Abstract
Heparan sulfate (HS) is a highly sulfated polysaccharide participated in essential physiological functions from regulating cell growth to blood coagulation. HS contains sulfated domains known as N-S domains and low sulfate domains known as N-Ac domains. The distribution of the domain structures is likely governed by the action of glucosaminyl N-deacetylase/N-sulfotransferase (NDST). Here, we sought to determine the substrate specificity of NDST using model substrates and recombinant NDST protein. We discovered that NDST-1 carries out the modification in a highly ordered fashion. The enzyme sulfates the substrate from the nonreducing end toward the reducing end consecutively, leading to the product with a cluster of N-sulfo glucosamine residues. Furthermore, a preexisting N-sulfo glucosamine residue prevents the action of NDST-1 at the residues immediately located at the nonreducing end, allowing the formation of an N-Ac domain. Our results provide the long sought evidence for understanding the formation of sulfated versus nonsulfated domains in the HS isolated from cells and tissues. The study demonstrates the regulating role of NDST-1 in mapping the sulfation patterns of HS.
Keywords: Carbohydrate, Carbohydrate Biosynthesis, Heparan Sulfate, Heparin, Sulfotransferase
Introduction
Heparan sulfate (HS)2 is a highly sulfated linear polysaccharide ubiquitously present on the cell surface and in the extracellular matrix. HS plays essential roles in a wide variety of biological processes, including assisting embryonic development, inflammatory response, viral and bacterial infection, and regulating blood coagulation (1). HS consists of a disaccharide repeating unit of glucuronic acid (GlcA) or iduronic acid (IdoA) and glucosamine, both capable of carrying sulfo groups. The location of the sulfo groups and IdoA plays a crucial role in determining the functions of HS (2). Structural analysis of HS isolated from natural sources indicates that the sulfated saccharide and IdoA residues are not evenly scattered along the polysaccharide, rather forming a cluster known as N-S domain (3). These N-S domains are separated by the unsulfated saccharide units known as N-Ac domain, referring to the domain containing a high level of GlcNAc residue (3). The N-S domain is believed to exhibit the high binding affinity to proteins, and the distribution of the N-S domain is proposed to be essential for HS to display its full range of biological properties (4). However, how to regulate the biosynthesis of HS with specific sulfation patterns and the distribution of the N-S domains remains unknown.
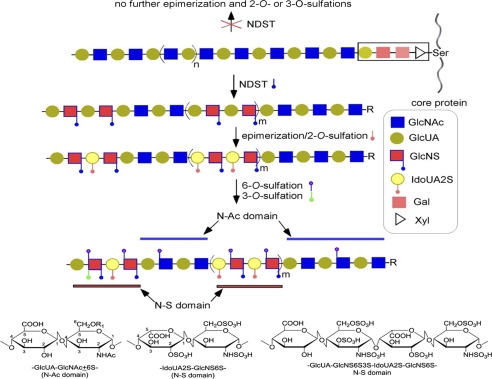
The biosynthesis of HS involves in building the polysaccharide backbone as well as introducing sulfo groups and IdoA residues. Multiple specialized enzymes, including HS polymerase, sulfotransferases and epimerase, participate in the process (Fig. 1). The synthesis is initiated from a tetrasaccharide (GlcA-Gal-Gal-Xyl) that is attached to the core protein. HS polymerase is responsible for building the polysaccharide backbone with a repeating unit of -GlcA-GlcNAc-. The backbone is then modified by N-deacetylase/N-sulfotransferase (NDST), C5-epimerase, 2-O-sulfotransferase, 6-O-sulfotransferase, and 3-O-sulfotransferase. The substrate specificities of the biosynthetic enzymes dictate the structures of HS product, including sulfation levels, the contents of IdoA units and the size of the polysaccharides (5). Understanding the biosynthetic mechanism of HS is not only important for delineating the contribution of structural selectivity of HS to the biological processes. The knowledge also aids us to develop an enzyme-based approach to synthesize HS-based therapeutics (6).
FIGURE 1.
