needs to be examined.
FIGURE 7.
Schematic diagram for the SCGB3A2-induced inhibition of the TGFβ signaling pathway. SCGB3A2 inhibits collagen production through STAT1 phosphorylation and increases expression of SMAD7, which inhibits TGFβ-mediated SMAD2/3 activation. A newly synthesized molecule X mediates the SCGB3A2-induced STAT1 phosphorylation.
Inflammation is the first to occur after BLM administration followed by fibrosis (3–6). In animal models the fibrosis stage starts ∼1 week after BLM administration (3). In our BLM model, SCGB3A2 was administered at the fibrosis period and exhibited anti-fibrotic activity through blocking the TGFβ signaling pathway. We previously demonstrated using mice-exogenously administered SCGB3A2 that SCGB3A2 exhibits anti-inflammatory activities. We do not know whether SCGB3A2, if administered at the beginning of BLM administration, exerts anti-inflammatory activities and reduces the damage otherwise caused by BLM. Furthermore, we do not know whether endogenous SCGB3A2 regulates pulmonary inflammation and/or fibrosis. However, the following evidence suggests a role for SCGB3A2 in inflammation in lung in vivo; in the ovalbumin-induced allergic airway inflammation model mouse, SCGB3A2 expression is reduced in the airways (24, 48, 49), and the plasma SCGB3A2 levels are significantly lower in severe asthmatics without oral corticosteroid treatment as compared with mild- or moderate-asthma patients and controls (50). In the current study, a rather strong focal SCGB3A2 expression was found in a part of epithelial cells, and overexpressed SCGB3A2 was accumulated in fibrotic foci of lungs of mice subjected to BLM. However, it should be noted that these lungs are already at late stages, having developed BLM-induced fibrosis. In BLM+SCGB3A2-treated lungs, SCGB3A2 expression is in similar patterns but is slightly up-regulated to those found in control mice. Whether SCGB3A2 is down-regulated right after BLM administration needs to be determined.
Although IFNγ has been used as a therapy for pulmonary fibrosis (5, 6, 12), it causes a number of potentially harmful side effects and thus is not useful for advanced fibrosis (nlm.nih.gov (13)). In this study SCGB3A2 signaling was found to cross-talk with the IFNγ except at the beginning of the pathway. This suggests that SCGB3A2 could be potentially useful in treating fibrosis, in particular pulmonary fibrosis. More importantly, SCGB3A2 can be administered intratracheally. SCGB3A2 is almost exclusively expressed in lung (14) and is present at relatively high levels in the BALF of normal lungs (24). Thus, delivering a protein into a place where the protein is naturally present in high amounts would probably not cause toxicities. Alternatively, SCGB3A2 can be administered intravenously. Indeed, SCGB3A2 growth factor activity was previously demonstrated in vivo by intravenous administration of SCGB3A2 to pregnant female mice through the tail vein resulting in advanced development of fetal lungs (25). In that study, no gross abnormalities were observed in any fetal organs after intravenous SCGB3A2 administration. Furthermore, histological examination of the dam's lung did not show any abnormalities. Based on these facts, it is likely that SCGB3A2 may potentially improve pulmonary fibrosis without harmful side effects as seen with IFNγ. It is possible that BLM treatment as a chemotherapy reagent may not damage the lung if SCGB3A2 is given at the same time. However, additional studies are necessary to determine the efficacy of SCGB3A2 in treating pulmonary fibrosis in humans.
In conclusion, the present study demonstrated that SCGB3A2 inhibits TGFβ signaling through increased STAT1 phosphorylation and expression of SMAD7 and decreased phosphorylation of SMAD2/3, leading to inhibition of myofibroblast differentiation. The inhibitory effect of SCGB3A2 on myofibroblast differentiation was reproduced using a BLM-induced lung fibrosis model mouse, in which the severity of lung fibrosis was reduced by SCGB3A2 administration.
Supplementary Material
Acknowledgments
We thank Drs. Lalage Wakefield, Kathleen Flanders, and Frank Gonzalez (NCI, National Institutes of Health) for advice and critical review of the manuscript and Michie Kobayashi for microarray analysis (DNA Chip Research Inc., Yokohama, Japan).
This work was supported by the National Institutes of Health, Center for Cancer Research (Intramural Research Program of the NCI; NIH 0010190305 and Z01 BC010449-06, to S. K.). This work was also supported by Grant-in-aid for Young Scientists (B) (21790207, to R. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and S2 and Figs. S1–S5.
All effective genes were submitted to the Gene Expression Omnibus (GEO: ID GSE21560, www.ncbi.nlm.nih.gov).
- BLM
- bleomycin
- SCGB
- secretoglobin
- CHX
- cycloheximide
- GO
- gene ontology
- MMPs
- metalloproteinases
- qRT
- quantitative RT
- BALF
- bronchoalveolar lavage fluid
- αSMA
- α smooth muscle actin
- rm
- recombinant mouse.
