Abstract
Mammalian polo-like kinase 1 (Plk1) plays a pivotal role during M-phase progression. Plk1 localizes to specific subcellular structures through the targeting activity of the C-terminal polo-box domain (PBD). Disruption of the PBD function results in improper bipolar spindle formation, chromosome missegregation, and cytokinesis defect that ultimately lead to the generation of aneuploidy. It has been shown that Plk1 recruits itself to centromeres by phosphorylating and binding to a centromere scaffold, PBIP1 (also called MLF1IP and CENP-U[50]) through its PBD. However, how PBIP1 itself is targeted to centromeres and what roles it plays in the regulation of Plk1-dependent mitotic events remain unknown. Here, we demonstrated that PBIP1 directly interacts with CENP-Q, and this interaction was mutually required not only for their stability but also for their centromere localization. Plk1 did not appear to interact with CENP-Q directly. However, Plk1 formed a ternary complex with PBIP1 and CENP-Q through a self-generated p-T78 motif on PBIP1. This complex formation was central for Plk1-dependent phosphorylation of PBIP1-bound CENP-Q and delocalization of the PBIP1-CENP-Q complex from mitotic centromeres. This study reveals a unique mechanism of how PBIP1 mediates Plk1-dependent phosphorylation event onto a third protein, and provides new insights into the mechanism of how Plk1 and its recruitment scaffold, PBIP1-CENP-Q complex, are localized to and delocalized from centromeres.
Keywords: Cell Cycle, Centromeres, Mitosis, Phosphorylation Enzymes, Protein Kinases, Kinetochores, Polo-like Kinase 1 (Plk1)
Introduction
Polo-like kinases (Plk)3 constitute a conserved subfamily of Ser/Thr protein kinases that play pivotal roles in cell proliferation (1–5). They are characterized by the presence of the polo-box domain (PBD) in the noncatalytic C-terminal region (6, 7) that functions as a phospho-Ser/Thr-binding module important for protein-protein interaction (8, 9). Unlike low eukaryotic organisms, mammalian cells appear to have at least four Plks (Plk1–4) that exhibit distinct expression patterns and functions (10). Among them, Plk1 has been most extensively studied not only because of its essential role during M-phase progression but also because of its strongly conserved function from budding yeast to human.
It has been shown that Plk1 localizes to centrosomes and centromeres/kinetochores at the late stage of the cell cycle, and remains at these locations until telophase. Plk1 localization to centrosomes is important to promote proper centrosome maturation and centrosome-based microtubule nucleation (11, 12). On the other hand, the role of Plk1 at centromeres/kinetochores remains largely elusive, although proper localization of Plk1 at these sites is thought to be important for proper M-phase progression (13–15). In anaphase, a fraction of Plk1 localizes to the spindle midzone (later, it becomes midbody) (16–18), an event that appears to be critical for proper cytokinesis.
We have shown that Plk1 recruits itself to centromeres by phosphorylating and binding to a centromere scaffold, PBIP1 (also called MLF1IP and CENP-U[50]; hereafter referred to as PBIP1 for simplicity) with an unusually high affinity and specificity (19). Although the physiological significance of PBIP1-dependent self-recruitment of Plk1 to interphase and mitotic centromeres is yet to be further determined, the importance of PBIP1 in this event suggests that PBIP1 serves as a crucial scaffold for Plk1-dependent mitotic regulation at this site. However, how PBIP1 itself is targeted to centromeres and whether PBIP1-bound Plk1 regulates the localization and delocalization of PBIP1 and its binding target(s) to and from the centromere are not known. Data obtained from the affinity purification of CENP-A-containing nucleosomes from human cultured cells suggest that PBIP1 is a component of the CENP-A nucleosome-associated complex (NAC) (13). Other studies show that PBIP1 closely interacts with a set of proteins called CENP-O class proteins, namely CENP-O, CENP-P, and CENP-Q (20, 21), all of which belong to CENP-A-nucleosome distal (CAD) components (13). These observations suggest that the interactions between PBIP1 and its binding targets at kinetochores are likely complex.
In this study, we demonstrated that a CENP-A NAC component, PBIP1, forms a binary complex with a CAD component, CENP-Q, to promote their mutual stability and subcellular localization to interphase and mitotic centromeres. We further showed that Plk1 forms a ternary complex with PBIP1 and CENP-Q by generating and binding to the p-T78 motif on PBIP1. This step was crucially required for Plk1 to phosphorylate PBIP1-bound CENP-Q and to induce delocalization of the complex from mitotic centromeres. This study reveals the first example of how a scaffold mediates Plk1-dependent phosphorylation onto a third protein and sheds light on the mechanism of how Plk1 interacts with the PBIP1-CENP-Q complex and regulates its dissociation from mitotic kinetochores.
EXPERIMENTAL PROCEDURES
Plasmid Construction
Recombinant adenoviruses were generated according to the procedure described previously (22), using a modified pShuttle-CMV-EGFP vector (pKM489) (15). Various pShuttle-CMV-EGFP-based constructs expressing full-length or truncated PBIP1 fragments (full-length [pKM282], T1 [pKL2987], T2 [pKL3181], T3 [pKL2988], T4 [pKL2989], T5 [pKL2990], T6 [pKL2993], T7 [pKL2994], T8 [pKL2991], or T9 [pKL2992]) were generated by inserting each of the respective KpnI-XhoI fragments into a pShuttle-CMV-EGFP vector digested with the corresponding enzymes. To generate pEGFP-C1 (Clontech, Palo Alto, CA) constructs containing T1 (pKM491), T2 (pKM492), T3 (pKM493), T4 (pKM494), T5 (pKM495), T6 (pKM497), T7 (pKM498), T8 (pKM499), or T9 (pKM500), BglII-HindIII fragments containing the respective PBIP1 forms were cloned into a pEGFP-C1 vector digested with the corresponding enzymes.
For the construction of EGFP-tagged PBIP1 WT (pKM504), PBIP1(L337P L344P) (pKM1490), PBIP1ΔNLS (pKM2437), PBIP1(K308A K316A) (pKM2475), CENP-Q WT (pKM1697), or CENP-Q(L179P) (pKM1670) plasmid, the respective PBIP1 or CENP-Q fragment digested with KpnI and XhoI was cloned into a modified pEGFP-Cl vector (15) digested with the same enzymes.
For the generation of Flag-tagged PBIP1 or CENP-Q constructs, a KpnI-XhoI fragment of PBIP1 WT (pKM530), PBIP1(L337P L344P) (pKM1485), CENP-Q WT (pKM1667), or CENP-Q(L179P) (pKM1668) was inserted into a pFlag-C1 (a pEGFP-C1 variant with a Flag epitope that replaces EGFP) vector (15) digested with the corresponding enzymes. Myc-tagged PBIP (pKM855) was constructed by inserting an AscI (end-filled)-BamHI fragment containing the full-length PBIP1 ORF into a pCS2-MT FA (a gift of Hongtao Yu, University of Texas Southwestern Medical Center, Dallas, TX) digested with Xho1 (end-filled) and BamHI. Similarly, a Myc-CENP-Q expression construct (pKM829) was created by inserting a FseI-AscI fragment containing the full-length CENP-Q ORF into the pCS2-MT FA vector digested with the corresponding enzymes. The pRcCMV-Myc-Plk1 construct was kindly provided by Erich A. Nigg (Max Planck Institute for Biochemistry, Martinsried, Germany).
To generate mammalian expression constructs for centromere proteins, a KpnI-XhoI fragment containing the entire open reading frame (ORF) of various centromere components was first amplified by PCR and then digested with KpnI and XhoI (CENP-A [BC002703], CENP-H [BC015355], CENP-M [NM_024053], CENP-N [BC007334], CENP-T [BC015202]; the XhoI site of the CENP-T gene was eliminated by a silence mutation, CENP-I [a gift of Tim Yen, Fox Chase Cancer Center, Philadelphia, PA]; CENP-K [BC005400], CENP-L [BC066658], CENP-O [BC002870], CENP-P [BC071726], CENP-Q [NM_018132], CENP-R [NM_014288], and CENP-S [BC029430]). The resulting KpnI-XhoI fragments were then inserted into a pFlag-C1 vector digested with the respective enzymes to generate pFlag-C1-based constructs containing CENP-A (pKM1430), CENP-H (pKM1432), CENP-M (pKM1433), CENP-N (pKM1434), CENP-T (pKM1435), CENP-I (pKM1436), CENP-K (pKM1437), CENP-L (pKM1438), CENP-O (pKM1439), CENP-P (pKM1440), CENP-Q (pKM1441), CENP-R (pKM1442), or CENP-S (pKM1443).