Biosynthetic pathway of HS. The biosynthesis of HS is initiated from a linkage region tetrasaccharide (GlcA-Gal-Gal-Xyl) that is attached to the core protein, where GlcA, Gal, and Xyl represents glucuronic acid, galactose, and xylose, respectively. The polysaccharide backbone is then modified by different HS biosynthetic enzymes as described in the text. The lack of modification by NDST results in the polysaccharide losing its biological activities because the missing of 2-O-sulfation, epimerization, and 3-O-sulfation. The chemical structures of the disaccharide repeating unit of HS are shown in the bottom. In the figure, R represents the core protein and the linkage region tetrasaccharide that is attached to the core protein, and R1 represents -H or -SO3.
NDST catalyzes the conversion from a GlcNAc residue to a N-sulfo glucosamine (GlcNS). NDST displays two activities: N-deacetylase activity removes the acetyl group from the GlcNAc residue to yield a GlcNH2 residue; N-sulfotransferase activity transfers a sulfo group to the primary amine to form a GlcNS residue. Total of four isoforms of NDST are present in human genome (7, 8). Different isoforms are believed to exhibit different substrate specificities, mainly differed in the ratio of GlcNS/GlcNH2 and the length of the GlcNS domain (8). Among them, NDST-1 exhibits profound physiological functions and is the most widely expressed isoform. The N-sulfation is known to be perquisite for the subsequent epimerization and 2-O-sulfation, and possibly for 3-O-sulfation as demonstrated by in vitro studies (9, 10, 12).3 In vivo, the functions of NDSTs were discerned by gene knock-out experiments. NDST-2-null mice appear to be normal, and the only abnormality seen was in connective tissue-type mast cells (13). The NDST-3 knock-out mice are fertile, and exhibit only minor hematological and behavioral phenotypes (14). Unlike other NDST isoforms, the absence of NDST-1 in mice led to respiratory distress, and subsequently neonatal death (15). Conditional knock-out of NDST-1 revealed a series of physiological and pathophysiological roles, including the clearance of lipoproteins (16), the development of lobuloalveolar of mammary gland (17), neutrophil trafficking (18), and inhibition of tumor angiogenesis (19). These in vivo effects of NDST-1 were attributed to the disruption of the N-sulfation, leading to dramatic changes in O-sulfations and epimerization of HS in genetically modified animals.
In this article, we studied NDST-1 using structurally defined oligosaccharide substrates followed by the structural analysis using electrospray ionization mass spectrometry (ESI-MS). Our results demonstrate that NDST-1 can act on a substrate in both random and nonrandom ways to yield the products with different sulfation patterns. It appears that NDST-1 can randomly choose to sulfate an internal GlcNAc residue. Once the sulfation starts, the enzymes reacts with the substrate in a highly ordered manner. NDST-1 sulfates consecutively from nonreducing toward the reducing end to yield a cluster of GlcNS residues, forming the anticipated N-S domain. The presence of N-sulfo group prevents the N-sulfation at the immediate nonreducing residue, permitting the formation of the N-Ac domain. Our results provide the long-sought biochemical evidence to understand the causal role of NDST to synthesize domain structures in HS.
EXPERIMENTAL PROCEDURES
Expression of NDST-1
The rat NDST-1 cDNA (R43-R882) was cloned into pYES2 vector (Invitrogen) using the XbaI and BglII sites to generate NDST-1 protein. The new recombinant plasmid was named pYES2-NDST-1. The coding region of expression plasmid was sequenced to confirm the reading frame and absence of deleterious mutations (University of North Carolina DNA sequencing facility). The pYES2-NDST-1 was transformed into the Saccharomyces cerevisiae InvSc1 (Invitrogen) using the S.c. Easy-Comp Transformation kit (Invitrogen) according to the manufacturer's instructions. NDST-1 was expressed and purified as previously described (20).
Modification of Oligosaccharides with NDST-1
As previously described (20), either heparosan or oligosaccharide substrates were incubated with NDST-1 in a buffer containing 50 mm MES buffer, pH 6.5, 10 mm MnCl2, and PAPS (50 μm) at 23 °C for 24 h.