REFERENCES
- 1. Coultas D. B., Zumwalt R. E., Black W. C., Sobonya R. E. (1994) Am. J. Resp. Crit. Care Med. 150, 967–972 [DOI] [PubMed] [Google Scholar]
- 2. Crystal R. G., Bitterman P. B., Mossman B., Schwarz M. I., Sheppard D., Almasy L., Chapman H. A., Friedman S. L., King T. E., Jr., Leinwand L. A., Liotta L., Martin G. R., Schwartz D. A., Schultz G. S., Wagner C. R., Musson R. A. (2002) Am. J. Respir. Crit. Care Med. 166, 236–246 [DOI] [PubMed] [Google Scholar]
- 3. Moeller A., Ask K., Warburton D., Gauldie J., Kolb M. (2008) Int. J. Biochem. Cell Biol. 40, 362–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cutroneo K. R., White S. L., Phan S. H., Ehrlich H. P. (2007) J. Cell. Physiol. 211, 585–589 [DOI] [PubMed] [Google Scholar]
- 5. Tzortzaki E. G., Antoniou K. M., Zervou M. I., Lambiri I., Koutsopoulos A., Tzanakis N., Plataki M., Maltezakis G., Bouros D., Siafakas N. M. (2007) Respir. Med. 101, 1821–1829 [DOI] [PubMed] [Google Scholar]
- 6. Wynn T. A. (2004) Nat. Rev. Immunol. 4, 583–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eickelberg O., Pansky A., Koehler E., Bihl M., Tamm M., Hildebrand P., Perruchoud A. P., Kashgarian M., Roth M. (2001) FASEB J. 15, 797–806 [DOI] [PubMed] [Google Scholar]
- 8. Ghosh A. K., Varga J. (2007) J. Cell. Physiol. 213, 663–671 [DOI] [PubMed] [Google Scholar]
- 9. Ulloa L., Doody J., Massagué J. (1999) Nature 397, 710–713 [DOI] [PubMed] [Google Scholar]
- 10. Zhang S., Fei T., Zhang L., Zhang R., Chen F., Ning Y., Han Y., Feng X. H., Meng A., Chen Y. G. (2007) Mol. Cell Biol. 27, 4488–4499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Scotton C. J., Chambers R. C. (2007) Chest 132, 1311–1321 [DOI] [PubMed] [Google Scholar]
- 12. Puente N. A., Aliotta J. M., Passero M. A. (2007) Med. Health R. I. 90, 43–45 [PubMed] [Google Scholar]
- 13. Selman M. (2003) Am. J. Respir. Crit. Care Med. 167, 945–946 [DOI] [PubMed] [Google Scholar]
- 14. Niimi T., Keck-Waggoner C. L., Popescu N. C., Zhou Y., Levitt R. C., Kimura S. (2001) Mol. Endocrinol. 15, 2021–2036 [DOI] [PubMed] [Google Scholar]
- 15. Reynolds S. D., Reynolds P. R., Pryhuber G. S., Finder J. D., Stripp B. R. (2002) Am. J. Respir. Crit. Care Med. 166, 1498–1509 [DOI] [PubMed] [Google Scholar]
- 16. Klug J., Beier H. M., Bernard A., Chilton B. S., Fleming T. P., Lehrer R. I., Miele L., Pattabiraman N., Singh G. (2000) Ann. N.Y. Acad. Sci. 923, 348–354 [DOI] [PubMed] [Google Scholar]
- 17. Shijubo N., Kawabata I., Sato N., Itoh Y. (2003) Curr. Pharm. Des. 9, 1139–1149 [DOI] [PubMed] [Google Scholar]
- 18. Wang S. Z., Rosenberger C. L., Bao Y. X., Stark J. M., Harrod K. S. (2003) J. Immunol. 171, 1051–1060 [DOI] [PubMed] [Google Scholar]
- 19. Mukherjee A. B., Zhang Z., Chilton B. S. (2007) Endocr. Rev. 28, 707–725 [DOI] [PubMed] [Google Scholar]
- 20. Mandal A. K., Zhang Z., Ray R., Choi M. S., Chowdhury B., Pattabiraman N., Mukherjee A. B. (2004) J. Exp. Med. 199, 1317–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Miele L. (2000) Ann. N.Y. Acad. Sci. 923, 128–140 [DOI] [PubMed] [Google Scholar]
- 22. Watson M. A., Fleming T. P. (1996) Cancer Res. 56, 860–865 [PubMed] [Google Scholar]
- 23. Culleton J., O'Brien N., Ryan B. M., Hill A. D., McDermott E., O'Higgins N., Duffy M. J. (2007) Int. J. Cancer 120, 1087–1092 [DOI] [PubMed] [Google Scholar]
- 24. Chiba Y., Kurotani R., Kusakabe T., Miura T., Link B. W., Misawa M., Kimura S. (2006) Am. J. Respir. Crit. Care Med. 173, 958–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kurotani R., Tomita T., Yang Q., Carlson B. A., Chen C., Kimura S. (2008) Am. J. Respir. Crit. Care Med. 178, 389–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bin L. H., Nielson L. D., Liu X., Mason R. J., Shu H. B. (2003) J. Immunol. 171, 924–930 [DOI] [PubMed] [Google Scholar]