For the generation of a lentivirus-based sh-RNA construct targeting CENP-Q (pKM1704), a double-stranded oligonucleotide fragment containing corresponding nt 510–528 (forward 5′-CCGGGTTAATGACTGGGAATATTCTCGAGAATATTCCCAGTCATTAACTCTTTTTG-3′ and reverse 5′-AATTCAAAAAGAGTTAATGACTGGGAATATTCTCGAGAATATTCCCAGTCATTAAC-3; targeting sequence is indicated in boldface type) was inserted into a pLKO.1-puro vector (a gift of Sheila A. Stewart, Washington University, St. Louis, MO) digested with AgeI and EcoRI. Lentivirus-based sh-RNAs targeting PBIP1 and Plk1 have been reported previously (15). A lentivirus construct expressing EGFP-fused CENP-Q (pKM1463) or PBIP1 (WT or the K308A K316A mutant; pKM542 or pKM2989, respectively) was cloned by inserting an AgeI (end-filled)-XhoI fragment containing EGFP-CENP-Q or a BglII-XhoI fragment containing PBIP1 (WT or the K308A K316A mutant) into a pHR′-CMV-SV-puro vector (a gift of Chou-Zen Giam, Uniformed Services University of the Health Sciences, Bethesda, MD) digested with BamHI (end-filled) and SalI or with BamHI and SalI, respectively.
A bacterial expression construct, pETDuet-1-His-MBP (pKM1647), was generated by inserting an MfeI-EcoRI fragment containing the maltose-binding protein (MBP) ORF into the EcoRI site of the pETDuet1 vector (Novagen, Madison, WI). For the expression of His-MBP-CENP-Q (pKM1648), an AscI-NotI fragment containing the full-length CENP-Q ORF was inserted into the pETDuet-1-His-MBP construct above. For the expression of the His-MBP-CENP-Q/His-PBIP1 (WT, T78A, or L337P L344P mutant; pKM1653, pKM1654, or pKM1655, respectively), an NdeI-XhoI fragment containing His-tagged PBIP1 (WT, the T78A, or the L337P L344P mutant) was additionally inserted into the pETDuet-1-His-MBP-CENP-Q at the corresponding sites. For the expression of His-Flag-MBP-CENP-Q (WT or the L179P mutant; pKM1666, pKM1815, respectively), a 33 residue long BamHI-Flag-BclI fragment was inserted into the BamHI site of the pETDuet-1-His-MBP-CENP-Q (WT or the L179P mutant).
To construct pGEX-4T-3-CENP-Q (pKM1645), a KpnI-XhoI fragment containing the full-length CENP-Q ORF was cloned into a multiple cloning site-modified pGEX-4T-3 (Amersham Biosciences, Piscataway, NJ) digested with the corresponding enzymes. The pMAL-c2-CENP-Q construct (pKM1646) was generated by inserting an XhoI (end-filled)-KpnI fragment containing the full-length CENP-Q ORF into the pMAL-c2 vector (New England Biolab, Ipswich, MA) digested with PstI (end-filled) and KpnI. All the PCR-generated DNA fragments used for the construction were confirmed by nucleotide sequencing.
Cell Culture, Transfection, Virus Generation, and Virus Infection
Cell lines were cultured as recommended by the American Type Culture Collection (Manassas, VA). HeLa cells were trapped in prometaphase by treating them with 200 nm of nocodazole (Sigma) for 20 h. For double-thymidine block and release experiments, cells were arrested for 16 h with 2.5 mm thymidine with a 9-h release interval between the thymidine treatments. To inhibit Plk1, cells were treated with 100 nm of BI 2536 (23) for the indicated length of time.
Transfection was routinely carried out with Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Various adenoviruses used in this study were generated as described previously (18). Lentivirus production was done as described previously (15). To select the lentivirus-integrated population, HeLa cells infected with indicated viruses were treated with 2 μg/ml of puromycin for 2–3 days.
Recombinant Protein Expression
GST-PBIP1-T7, GST-CENP-Q, and MBP-CENP-Q were expressed in Escherichia coli BL21(DE3) and purified with glutathione (GSH)-agarose (Sigma) and amylose resin (New England Biolab, Ipswich, MA). His-tagged proteins (His-MBP-CENP-Q, the His-MBP-CENP-Q/His-PBIP1 [WT or T78A] complex, or His-Flag-MBP-CENP-Q [WT or L179P]) were expressed in E. coli (BL21[DE3]) and purified with Ni-affinity resin (Sigma).
Antibody Production and Purification
Bacterially expressed GST-fused CENP-Q (full-length) was purified using GSH-agarose (Amersham Biosciences, Piscataway, NJ), and then injected into rabbits to raise polyclonal anti-CENP-Q antisera (through collaboration between NCI and Rockland Immunologicals, Gilbertsville, PA). To affinity-purify the anti-CENP-Q antibody, immunized sera were purified, using bacterially expressed MBP-CENP-Q immobilized with Affi-Gel-10 (Bio-Rad). Similarly, a PBIP1 N-terminal antibody was generated against bacterially expressed GST-PBIP1 T2 fragment (residues 1–199) and affinity-purified before use.
Immunoprecipitation, In Vitro Binding, Immunoblotting, and In Vitro Kinase Assay
Immunoprecipitation was carried out essentially as described previously (17). Briefly, cells were lysed in TBSN buffer (20 mm Tris-Cl, pH 8.0, 150 mm NaCl, 0.5% Nonidet P-40, 5 mm EGTA, 1.5 mm EDTA, 0.5 mm Na3VO4, and 20 mm p-nitrophenylphosphate (PNPP)). The resulting lysates were clarified by centrifugation at 15,000 × g for 20 min at 4 °C before subjecting them to immunoprecipitation with the specified antibody. Immunoprecipitated proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to polyvinylidene difluoride (PVDF) (Immobilon-P; Millipore, Bedford, MA) membrane, and then detected by immunoblotting with the indicated antibodies. In vitro protein-protein interaction experiments were performed in an in vitro binding buffer (50 mm Tris-Cl, pH 7.4, 150 mm NaCl, 1 mm EDTA, and 0.1% Nonidet P-40).
Immunoblotting analyses were performed using the indicated primary antibodies and appropriate horseradish peroxidase (HRP)-conjugated secondary antibodies. Proteins that interact with antibodies were detected by the enhanced chemiluminescence (ECL) detection system (Pierce).
In vitro kinase assays were carried out essentially as described previously (17) in a kinase mixture (50 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 5 mm dithiothreitol, 2 mm EGTA, and 0.5 mm Na3VO4) supplemented with the indicated substrate and 10 μm ATP (5 μCi of [γ-32P]ATP; 1 Ci = 37 GBq). To generate the self-primed binding site for Plk1 on His-PBIP1, the first reaction was carried out in the presence of 100 μm ATP for 1 h. The resulting samples were then subjected to the second reaction in the presence of 10 μCi of [γ-32P]ATP for the indicated lengths of time. All the reactions were done at 30 °C and terminated by the addition of 5× SDS sample buffer. Proteins were separated on 10% SDS-PAGE, and 32P was detected by autoradiography, where appropriate. Both WT and kinase-inactive forms of HA-Plk1(K82M) were purified from Sf9 cells.
Indirect Immunofluorescence Microscopy and Quantification
Indirect immunostaining was carried out as described previously (18). All the appropriate secondary antibodies (Alexa Fluor 488 [green] or Texas red [red]-conjugated antibodies) were purchased from Invitrogen. Confocal images were acquired using a Zeiss LSM 510 system mounted on a Zeiss Axiovert 100 m microscope (Carl Zeiss MicroImaging, Inc., Thornwood, NY). For the quantification of the fluorescence signal intensities, images of unsaturated fluorescence signals were acquired with the same laser intensity at 512 × 512 pixels and 12-bit resolution. Fluorescence intensity for a particular subcellular signal was determined after subtraction of the background signal intensity using Zeiss AIM confocal software.