Preparation of Size-defined N-Acetyl Heparosan Oligosaccharides
N-Acetyl heparosan polysaccharide was purified from the overnight culture of E. coli K5 using a DEAE column (21). As described by Sigulinsky et al. (22), the size-defined N-acetyl heparosan oligosaccharides were prepared by partial digestion using heparin lyase III. Heparosan polysaccharide (5 mg) was incubated in 25 mm NaH2PO4, pH 7.0 at 37 °C for 24 h, using 2 μg/ml heparin lyases III. Then, the N-acetyl heparosan oligosaccharides of varying degree of polymerization were separated and purified by fractionation on Bio-Gel P10, which was eluted with a buffer containing 25 mm Tris (pH 7.2) and 1000 mm NaCl at a flow rate of 6 ml/h. The eluent was monitored by UV 232 nm. Fractions containing size-defined N-acetyl heparosan oligosaccharides were pooled and desalted by dialysis. The purity of the oligosaccharides was confirmed by ESI-MS.
HPLC Analysis
To confirm the identity of NDST-1, the recombinant protein was incubated with heparosan and PAPS. The product was digested with heparin lyase III followed by a disaccharide analysis using reverse phase ion pairing HPLC to demonstrate the formation of the GlcNS-containing disaccharides. The method was described in a report by Duncan et al. (21). N-Sulfo oligosaccharides were analyzed and separated on a DEAE column (4.6 mm ID × 7.5 cm, Tosoh Bioscience), which was eluted with a linear gradient of from 0 to 1 m NaCl in 20 mm Tris-HCl (pH 7.0) over 80 min at a flow rate of 0.75 ml/min.
Microdialysis of Oligosaccharide
The oligosaccharides were subjected to microdialysis prior to the MS analysis. The dialysis was carried out using hollow fiber dialysis tubing (13,000 MWCO; Spectrum) against 20 mm ammonium acetate.
Mass Spectrometry Analysis
MS analyses were performed on a Thermo LCQ-Deca. The oligosaccharides were dissolved in 50% methanol. A syringe pump (Harvard Apparatus) was used to introduce the sample via direct infusion (35 μl/min) into the instrument. Experiments were performed in negative ionization mode with a spray voltage at 3 KV and a capillary temperature of 275 °C. The automatic gain control was set to 1 × 107 for full scan MS and 2 × 107 for MS/MS experiments. For MS/MS experiments, selection of each precursor ion was achieved using isolation width of 3 Da and the activation energy was 40% normalized collision energy. The product ions in MS/MS data were labeled according to the Domon-Costello nomenclature. The MS and MS/MS data were acquired and processed using Xcalibur 1.3 software.
Preparation of Hexasaccharide 9, Decasaccharide 10, and Dodecasaccharide 11
The hexasaccharide backbone with a structure of GlcA-GlcNTFA-GlcA-GlcNAc-GlcA-AnMan was elongated from a disaccharide, GlcA-AnMan, as described previously (12), where GlcNTFA represents N-trifluoroacetylated glucosamine. The elongation as completed with UDP- monosaccharide donors (UDP-GlcNAc, UDP-GlcA, or UDP-N-trifluoroacetylated glucosamine (GlcNTFA)) and bacterial glycosyltransferases (KfiA or pmHS2) in the buffer containing 25 mm Tris-HCl (pH 7.2) and 10 mm MnCl2. The reaction was incubated at room temperature for 16 h and analyzed by a polyamine-based HPLC column (from Waters) to ensure that >95% of UDP-monosaccharide donors was converted to UDP. After elongating two monosaccharides, the reaction mixture was resolved on a Bio-Gel P-2 column which was equilibrated with 0.1 m ammonium bicarbonate at a flow rate of 3 ml/h. The elution position of the product was determined by MS analysis. The reaction cycles was repeated two times to prepare the hexasaccharide backbone.
The N-sulfation of the hexasaccharide backbone was completed by detrifluoroacetylation followed by N-sulfation. The hexasaccharide backbone (5 μmol) was dried and resuspended in a solution (20 ml) containing CH3OH, H2O, and (C2H5)3N (v/v/v, 2:2:1). The reaction was incubated at 37 °C for 6 h. The samples were dried and reconstituted in H2O. The de-N-trifluoroacetylated hexasaccharide was incubated with N-sulfotransferase (180 μg/ml) and 0.75 mm PAPS in a total volume of 20 ml to yield hexasaccharide 9. The product was confirmed by ESI-MS analysis. Further elongation of hexasaccharide 9 to decasaccharide 10 and dodecasaccharide 11 was achieved by KfiA and pmHS2 following essentially the same procedure.