- 27. Maeng H. G., Lim H., Jeong Y. J., Woo A., Kang J. S., Lee W. J., Hwang Y. I. (2009) Immunobiology 214, 311–320 [DOI] [PubMed] [Google Scholar]
- 28. Lin Y. C., Huang D. Y., Chu C. L., Lin W. W. (2010) Mol. Immunol. 47, 1569–1578 [DOI] [PubMed] [Google Scholar]
- 29. Asazuma-Nakamura Y., Dai P., Harada Y., Jiang Y., Hamaoka K., Takamatsu T. (2009) Exp. Cell Res. 315, 1190–1199 [DOI] [PubMed] [Google Scholar]
- 30. Kurotani R., Yoshimura S., Iwasaki Y., Inoue K., Teramoto A., Osamura R. Y. (2002) J. Endocrinol. 172, 477–487 [DOI] [PubMed] [Google Scholar]
- 31. Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G. (2000) Nat. Genet. 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Doniger S. W., Salomonis N., Dahlquist K. D., Vranizan K., Lawlor S. C., Conklin B. R. (2003) Genome Biol. 4, R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tomasek J. J., Gabbiani G., Hinz B., Chaponnier C., Brown R. A. (2002) Nat. Rev. Mol. Cell Biol. 3, 349–363 [DOI] [PubMed] [Google Scholar]
- 34. Shoulders M. D., Raines R. T. (2009) Annu. Rev. Biochem. 78, 929–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Weng H., Mertens P. R., Gressner A. M., Dooley S. (2007) J. Hepatol. 46, 295–303 [DOI] [PubMed] [Google Scholar]
- 36. Yu H., Pardoll D., Jove R. (2009) Nat. Rev. Cancer 9, 798–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Luppi F., Losi M., D'Amico R., Fabbri L. M., Richeldi L. (2009) Sarcoidosis Vasc. Diffuse Lung Dis. 26, 64–68 [PubMed] [Google Scholar]
- 38. Azuma A., Li Y. J., Abe S., Usuki J., Matsuda K., Henmi S., Miyauchi Y., Ueda K., Izawa A., Sone S., Hashimoto S., Kudoh S. (2005) Am. J. Respir. Cell Mol. Biol. 32, 93–98 [DOI] [PubMed] [Google Scholar]
- 39. Senft A. P., Taylor R. H., Lei W., Campbell S. A., Tipper J. L., Martinez M. J., Witt T. L., Clay C. C., Harrod K. S. (2010) Am. J. Respir. Cell Mol. Biol. 42, 404–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Papatheodoridis G. V., Petraki K., Cholongitas E., Kanta E., Ketikoglou I., Manesis E. K. (2005) J. Viral. Hepat. 12, 199–206 [DOI] [PubMed] [Google Scholar]
- 41. Tamai M., Kawakami A., Tanaka F., Miyashita T., Nakamura H., Iwanaga N., Izumi Y., Arima K., Aratake K., Huang M., Kamachi M., Ida H., Origuchi T., Eguchi K. (2006) J. Lab. Clin. Med. 147, 182–190 [DOI] [PubMed] [Google Scholar]
- 42. Seo J. Y., Kim D. Y., Lee Y. S., Ro J. Y. (2009) Cytokine 46, 51–60 [DOI] [PubMed] [Google Scholar]
- 43. Tomita T., Kido T., Kurotani R., Iemura S., Sterneck E., Natsume T., Vinson C., Kimura S. (2008) J. Biol. Chem. 283, 25617–25627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mukherjee A. B., Kundu G. C., Mantile-Selvaggi G., Yuan C. J., Mandal A. K., Chattopadhyay S., Zheng F., Pattabiraman N., Zhang Z. (1999) Cell. Mol. Life Sci. 55, 771–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee Y. J., Benveniste E. N. (1996) J. Immunol. 157, 1559–1568 [PubMed] [Google Scholar]
- 46. Cheng M., Nguyen M. H., Fantuzzi G., Koh T. J. (2008) Am. J. Physiol. Cell Physiol. 294, C1183–C1191 [DOI] [PubMed] [Google Scholar]
- 47. Krämer O. H., Heinzel T. (2010) Mol. Cell Endocrinol. 315, 40–48 [DOI] [PubMed] [Google Scholar]
- 48. Chiba Y., Kusakabe T., Kimura S. (2004) Am. J. Physiol. Lung Cell Mol. Physiol. 287, L1193–L1198 [DOI] [PubMed] [Google Scholar]
- 49. Chiba Y., Srisodsai A., Supavilai P., Kimura S. (2005) Immunol. Lett. 97, 123–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Inoue K., Wang X., Saito J., Tanino Y., Ishida T., Iwaki D., Fujita T., Kimura S., Munakata M. (2008) Allergol. Int. 57, 57–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.