RESULTS
Requirement of the C-terminal Domain in Proper Localization of PBIP1 to Centromeres
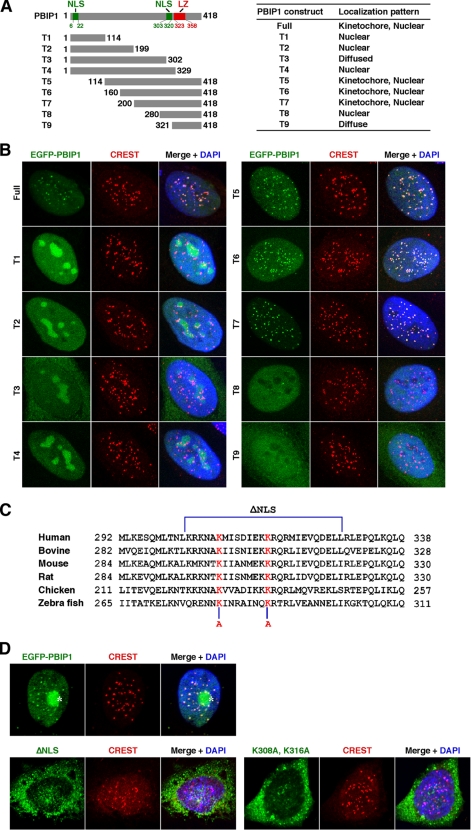
To investigate the mechanism of how PBIP1 localizes to centromeres, we first determined the ability of various EGFP-fused PBIP1 truncation mutants to localize to these sites. To this end, HeLa cells infected with adenoviruses expressing various EGFP-PBIP1 truncations were fixed and immunostained for confocal microscopy analyses. Results showed that the full-length EGFP-PBIP1 efficiently localized to interphase centromeres (Fig. 1, A and B) and also somewhat weakly to mitotic kinetochores (data not shown). This localization pattern closely parallels that of endogenous PBIP1 (15), suggesting that EGFP-PBIP1 is fully functional. Under these conditions, three truncation mutants (T5-T7) containing the C-terminal half of the protein localized to centromeres as proficiently as the full-length PBIP1, whereas other truncated forms (T1-T4 and T8-T9) failed to localize to these sites (Fig. 1, A and B, data not shown). Interestingly, among the N-terminal constructs (T1-T4), the shorter forms (T1 and T2) bearing the putative N-terminal nuclear localization signal (NLS) motif (residues 6–22) efficiently translocated into the nucleus. Interestingly, however, the T3 form bearing the N-terminal NLS motif but lacking its neighboring C-terminal NLS motif (residues 303–320) failed to localize to the nucleus, whereas the T4 form bearing both the N- and C-terminal NLS motifs was competent in nuclear translocation (Fig. 1B). On the other hand, all the C-terminal truncation mutants bearing the C-terminal NLS motif efficiently localized to the nucleus, whereas the T9 form lacking the C-terminal NLS motif exhibited only diffused signals (Fig. 1B). These observations suggest that the C-terminal NLS motif may play a more pivotal role in the nuclear translocation of PBIP1 than the N-terminal motif, which exhibits a limited nuclear localization capacity only for the N-terminal PBIP1 fragments.
FIGURE 1.
Analyses of the subcellular localization of PBIP1. A and B, HeLa cells infected with adenoviruses expressing either EGFP-fused full-length or truncated PBIP1 constructs were subjected to immunostaining analyses. Localization patterns of various PBIP1 constructs are summarized in A, right. Representative images for each of the constructs are provided in B. Numbers indicate amino acid residues. NLS, putative nuclear localization sequences; LZ, putative leucine zipper domain. Anti-CREST signals serve as kinetochore markers. C, sequences from various PBIP1 orthologs are aligned. The ΔNLS mutation deletes amino acid residues from 303 to 328. Mutations of the two conserved Lys residues (Lys-308 and Lys-316) to Ala are indicated in red. D, HeLa cells transfected with the indicated EGFP-fused PBIP1 constructs were immunostained and analyzed. Asterisk, transfected EGFP-PBIP1 aggregate.
To directly test whether the C-terminal NLS motif is required for proper PBIP1 translocation into the nucleus, we generated a PBIP1ΔNLS mutant lacking the residue 303–328 region and a PBIP1(K308A K316A) mutant bearing mutations at the two conserved Lys residues within the NLS (Fig. 1C). Analyses of these mutants showed that they are largely defective in nuclear translocation (Fig. 1D). Thus, the C-terminal NLS motif plays a crucial role in determining the capacity of PBIP1 to localize into the nucleus. These observations suggest that the two previously described putative NLS motifs (residues 127–134 and residues 218–223) (24) present within the T3 form do not play a significant role in determining the PBIP1 localization.
Differential Requirement of the N-terminal and C-terminal Domains of PBIP1 in the Interactions with Plk1 and CENP-Q, Respectively
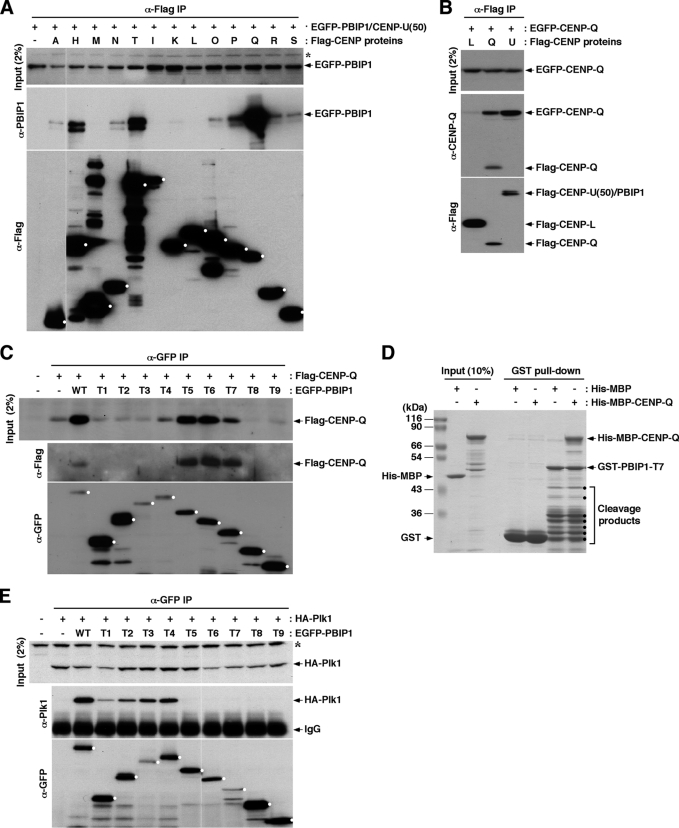
To gain new insights into the mechanism underlying PBIP1 recruitment to centromeres, we examined the interactions between PBIP1 and various centromere components using transfected lysates. We observed that immunoprecipitation of CENP-Q efficiently co-immunoprecipitated EGFP-PBIP1, while immunoprecipitation of CENP-H, CENP-T, or CENP-P co-immunoprecipitated the latter at a somewhat reduced level (Fig. 2A). A strong interaction between a CENP-A NAC component, PBIP1, and a CAD component, CENP-Q, was unexpected. This observation was confirmed by a reciprocal immunoprecipitation analysis that showed that immunoprecipitation of Flag-PBIP1/CENP-U(50) co-immunoprecipitated CENP-Q (Fig. 2B). Notably, immunoprecipitation of Flag-CENP-Q efficiently co-immunoprecipitated EGFP-CENP-Q, whereas immunoprecipitation of Flag-CENP-L, a component of an apparently distinct CENP-H, I, K, L complex (21), did not (Fig. 2B). This observation is in line with the previous finding that CENP-Q forms a homo-octameric complex in vitro (25). Unlike the homomeric nature of CENP-Q, PBIP1 did not appear to significantly interact with another PBIP1 molecule under the same conditions (supplemental Fig. S1).
FIGURE 2.
In vivo interactions among PBIP1, CENP-Q, and Plk1. A–C, HeLa cells transfected with the indicated constructs were subjected to co-immunoprecipitation analyses. Alphabetical letters in A and B denote centromeric proteins from CENP-A to CENP-S. PBIP1, which is also called CENP-U (13) or CENP-50 (14), is denoted as “U” for simplicity. Dots in A and C mark the full-length form of each ligand immunoprecipitated. Asterisks, cross-reacting proteins. D, purified bacterial proteins were subjected to GST pull-downs. Samples were separated by 10% SDS-PAGE and stained with Coomassie. Dots, cleavage products of GST-PBIP1-T7. E, HeLa cells were transfected and immunoprecipitated as in A–C. Dots mark the full-length form of each ligand immunoprecipitated. Asterisk, a cross-reacting protein.