RESULTS
Our efforts for determining the substrate specificity of NDST-1 included the expression of recombinant NDST-1 protein and preparation of structurally defined oligosaccharide substrates. A catalytic domain NDST-1, comprising of the activities of N-deacetylase and N-sulfotransferase, was successfully expressed in S. cerevisiae following the report by Saribas et al. (20). Structurally defined oligosaccharide substrates were isolated from partially depolymerized heparosan, a polysaccharide with a repeating disaccharide of -GlcA-GlcNAc- (23). To this end, heparosan was incubated with a limited amount of heparin lyase III, and the depolymerized products were resolved on size exclusion chromatography, Bio-Gel P-10 column. Total of five oligosaccharide substrates ranging from hexasaccharide to dodecasaccharide, with a generic structure of ΔUA-(GlcNAc-GlcA)n-GlcNAc, were obtained. The structures of the oligosaccharide substrates were confirmed by ESI-MS (supplemental Table S1).
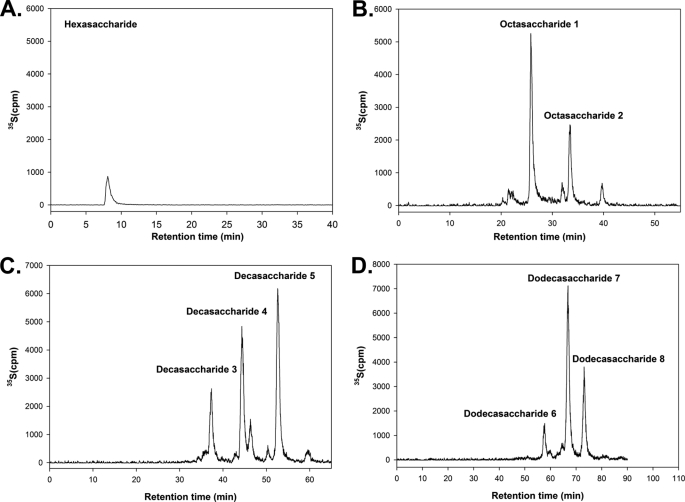
The availability of the structurally defined oligosaccharides enabled us to determine the minimum size of the substrate for NDST-1. The oligosaccharide substrates were incubated with NDST-1and [35S]PAPS, and the products were analyzed by DEAE-HPLC. Several 35S-labeled peaks were detected as analyzed by DEAE-HPLC when octa-, deca-, and dodecasaccharides were utilized, suggesting that these oligosaccharides are the substrates for NDST-1 (Fig. 2, B–D). In contrast, only a very small 35S-labeled peak was observed from the hexasaccharide sample (Fig. 2A), suggesting that hexasaccharide is not a good substrate for NDST-1. Our results also suggest that octasaccharide is likely to be the minimum size for NDST-1 to modify.
FIGURE 2.
DEAE-HPLC profiles of NDST-1-modified oligosaccharides. The oligosaccharide substrates were incubated with NDST-1 and [35S]PAPS to yield the 35S-labeled products. The products were purified by Bio-Gel P-10 and then subjected to the analysis by high resolution DEAE-HPLC. Panel A shows the profile of using a hexasaccharide substrate. Panel B shows the profile of using an octasaccharide substrate. Panel C shows the profile of using a decasaccharide. Panel D shows the profile of using a dodecasaccharide.
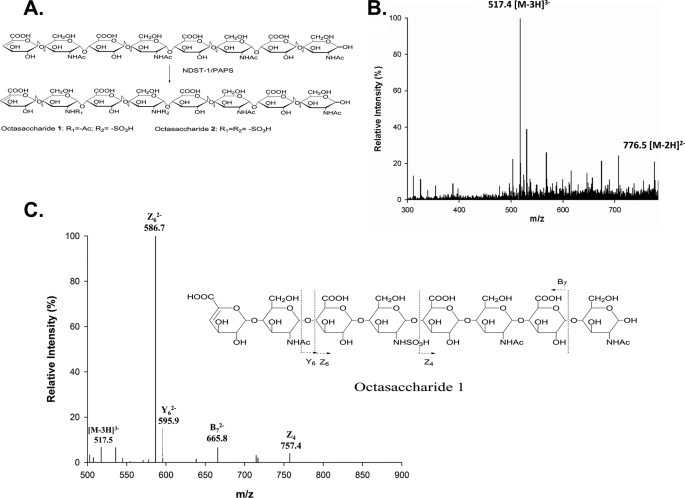
Using mass spectrometric techniques, we proved the structures of the NDST-1-modified oligosaccharides. We observed two 35S-labeled peaks for octasaccharide eluted at 27.5 and 33.5 min (Fig. 2B), three for decasaccharide eluted at 36.5, 43.5, and 54.0 min (Fig. 2C), and three for dodecasaccharide eluted at 57.5, 66.0, and 75.0 min (Fig. 2C). These oligosaccharide products were purified to homogeneity using DEAE-HPLC. An example for proving the structure of octasaccharide 1 is shown in Fig. 3 using ESI-MS and MS/MS techniques. The molecular mass of octasaccharide 1 was determined to be 1555.1 ± 0.1 Da, very close to the calculated molecular mass for an octasaccharide carrying a single sulfo group (1555.3 Da) (Fig. 3B). The position of the N-sulfo group in octasaccharide 1 was proved by using a MS/MS technique, a condition that fragments the octasaccharide at the glycolytic linkages. This MS/MS technique was successfully used to characterize the structure of N-sulfated octasaccharides in our previous publication (12). For example, two characteristic fragmented ions, Z4 and Z62−, at the m/z values of 757.4 and 586.7 provided the structural evidence that the GlcNS residue is located at the designated position of octasaccharide 1. Using similar techniques, we proved the structures of remaining seven oligosaccharide products generated by NDST-1 (Table 1 and supplemental Figs. S1–S7). Because only very small amount of 35S-labeled hexasaccharide was observed, we did not pursue to analyze its structure.
FIGURE 3.
Structural analysis of N-sulfo octasaccharide 1. A, reaction involved in the preparation of octasaccharide 1 and 2 by NDST-1. Non-radioactively labeled octasaccharide 1 and 2 were prepared by using unlabeled PAPS and separated by DEAE-HPLC. The separation of octasaccharide 1 and 2 was achieved by DEAE-HPLC as described in Fig. 2. B, ESI-MS spectrum of octasaccharide 1. C, MS/MS of octasaccharide 1. Precursor ion selection was at [M-3H]3−, m/z 517.4. The fragmentation pattern is depicted on the right. The conditions for MS and tandem MS analysis are described under “Experimental Procedures.” The product ions in the MS/MS data were labeled according to the Domon-Costello nomenclature (11).
TABLE 1.
Summary of the structures of NDST-1-modified products
| Substrates | Structures of the products | Measured | Calculated |
|---|---|---|---|
| MW (Da) | MW (Da) | ||
| ΔUA-(GlcNAc-GlcA)n-GlcNAca | |||
| n = 2 | No products | N/A | N/A |
| n = 3 | |||
| Octasaccharide 1 | ΔUA-GlcNAc-GlcA-GlcNS-GlcA-GlcNAc-GlcA-GlcNAc | 1555.1 ± 0.1 | 1555.3 |
| Octasaccharide 2 | ΔUA-GlcNS-GlcA-GlcNS-GlcA-GlcNAc-GlcA-GlcNAcb | 1593.9 ± 0.9 | 1593.3 |
| n = 4 | |||
| Decasaccharide 3 | ΔUA-GlcNAc-GlcA-GlcNAc-GlcA-GlcNS-GlcA-GlcNAc-GlcA-GlcNAc | 1934.6 ± 0.1 | 1934.6 |
| Decasaccharide 4 | ΔUA-GlcNAc-GlcA-GlcNS-GlcA-GlcNS-GlcA-GlcNAc-GlcA-GlcNAc | 1972.3 ± 0.6 | 1972.6 |
| Decasaccharide 5 | ΔUA-GlcNS-GlcA-GlcNS-GlcA-GlcNS-GlcA-GlcNAc-GlcA-GlcNAc | 2010.7 ± 0.1 | 2010.7 |
| n = 5 | |||
| Dodecasaccharide 6 | ΔUA-GlcNAc-GlcA-GlcNAc-GlcA-GlcNS-GlcA-GlcNS-GlcA-GlcNAc-GlcA-GlcNAc | 2351.8 ± 0.8 | 2352.0 |
| Dodecasaccharide 7 | ΔUA-GlcNAc-GlcA-GlcNS-GlcA-GlcNS-GlcA-GlcNS-GlcA-GlcNAc-GlcA-GlcNAc | 2390.1 ± 0.3 | 2390.9 |
| Dodecasaccharide 8 | ΔUA-GlcNS-GlcA-GlcNS-GlcA-GlcNS-GlcA-GlcNS-GlcA-GlcNAc-GlcA-GlcNAc | 2428.2 ± 0.2 | 2428.0 |
a The oligosaccharide substrates were purified by a Bio-Gel P-10 column from partially heparin lyase III-digested heparosan. The structures of the substrates were confirmed by ESI-MS.