Next, we investigated which domain of PBIP1 is responsible for the interaction with CENP-Q. Results showed that immunoprecipitation of EGFP-T5, -T6, or -T7 not only greatly stabilized transfected CENP-Q but also efficiently co-precipitated the latter (Fig. 2C), suggesting that the C-terminal half of PBIP1 (residues 200–418) is sufficient to form a heteromeric complex with CENP-Q, and that the complex formation is important for the stability of these two proteins. As expected if the stabilization of CENP-Q is a crucial step for PBIP1 localization, EGFP-T5, -T6, and -T7 truncations localized to centromeres as efficiently as the EGFP-PBIP1 full-length in cells depleted of endogenous PBIP1 (supplemental Fig. S2). In a second experiment using purified bacterial proteins, precipitation of T7 co-precipitated CENP-Q as efficiently as the T7 ligand itself (Fig. 2D), thus strongly suggesting the formation of a tight complex between these two proteins. In contrast to this finding, Plk1 interacted with the N-terminal PBIP1 truncations (T1-T4), but failed to interact with the C-terminal PBIP1 truncation mutants lacking residues as small as the N-terminal residues 1–114 (Fig. 2E). This finding is consistent with the earlier observation that Plk1 interacts with PBIP1 through the T78 motif (15). Thus, PBIP1 interacts with both Plk1 and CENP-Q, but through its two distinct N-terminal and C-terminal domains, respectively (see summary in supplemental Fig. S3).
Mutual Requirement of the PBIP1 Leucine Zipper Domain and CENP-Q Coiled-coil Domain in the Formation and Subcellular Localization of the PBIP1-CENP-Q Complex
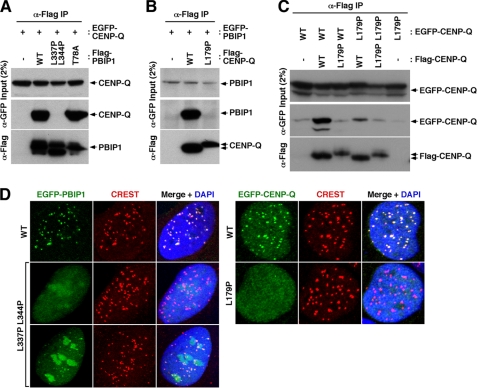
To closely investigate the nature of the PBIP1-CENP-Q interaction, we used the 2ZIP-Server program (26) to better define domains responsible for the interaction. This search revealed that PBIP1 bears one putative leucine zipper domain from residue 323 to residue 358, while CENP-Q contains one putative coiled-coil domain (which does not correspond to a leucine zipper) from residue 155 to residue 203. Mutations of two conserved Leu residues (Leu-337 and Leu-344) within the PBIP1 leucine zipper domain to Pro completely disrupted the PBIP1-CENP-Q interaction (Fig. 3A). Similarly, mutation of a conserved Leu residue (Leu-179) within the CENP-Q coiled-coil domain to Pro also eliminated the PBIP1-CENP-Q interaction (Fig. 3B). These observations suggest that the leucine zipper domain of PBIP1 may directly interact with the coiled-coil domain of CENP-Q. Alternatively, since CENP-Q forms a homo-octameric complex, the coiled-coil domain could be required for the formation of the homomeric CENP-Q complex, and this step might be important for its subsequent interaction with PBIP1. To distinguish these two possibilities, we examined homomeric interactions using CENP-Q WT and the L179P mutant. Results showed that the presence of the L179P mutation in one part of the two CENP-Q constructs used for the co-immunoprecipitation experiment was sufficient to disrupt the homomeric CENP-Q complex formation (Fig. 3C), suggesting that the latter possibility is more likely.
FIGURE 3.
The leucine zipper domain of PBIP1 and the coiled-coil domain of CENP-Q are mutually required for both the formation of the PBIP1-CENP-Q complex and the subcellular localization of the complex to centromeres. A–C, HeLa cells transfected with the indicated constructs were immunoprecipitated and immunoblotted. L337P L344P, a PBIP1 leucine zipper mutant; T78A, a Plk1-binding defective PBIP1 mutant (15); L179P, a CENP-Q coiled-coil mutant. D, HeLa cells were transfected with EGFP-PBIP1, EGFP-CENP-Q, or their respective leucine zipper/coiled-coil mutants, and then immunostained. Note that cells expressing a high level of EGFP-PBIP1(L337P L344P) frequently exhibited a low level of centrosome-localized fluorescence. Anti-CREST signals serve as centromere markers.
Next, we examined whether the formation of the PBIP1-CENP-Q complex is important for the subcellular localization of the complex to centromeres. HeLa cells transfected with EGFP-fused PBIP1 showed that the L337P L344P mutations greatly impaired the ability of PBIP1 to localize to centromeres (Fig. 3D). However, a small fraction of the cells with highly expressed EGFP-PBIP1(L337P L344P) exhibited a low level of localized fluorescence at the centromeres (Fig. 3D, third panel on left). Because the double mutations appeared to eliminate the PBIP1-CENP-Q interaction in Fig. 3A, the residual ability of the L337P L344P mutant to localize to the centromere could be due to its ability to interact with other centromere component(s), such as CENP-H and/or CENP-T, as shown in Fig. 2A. The PBIP1-binding incompetent CENP-Q(L179P) mutant failed to localize to centromeres under similar conditions (Fig. 3D). These results strongly suggest that the formation of the PBIP1-CENP-Q complex is a crucial step required for proper localization of the complex to centromeres. Consistent with this view, localization of ectopically expressed EGFP-CENP-Q was disrupted in HeLa cells depleted of PBIP1 (si-PBIP1) or in HeLa si-PBIP1 cells expressing a nuclear localization-defective PBIP1(K308A K316A) mutant (supplemental Fig. S4).
PBIP1 Mediates the Formation of a Ternary Plk1-PBIP1-CENP-Q Complex and the in Vivo Phosphorylation of CENP-Q by Plk1
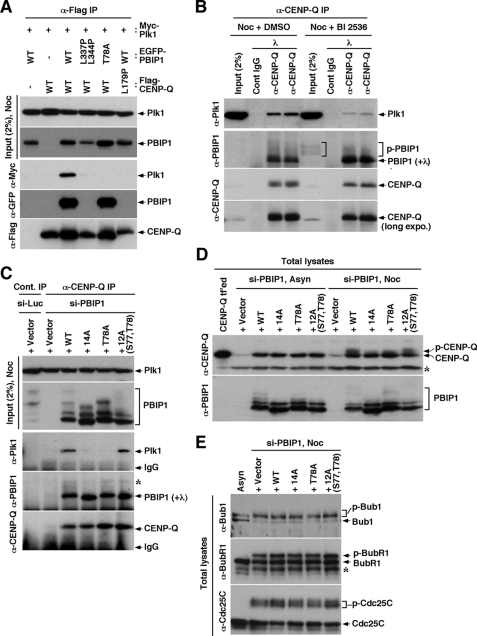
Because PBIP1 interacts with both Plk1 and CENP-Q via its distinct domains, we next examined whether these three proteins form a ternary complex. Using HeLa cells co-transfected with various constructs, we observed that immunoprecipitation of CENP-Q co-precipitated both PBIP1 and Plk1 (Fig. 4A). Immunoprecipitation of CENP-Q also co-precipitated PBIP1(T78A) efficiently, but failed to co-precipitate Plk1. This is consistent with the previous finding that the Thr-78 residue is critical for the PBIP1-Plk1 interaction (15). Disruption of the PBIP1-CENP-Q interaction, by either the leucine zipper mutations (L337P L344P) in PBIP1 or the coiled-coil mutation (L179P) in CENP-Q, completely deprived the ability of CENP-Q to co-precipitate Plk1 (Fig. 4A). These findings suggest that CENP-Q interacts with PBIP1 independently of Plk1, and that CENP-Q associates with Plk1 only through PBIP1. In support of this notion, immunoprecipitation of CENP-Q co-precipitated PBIP1, regardless of the presence or absence of Plk1 activity at the endogenous level (Fig. 4B). Consistent with the previous observation that Plk1 self-primes and binds to PBIP1 (15), inhibition of Plk1 activity by BI 2536 significantly diminished the level of Plk1 co-precipitating with the PBIP1-CENP-Q complex without influencing the PBIP1-CENP-Q interaction (Fig. 4B).
FIGURE 4.
The Plk1 interacts with and phosphorylates CENP-Q via the PBIP1 T78 motif. A and B, HeLa cells transfected with the indicated constructs were subjected to immunoprecipitation analyses. Where indicated, cells were treated with nocodazole for 18 h and BI 2536 for 4 h before harvest. Immunoprecipitates were reacted with λ phosphatase to convert all the phosphorylated, slow-migrating forms to non-phosphorylated form for easy detection by immunoblotting analysis. Bracket, hyperphosphorylated PBIP1 forms. C, HeLa cells expressing the indicated RNAi-insensitive constructs were depleted of control luciferase (si-Luc) or endogenous PBIP1 (si-PBIP1), and then treated with nocodazole for 18 h before harvest. Samples were immunoprecipitated with control IgG (first lane) or anti-CENP-Q antibody and the immunoprecipitates were treated with λ phosphatase before immunoblotting analyses. Asterisk, a cross-reacting protein. Note that cells depleted of PBIP1 exhibited a greatly diminished level of CENP-Q immunoprecipitates due to its instability in the absence of PBIP1 (lane 2). D and E, The cells used in C were depleted of endogenous PBIP1 (si-PBIP1), and then either left untreated or treated with nocodazole before harvest. Samples were separated 10% SDS-PAGE and immunoblotted. Transfected CENP-Q (no tag) in lane 1 denotes the position of endogenous CENP-Q. Note that the T78-dependent PBIP1 function is required for mitotic-specific phosphorylation of CENP-Q (D), but not for other Plk1 substrates, such as Bub1, BubR1, and Cdc25C (E). Asterisks, cross-reacting proteins.
To confirm these findings, we then utilized the previously established cell lines expressing various Plk1 binding-competent (i.e. WT and the 12A mutant containing the Plk1 self-priming and binding S77-T78 motif) or -incompetent (i.e. the T78A and 14A mutants lacking the S77-T78 motif) PBIP1 forms at the level of endogenous PBIP1 (15). Consistent with the results shown above, immunoprecipitation of CENP-Q co-precipitated both PBIP1 WT and various mutants equally well. However, only the cells expressing PBIP1 WT or the 12A mutant bearing the Plk1-binding S77-T78 motif co-precipitated Plk1 (Fig. 4C).
Because Plk1 associates with the PBIP1-CENP-Q complex via the T78 motif of PBIP1, we next investigated whether Plk1 phosphorylates CENP-Q in a T78-dependent manner. We observed that cells expressing PBIP1 WT and the Plk1 binding-competent 12A mutant exhibited phosphorylated and slow-migrating CENP-Q when arrested with nocodazole (M phase) (Fig. 4D). The phosphorylated CENP-Q form was not easily detectable in asynchronously (Asyn) growing cells, likely because of a low fraction of mitotic cells under these conditions (Fig. 4D). In contrast, cells expressing the Plk1 binding-incompetent 14A or T78A mutant failed to induce CENP-Q phosphorylation in vivo (Fig. 4D). As expected if Plk1 kinase activity were responsible for CENP-Q phosphorylation, either inhibition or depletion of Plk1 significantly diminished the level of slow-migrating CENP-Q form (supplemental Fig. S5). On the other hand, Bub1, BubR1, and Cdc25C, which are shown to directly interact with Plk1 through the PBD (27–29), were efficiently phosphorylated in a manner that requires Plk1 (supplemental Fig. S5B), but not the T78-dependent PBIP1-Plk1 interaction (Fig. 4E). Taken together, these observations suggest that T78-dependent PBIP1-Plk1 interaction is specifically required for proper phosphorylation of CENP-Q during mitosis.
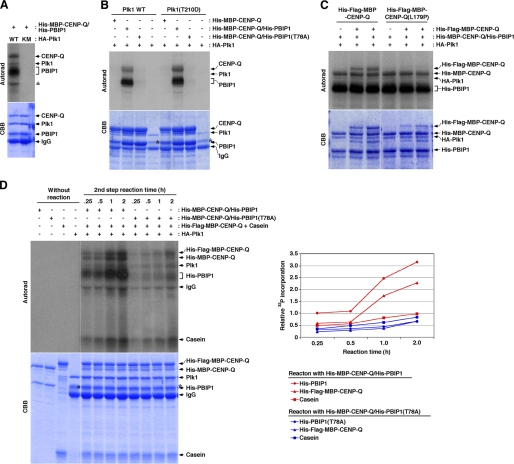
In Vitro Reconstitution of CENP-Q Phosphorylation by PBIP1-bound Plk1
To directly demonstrate the requirement of PBIP1 in Plk1-dependent CENP-Q phosphorylation, we reconstituted PBIP1-Plk1-dependent CENP-Q phosphorylation in vitro, using purified recombinant proteins from bacterial cells. Expression of PBIP1 alone (and also CENP-Q to some extent; supplemental Fig. S6A) was difficult because of its instability in the absence of CENP-Q (data not shown). Thus, we simultaneously expressed PBIP1 and CENP-Q, using a co-expression system, and purified the complex using a His affinity column. In in vitro kinase assays using the PBIP1/CENP-Q complex as substrates, Plk1 WT, but not the respective kinase-inactive K82M mutant (17), phosphorylated PBIP1 efficiently and also phosphorylated CENP-Q at a somewhat reduced level (Fig. 5A). We then examined whether the PBIP1 T78 function is required for Plk1-dependent CENP-Q phosphorylation using the PBIP1(T78A)/CENP-Q complex as substrates. Strikingly, the single T78A mutation greatly diminished not only Plk1-dependent PBIP1 phosphorylation but also Plk1-dependent CENP-Q phosphorylation (Fig. 5B). These observations suggest that Plk1-dependent PBIP1 phosphorylation at Thr-78 (and, therefore, subsequent Plk1 binding to the resulting phospho-T78 motif, as demonstrated in Fig. 4, A–C) is a prerequisite for Plk1-dependent CENP-Q phosphorylation. Plk1 binding to a phosphorylated target has been shown to activate its kinase activity (9), raising the possibility that an increased Plk1 activity through PBIP1 binding may have resulted in Plk1-dependent CENP-Q phosphorylation. However, this is unlikely, since a constitutively active Plk1(T210D) mutant (30) insensitive to PBD binding-induced regulation of its kinase activity (31) also failed to phosphorylate the PBIP1(T78A)/CENP-Q complex. Hence, Plk1-dependent CENP-Q phosphorylation is mediated by T78-dependent PBIP1 function.
FIGURE 5.
In vitro phosphorylation of CENP-Q by Plk1 is enhanced by the PBIP1-Plk1 interaction. A, His-MBP-CENP-Q/His PBIP1 complex purified from bacterial cells was reacted with HA-Plk1 in the presence of [32P]ATP. Samples were separated by 8% SDS-PAGE, stained with Coomassie (CBB), and then exposed. Asterisk, a nonspecific signal. B, Plk1 WT or a constitutively active Plk1(T210D) was reacted with the indicated His-MBP-CENP-Q/His-PBIP1 complex as in vitro substrates. Co-expression of both CENP-Q and PBIP1 was necessary to stably produce PBIP1 and, to some degree, CENP-Q (see supplemental Fig. S6). Asterisks, a contaminated protein. Note that Plk1(T210D), which is refractory to PBD binding-induced regulation of its own kinase activity (31), still requires T78-dependent PBIP1-Plk1 interaction to phosphorylate CENP-Q. C, in vitro Plk1 kinase reaction was carried out as in A using the His-MBP-CENP-Q/His-PBIP1 complex and either His-Flag-MBP-CENP-Q or His-Flag-MBP-CENP-Q(L179P) as substrates. Note that Plk1 phosphorylates the PBIP1 binding-competent His-MBP-CENP-Q and His-Flag-MBP-CENP-Q, but not the PBIP1 binding-incompetent His-Flag-MBP-CENP-Q(L179P) mutant in the same reaction tube. D, either the His-MBP-CENP-Q/His-PBIP1 or the His-MBP-CENP-Q/His-PBIP1(T78A) complex was first reacted with Plk1 in the presence of 100 μm ATP for 1 h; then each reaction was divided into four tubes. The second reaction was carried out for the indicated lengths of time after the addition of His-Flag-MBP-CENP-Q, casein, and 10 μCi [32P]ATP. Samples were separated by 8% SDS-PAGE, stained with Coomassie (CBB), and then autoradiographed. Casein, which does not required PBD-dependent binding for phosphorylation, was also included to compare with PBIP1 and CENP-Q. The level of phosphorylation onto each substrate was quantified (graph).