b The position of the N-sulfo glucosamine in the products was determined by MS/MS as demonstrated in Fig 3 and supplemental Figs. S1 to S7. The N-sulfation sites are presented in bold face.
Analysis of the structures of NDST-1-modified oligosaccharides revealed unique N-sulfation patterns. First, the N-sulfation site occurs five saccharide units from the reducing end. NDST-1 does not appear to prefer to a specific GlcNAc in the substrate as long as it is located at certain distant from the reducing end. By choosing different internal GlcNAc sites to start the N-sulfation, NDST-1 modification resulted in multiple products. Second, a cluster of N-sulfation was formed. Regardless of the size of the oligosaccharide substrate, the products contained a cluster of the repeating unit of -GlcA-GlcNS- unless only a single unit of -GlcA-GlcNS- was present. At present time, the maximum size of the cluster of N-sulfation domain generated by NDST-1 could not be determined due to the technical difficulty in analyzing the structures larger than dodecasaccharide by MS. Third, the N-sulfation by NDST-1 appears to occur in a direction from the nonreducing end toward the reducing end. This conclusion was drawn from our attempt to introduce additional N-sulfo groups to those partially N-sulfated oligosaccharides, i.e. conversion of octasaccharide 1 to octasaccharide 2. Continuing incubation of octasaccharide 1 with fresh batches of NDST-1 and PAPS failed to yield octasaccharide 2, suggesting that octasaccharide 1 is no longer a substrate for NDST-1. Similarly, conversion of decasaccharide 3 and 4, and dodecasaccharide 6 and 7 to decasaccharide 5 and dodecasaccharide 8, respectively were also unsuccessful. Our results suggest that the presence of a GlcNS residue prevents the NDST-1 modification of the residues immediately at its nonreducing end. This conclusion is further strengthened by using a synthetic oligosaccharide carrying an N-sulfo group as described below.
HS also contains N-Ac domain, a stretch of repeating disaccharide of -GlcA-GlcNAc-, which is located between the N-sulfated domains. Our next goal was to determine if NDST-1 is capable of generating the gap of N-Ac domain between two GlcNS residues and the size of the nonsulfated gap. To achieve this goal, we synthesized a hexasaccharide substrate, hexasaccharide 9, with a structure of GlcA-GlcNS-GlcA-GlcNAc-GlcA-AnMan. This hexasaccharide contained a GlcNS residue near the nonreducing end that was synthesized using a chemoenzymatic approach as described previously (12). Hexasaccharide 9 was then elongated to a decasaccharide (decasaccharide 10) and a dodecasaccharide (dodecasaccharide 11) using two glycosyl transferases: KfiA of Escherichia coli, a N-acetyl glucosaminyl transferase (24) and heparosan synthase 2 of Pasteurella multocida (pmHS2), an enzyme possessing a GlcA transferase activity (25) (Fig. 4A). KfiA transfers a GlcNAc residue to the reducing end of hexasaccharide 9 primer to form an α1→4 glycosylic linkage to yield a heptasaccharide. Further, pmHS2 transfers a GlcA to the reducing end of the heptasaccharide to form a β1→4 glycosylic linkage. Repeating the process resulted in milligrams of decasaccharide 10 and dodecasaccharide 11. The success for the synthesis of hexasaccharide 9, decasaccharide 10, and dodecasaccharide 11 was confirmed by ESI-MS (supplemental Table S1).