Next, we investigated whether the complex formation between PBIP1 and CENP-Q is required for Plk1-dependent CENP-Q phosphorylation, by providing additional CENP-Q WT or the PBIP1 binding-defective CENP-Q(L179P) mutant (larger in molecular size because of the addition of four copies of Flag tag) into the reaction mixture containing Plk1 and the PBIP1/CENP-Q complex. Results showed that Plk1 phosphorylated the PBIP1/CENP-Q complex and the PBIP1 binding-competent Flag-CENP-Q, presumably because Flag-CENP-Q was able to incorporate into the homo-octameric CENP-Q complex and associate with PBIP1 (Fig. 5C). In contrast, Plk1 failed to phosphorylate the PBIP1-binding defective Flag-CENP-Q(L179P) mutant under the same conditions (Fig. 5C). We were not able to examine the PBIP1(L337P L344P) mutant that also disrupted the PBIP1-CENP-Q interaction, because bacterially expressed His-PBIP1(L337P L344P) was much more unstable than the Flag-CENP-Q(L179P) mutant (supplemental Fig. S6B).
To closely determine the significance of the Plk1-PBIP1-CENP-Q complex formation in Plk1-dependent phosphorylation events, we performed in vitro kinase reactions after providing both Flag-CENP-Q and casein as additional substrates. Reactions were terminated at different time points to determine the efficiency of phosphorylation onto the substrates in a time-dependent manner. Similarly as above, Plk1 efficiently phosphorylated PBIP1 binding-competent Flag-CENP-Q only in the reactions containing the PBIP1/CENP-Q complex, but not in the reactions containing the PBIP1(T78A)/CENP-Q complex (Fig. 5D). In contrast, Plk1 phosphorylated casein at moderate levels under both conditions (Fig. 5D). A mildly increased level of casein phosphorylation in the presence of the PBIP1/CENP-Q complex could be attributable to a partial activation of Plk1 upon binding to the p-T78 motif of the PBIP1/CENP-Q complex, as reported previously (9). These results demonstrate that Plk1-dependent CENP-Q phosphorylation, but not the PBD binding-deficient casein, requires the presence of T78-containing PBIP1.
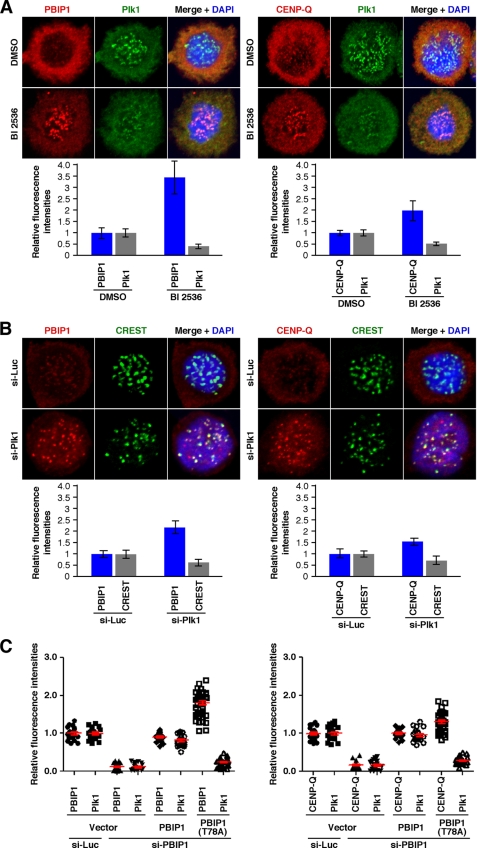
Plk1 Kinase Activity Induces Delocalization of the PBIP1-CENP-Q Complex from Mitotic Kinetochores
To investigate the physiological significance of Plk1-dependent PBIP1 and CENP-Q phosphorylation in vivo, HeLa cells were arrested at the G1/S boundary by double thymidine treatment (G1/S block) and released into a nocodazole-containing medium. As cells entered mitosis, cells were treated with BI 2536 to acutely inhibit Plk1 activity and then immunostained. As observed previously (23), BI 2536 treatment induced Plk1 delocalization from early mitotic kinetochores (e.g. prometaphase kinetochores), presumably due to the lack of Plk1 recruitment to these sites through a self-priming and binding mechanism. Interestingly, this treatment resulted in a drastic accumulation of PBIP1 and, to a somewhat lesser extent, CENP-Q at these locations (Fig. 6A). On the other hand, control cells untreated with BI 2536 exhibited an undetectable or very low level of kinetochore-localized PBIP1 and CENP-Q under these conditions (Fig. 6A). Depletion of Plk1 resulted in significantly increased levels of PBIP1 and CENP-Q at mitotic kinetochores (Fig. 6B). The level of the kinetochore marker, CREST, exhibited a somewhat diminished level in cells depleted of Plk1, which could be in part attributable to the detrimental effect of Plk1 knockdown. These findings suggest that the formation of the Plk1-PBIP1-CENP-Q complex and subsequent Plk1-dependent CENP-Q phosphorylation in early mitosis ultimately induces the dissociation of the PBIP1-CENP-Q complex from mitotic kinetochores. In support of this view, the level of the Plk1 binding-defective PBIP1(T78A) mutant localized at mitotic kinetochores was ∼80% higher than that of Plk1 binding-competent PBIP1 WT (Fig. 6C and supplemental Fig. S7). Although somewhat less dramatic than PBIP1, the level of kinetochore-localizing CENP-Q was also ∼30% higher in PBIP1(T78A) cells than in PBIP1 WT cells. Taken together, these observations suggest that Plk1 regulates the localization and delocalization of the PBIP1-CENP-Q complex by directly interacting with and phosphorylating the latter in a T78-dependent manner.
FIGURE 6.
Plk1 induces dissociation of the PBIP1-CENP-Q complex from mitotic kinetochores in a kinase activity-dependent manner. A, HeLa cells arrested at a G1/S block were released into nocodazole-containing medium. The cells were treated with a Plk1 inhibitor, BI 2536, 9 h after release and fixed 15 h after release. The resulting samples were immunostained and fluorescent signal intensities were quantified using Zeiss AIM confocal software from the kinetochores of greater than 20 cells. Representative confocal images are shown. The reduction in the level of kinetochore-associated Plk1 is in part due to the lack of self-recruited Plk1 to this location through a self-priming and binding mechanism (15, 23). Bars, standard deviation. B, HeLa cells silenced for control luciferase (si-Luc) or Plk1 (si-Plk1) were treated with nocodazole for 18 h, fixed, and immunostained. Fluorescent signal intensities were quantified as in A from the kinetochores of greater than 20 cells. Why the level of CREST is diminished in the si-Plk1 cells is not clear at present. Bars, standard deviation. C, HeLa cells expressing the indicated RNAi-insensitive PBIP1 constructs were depleted of control luciferase (si-Luc) or endogenous PBIP1 (si-PBIP1), and then subjected to immunostaining analyses (see supplemental Fig. S7). Relative fluorescence intensities quantified from metaphase cells were plotted. Bars (red) indicate the averages of signal intensities with standard error of the mean obtained from greater than 20 cells.
DISCUSSION
Identification of CENP-Q as a Primary Binding Target of PBIP1
We have shown that Plk1 recruits itself to the PBIP1-loaded kinetochore through a self-priming and binding mechanism (7, 15, 32), suggesting that PBIP1 serves as a scaffold for proper recruitment of Plk1 to this site. In this study, we investigated the mechanism of how PBIP1 itself is targeted to kinetochores. Previous studies have shown that PBIP1 belongs to CENP-A NAC, which is recruited to CENP-A-containing nucleosomes (13). Interestingly, however, PBIP1 is not required for the localization of CENP-A NAC components, but rather is required for the localization of the CAD components such as CENP-O, CENP-P, and CENP-Q at interphase and mitotic centromeres (13, 21). In good agreement with these observations, our co-immunoprecipitation studies using co-transfected lysates showed that PBIP1 interacted strongly with CENP-Q. PBIP1 also interacted with CENP-P and at least two CENP-A NAC components, CENP-H and CENP-T, albeit at somewhat diminished levels. These observations suggest that the primary binding target of PBIP1 is likely CENP-Q. Consistent with this finding, among the four CENP-O class proteins (CENP-O, CENP-P, CENP-Q, and PBIP1/CENP-50[U]) mutually required for their stability in chicken DT40 cells, loss of either PBIP1 or CENP-Q is particularly detrimental to the stability of CENP-Q or PBIP1, respectively (20). Taken together, our results support the previous observation that PBIP1 interacts with various proteins in the CENP-A NAC and CAD (13), and further suggest that PBIP1 and CENP-Q form a stable binary subcomplex. Consistent with this view, we found that the unstable nature of bacterially expressed PBIP1 could be fully remedied by the co-expression of CENP-Q in the same cell (supplemental Fig. S6).
Requirement of the PBIP1-CENP-Q Interaction in Their Localization to Centromeres
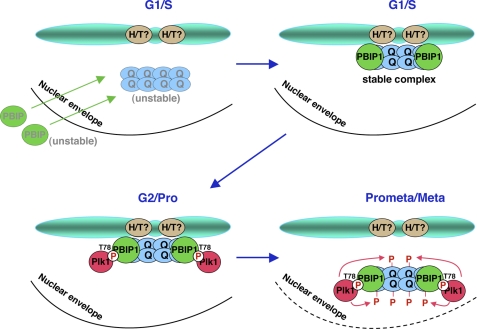
Hori et al. (20) showed that PBIP1 and CENP-Q are mutually required to localize to interphase and mitotic centromeres. However, this could be largely due to the unstable nature of each of these proteins in the absence of the other. We observed that either mutations in the putative leucine zipper domain of PBIP1 or a mutation in the putative coiled-coil domain of CENP-Q completely disrupted the PBIP1-CENP-Q interaction. Interestingly, these mutants were crippled in their localizations to centromeres, even though they were detectably expressed (Fig. 3D), hinting that a prior formation of the PBIP1-CENP-Q complex is crucial for proper localization of these two proteins at centromeres. Consistent with this notion, all of the PBIP1 fragments capable of interacting with CENP-Q localized efficiently to centromeres (Fig. 2C and supplemental Figs. S2 and S3). Furthermore, expression of a nuclear localization-defective PBIP1 mutant in si-PBIP1 cells failed to support CENP-Q localization to centromeres (supplemental Fig. S4). Unlike CENP-Q, which associates with centromeres throughout the cell cycle, a large fraction of PBIP1 was found in cytosol (data not shown). Therefore, proper translocation of PBIP1 into the nucleus is an important step for the formation of the PBIP1-CENP-Q complex and subsequent localization of the complex to centromeres (Fig. 7).
FIGURE 7.
A model illustrating the formation of the PBIP1-CENP-Q complex in the nucleus and subsequent Plk1-dependent delocalization of the complex from mitotic kinetochores. Early in the cell cycle (e.g. G1/S), monomeric PBIP1 translocates into the nucleus and binds to octameric CENP-Q (25) to stabilize the latter by forming the PBIP1-CENP-Q complex. Disruption of the PBIP1-CENP-Q complex by either the mutation of the leucine zipper domain of PBIP1 or the coiled-coil domain of CENP-Q delocalizes both PBIP1 and CENP-Q from kinetochores, suggesting that the complex formation is a prerequisite not only for their stability but also for their subcellular localization. Late in the cell cycle (e.g. G2 or early Prophase), Plk1 phosphorylates PBIP1 at T78 and recruits itself to the centromere-associated PBIP1-CENP-Q complex. As Plk1 becomes activated in early mitosis, p-T78 PBIP1-bound Plk1 phosphorylates CENP-Q and dissociates the complex from the kinetochores. This event may lead to the degradation of the PBIP1-CENP-Q complex, thus allowing Plk1 to interact with other binding targets at kinetochores important for proper M phase progression.
The mechanism underlying the PBIP1-CENP-Q complex-dependent subcellular localization remains elusive. We found that PBIP1 does not appear to form a homomeric complex (supplemental Fig. S1), suggesting that the impaired capacity of the PBIP1 leucine zipper mutant to localize to centromeres could be largely due to its inability to interact with endogenous CENP-Q. On the other hand, because CENP-Q is shown to form a homo-octameric complex (25), the localization defect associated with the CENP-Q(L179P) mutant could be the result of a failure to form either a homo-octameric CENP-Q complex or a larger complex with PBIP1. However, we failed to detect a homomeric interaction for the L179P mutant (Fig. 3C), suggesting that a defect in the formation of the homomeric CENP-Q complex resulted in the loss of the CENP-Q-PBIP1 interaction and their localization to centromeres. At present, how PBIP1 forms a larger complex with the homo-octameric CENP-Q complex is not known. Nonetheless, the mutual requirement of PBIP1 and CENP-Q for both protein stability and subcellular localization suggests that the PBIP1-CENP-Q complex serves as a functional unit for the assembly of kinetochore components.
It should be noted that, in addition to the octameric CENP-Q complex, PBIP1 appears to bind to some of the CENP-A NAC components, such as CENP-H or CENP-T (Fig. 1) (13), hinting that these latter interactions may also contribute, at least in part, to the recruitment of the PBIP1-CENP-Q complex to centromeres (Fig. 7). The presence of these additional interactions may help explain why the CENP-Q binding-incompetent PBIP1 leucine zipper mutant still exhibits a low level of localization capacity to centromeres.
Mechanism Underlying the Formation of the CENP-Q-PBIP1-Plk1 Complex and Plk1-dependent CENP-Q Phosphorylation by PBIP1-T78-bound Plk1
Our results demonstrate that PBIP1 has two distinct interaction domains: the N-terminal Plk1-binding domain and the C-terminal CENP-Q-binding domain. Plk1 interacted with the PBIP1-CENP-Q complex by binding to the T78 motif of PBIP1 through the self-priming and binding mechanism (15, 32). However, Plk1 did not appear to interact with CENP-Q directly under various experimental conditions. PBIP1(T78A) defective in Plk1 binding interacted with CENP-Q normally. Taken together, these observations suggest that Plk1, PBIP1, and CENP-Q form a ternary complex in which the PBIP1-CENP-Q interaction occurs independently of the PBIP1-Plk1 interaction.
It is well appreciated that the PBD of Plk1 binds to a phosphorylated motif on a protein that has been phosphorylated beforehand by a priming kinase. This binding step is thought to position the catalytic domain of Plk1 to phosphorylate the same protein bound to its PBD. Numerous Plk1 substrates following this so-called “processive phosphorylation” mechanism have been reported (7, 33). In contrast to this prevailing mechanism, however, our results provided here demonstrate that PBIP1 bridges the interaction between Plk1 and CENP-Q, and PBIP1-bound Plk1 phosphorylates CENP-Q. Loss of either the T78-dependent Plk1-PBIP1 interaction or the PBIP1-CENP-Q interaction was sufficient to disrupt Plk1-dependent CENP-Q phosphorylation. This unique mechanism of phosphorylating a third protein associating with the PBD-binding target is the first example of the Plk1 PBD-dependent “distributive phosphorylation” model originally proposed by Lowery et al. (33).
At present, the significance of the formation of the Plk1-PBIP1-CENP-Q ternary complex remains elusive. Biochemically, the PBIP1-CENP-Q interaction may help promote Plk1-dependent CENP-Q phosphorylation by bringing CENP-Q in close proximity to PBIP1-bound Plk1. However, the fact that Plk1 fails to phosphorylate CENP-Q in the absence of the PBIP1-CENP-Q interaction suggests that the ternary complex formation is an obligatory step for Plk1-dependent CENP-Q phosphorylation. One possibility is that this mechanism may be important to limit the level of CENP-Q phosphorylation to the amount of PBIP1-bound Plk1, thus allowing Plk1 to tightly regulate the phosphorylation-dependent CENP-Q function at centromeres. Because the PBIP1-Plk1 interaction occurs during late G2 or early mitosis, this mechanism ensures that Plk1-dependent CENP-Q phosphorylation occurs only at this stage. Alternatively, the formation of the PBIP1-CENP-Q complex may be required to properly expose Plk1-dependent phosphorylation site(s) on CENP-Q. These possibilities are not mutually exclusive. Highlighting the specificity of the Plk1-PBIP1 complex-dependent CENP-Q phosphorylation, Plk1-dependent phosphorylation onto Bub1, BubR1, or Cdc25C does not require the PBIP1-Plk1 interaction.
Plk1-dependent Regulation of the PBIP1-CENP-Q Complex at Mitotic Kinetochores
Loss of T78-dependent PBIP1 function in human cultured cells results in a mitotic block as a result of improper chromosome alignment and spindle checkpoint activation (15). PBIP1/CENP-50 is also shown to be required for proper recovery from nocodazole-induced spindle damage in chicken DT40 cells (14, 20). These observations suggest that PBIP1, and likely the Plk1-PBIP1-CENP-Q complex as described above, plays an important role in the generation and/or maintenance of stable MT-kinetochore interaction and proper chromosome segregation. PBIP1 has been proposed to function as a temporal scaffold for proper recruitment and delivery of Plk1 to other Plk1-binding targets at kinetochores (34), thus allowing Plk1 to induce a previously uncharacterized downstream event(s) important to promote metaphase-to-anaphase transition. Intriguingly, we observed that loss of PBIP1 T78-dependent Plk1 function substantially increased the amounts of PBIP1 and CENP-Q at mitotic kinetochores (Fig. 6). Given that Plk1 phosphorylates CENP-Q in vitro, we propose that Plk1 induces dissociation of the PBIP1-CENP-Q complex from kinetochores by directly phosphorylating CENP-Q (Fig. 7). At present, however, we cannot exclude the possibility that PBIP1-bound Plk1 also phosphorylates other neighboring proteins to promote this event.
Although PBIP1 appears to be the major scaffold for the initial recruitment of Plk1 to interphase and early mitotic centromeres (34), the role of the PBIP1-Plk1 interaction has been elusive. Here we show that Plk1 generates a ternary complex with PBIP1 and CENP-Q, and this complex formation is critical for PBIP1-bound Plk1 to phosphorylate CENP-Q and to dissociate the PBIP1-CENP-Q complex from mitotic kinetochores in the mid/late stages of mitosis. In this regard, determination of Plk1-dependent CENP-Q phosphorylation sites and investigation of the significance of this event is likely an important step to better comprehend the function of the Plk1-PBIP1-CENP-Q complex in the regulation of Plk1-mediated mitotic events.
Supplementary Material
Acknowledgments
We thank Chou-Zen Giam, Erich A. Nigg, Sheila A. Stewart, Michael B. Yaffe, Tim Yen, and Hongtao Yu for reagents. We are also indebted to Susan Garfield for assistance in confocal microscopy.
This work was supported, in whole or in part, by NCI, National Institutes of Health intramural Grant Z01 BC 010520 (to K. S. L.), the Korea Basic Science Institute's high field NMR Research Program Grant T3022B (to J. K. B.), and the World Class Institute research program of the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology of Korea (to B. Y. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7.
- Plk
- polo-like kinase
- PBD
- polo-box domain
- NAC
- nucleosome-associated complex
- CAD
- CENP-A-nucleosome distal.
REFERENCES
- 1. Archambault V., Glover D. M. (2009) Nat. Rev. Mol. Cell Biol. 10, 265–275 [DOI] [PubMed] [Google Scholar]
- 2. Barr F. A., Silljé H. H., Nigg E. A. (2004) Nat. Rev. Mol. Cell Biol. 5, 429–440 [DOI] [PubMed] [Google Scholar]
- 3. Petronczki M., Lénárt P., Peters J. M. (2008) Dev. Cell 14, 646–659 [DOI] [PubMed] [Google Scholar]
- 4. van de Weerdt B. C., Medema R. H. (2006) Cell Cycle 5, 853–864 [DOI] [PubMed] [Google Scholar]
- 5. Takaki T., Trenz K., Costanzo V., Petronczki M. (2008) Curr. Opin. Cell Biol. 20, 650–660 [DOI] [PubMed] [Google Scholar]
- 6. Lowery D. M., Lim D., Yaffe M. B. (2005) Oncogene 24, 248–259 [DOI] [PubMed] [Google Scholar]
- 7. Park J. E., Soung N. K., Johmura Y., Kang Y. H., Liao C., Lee K. H., Park C. H., Nicklaus M. C., Lee K. S. (2010) Cell. Mol. Life Sci. 67, 1957–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cheng K. Y., Lowe E. D., Sinclair J., Nigg E. A., Johnson L. N. (2003) EMBO J. 22, 5757–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Elia A. E., Rellos P., Haire L. F., Chao J. W., Ivins F. J., Hoepker K., Mohammad D., Cantley L. C., Smerdon S. J., Yaffe M. B. (2003) Cell 115, 83–95 [DOI] [PubMed] [Google Scholar]
- 10. Winkles J. A., Alberts G. F. (2005) Oncogene. 24, 260–266 [DOI] [PubMed] [Google Scholar]
- 11. Lane H. A., Nigg E. A. (1996) J. Cell Biol. 135, 1701–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Cárcer G., do Carmo Avides M., Lallena M. J., Glover D. M., González C. (2001) EMBO J. 20, 2878–2884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Foltz D. R., Jansen L. E., Black B. E., Bailey A. O., Yates J. R., 3rd, Cleveland D. W. (2006) Nat. Cell Biol. 8, 458–469 [DOI] [PubMed] [Google Scholar]
- 14. Minoshima Y., Hori T., Okada M., Kimura H., Haraguchi T., Hiraoka Y., Bao Y. C., Kawashima T., Kitamura T., Fukagawa T. (2005) Mol. Cell. Biol. 25, 10315–10328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kang Y. H., Park J. E., Yu L. R., Soung N. K., Yun S. M., Bang J. K., Seong Y. S., Yu H., Garfield S., Veenstra T. D., Lee K. S. (2006) Mol. Cell 24, 409–422 [DOI] [PubMed] [Google Scholar]
- 16. Golsteyn R. M., Mundt K. E., Fry A. M., Nigg E. A. (1995) J. Cell Biol. 129, 1617–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee K. S., Yuan Y. L., Kuriyama R., Erikson R. L. (1995) Mol. Cell. Biol. 15, 7143–7151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Seong Y. S., Kamijo K., Lee J. S., Fernandez E., Kuriyama R., Miki T., Lee K. S. (2002) J. Biol. Chem. 277, 32282–32293 [DOI] [PubMed] [Google Scholar]
- 19. Yun S. M., Moulaei T., Lim D., Bang J. K., Park J. E., Shenoy S. R., Liu F., Kang Y. H., Liao C., Soung N. K., Lee S., Yoon D. Y., Lim Y., Lee D. H., Otaka A., Appella E., McMahon J. B., Nicklaus M. C., Burke T. R., Jr., Yaffe M. B., Wlodawer A., Lee K. S. (2009) Nat. Struct. Mol. Biol. 16, 876–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hori T., Okada M., Maenaka K., Fukagawa T. (2008) Mol. Biol. Cell 19, 843–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Okada M., Cheeseman I. M., Hori T., Okawa K., McLeod I. X., Yates J. R., 3rd, Desai A., Fukagawa T. (2006) Nat. Cell Biol. 8, 446–457 [DOI] [PubMed] [Google Scholar]
- 22. He T. C., Zhou S., da Costa L. T., Yu J., Kinzler K. W., Vogelstein B. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lénárt P., Petronczki M., Steegmaier M., Di Fiore B., Lipp J. J., Hoffmann M., Rettig W. J., Kraut N., Peters J. M. (2007) Curr. Biol. 17, 304–315 [DOI] [PubMed] [Google Scholar]
- 24. Hanissian S. H., Akbar U., Teng B., Janjetovic Z., Hoffmann A., Hitzler J. K., Iscove N., Hamre K., Du X., Tong Y., Mukatira S., Robertson J. H., Morris S. W. (2004) Oncogene. 23, 3700–3707 [DOI] [PubMed] [Google Scholar]
- 25. Amaro A. C., Samora C. P., Holtackers R., Wang E., Kingston I. J., Alonso M., Lampson M., McAinsh A. D., Meraldi P. (2010) Nat. Cell Biol. 12, 319–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bornberg-Bauer E., Rivals E., Vingron M. (1998) Nucleic Acids Res. 26, 2740–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Elowe S., Hümmer S., Uldschmid A., Li X., Nigg E. A. (2007) Genes Dev. 21, 2205–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Qi W., Tang Z., Yu H. (2006) Mol. Biol. Cell 17, 3705–3716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Elia A. E., Cantley L. C., Yaffe M. B. (2003) Science 299, 1228–1231 [DOI] [PubMed] [Google Scholar]
- 30. Lee K. S., Erikson R. L. (1997) Mol. Cell. Biol. 17, 3408–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jang Y. J., Lin C. Y., Ma S., Erikson R. L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 1984–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee K. S., Park J. E., Kang Y. H., Zimmerman W., Soung N. K., Seong Y. S., Kwak S. J., Erikson R. L. (2008) Cell Cycle 7, 141–145 [DOI] [PubMed] [Google Scholar]
- 33. Lowery D. M., Mohammad D. H., Elia A. E., Yaffe M. B. (2004) Cell Cycle. 3, 128–131 [PubMed] [Google Scholar]
- 34. Lee K. S., Oh D. Y., Kang Y. H., Park J. E. (2008) Cell Div. 3, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.