FIGURE 4.
Determination of the gap between GlcNS residues. A, scheme for the synthesis of decasaccharide 10 and dodecasaccharide 11 from hexasaccharide 9. Hexasaccharide 9 was prepared using a chemoenzymatic approach as described under “Experimental Procedures”). The elongation of hexasaccharide 9 to decasaccharide 10 and dodecasaccharide 11 were completed using KfiA and pmHS2. Both decasaccharide 10 and dodecasaccharide 11 were incubated with NDST-1 and [35S]PAPS, and the products were analyzed by BioGel P-10. B, shows the elution profile of the reaction without an oligosaccharide substrate. C, shows the elution profile of the reaction using decasaccharide 10 as a substrate. D, shows that elution profile of the reaction using dodecasaccharide 11 as a substrate.
Using the synthetic oligosaccharide substrates, we identified the size of the N-Ac domain generated by NDST-1. Both decasaccharide 10 and dodecasaccharide 11 were incubated with NDST-1 and [35S]PAPS, and the products were analyzed by Bio-Gel P10 (Fig. 4, C and D). Two substrates provided stark contrast elution profiles from the P10 column. A clear 35S-labeled component was eluted at higher molecular weight range in the reaction using dodecasaccharide 11 as a substrate (Fig. 4D). The 35S-labeled product was absent in the reaction using decasaccharide 10 as a substrate (Fig. 4C), suggesting that no reaction occurred.
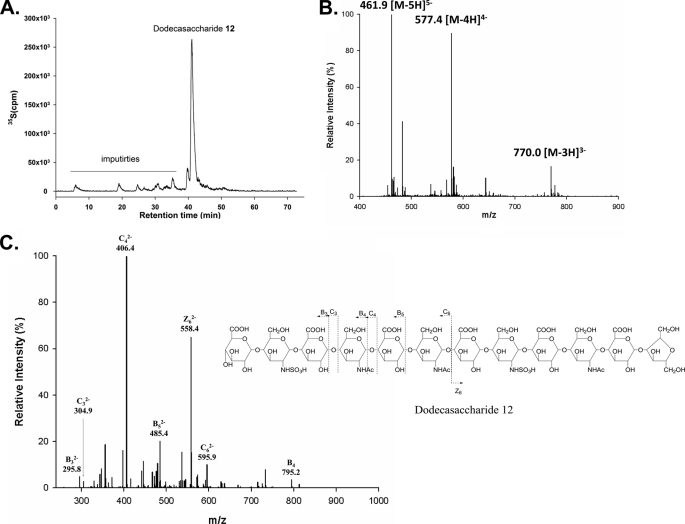
The structure of NDST-1-modified dodecasaccharid 12 was determined by ESI-MS and MS/MS. First, DEAE-HPLC analysis of NDST-1-modified dodecasaccharide revealed that the product was eluted as a single symmetric peak, suggesting only one product was formed after NDST-1 modification (Fig. 5). Analysis of NDST-1-modified dodecasaccharide 11 using MS and MS/MS confirmed its structure to be a dodecasaccharide carrying two sulfo groups with a structure of GlcA-GlcNS-GlcA-GlcNAc-GlcA-GlcNAc-GlcA-GlcNS-GlcA-GlcNAc-GlcA-AnMan (Fig. 5). For example, ESI-MS analysis revealed the molecular weight of dodecasaccharide 12 to be 2313.7 ± 0.7 Da, very close to the calculated molecular of dodecasaccharide carrying two sulfo groups (calculated 2312.9 Da) (Fig. 5B). Meanwhile, two characteristic fragmented ions of the MS/MS analysis, B32− (m/z, 295.8) and C32− (m/z, 304.9) were observed. The results suggest that the newly formed GlcNS residue was located at the nonreducing end, and thus, the structure of dodecasaccharide 12 was proven to be GlcA-GlcNS-GlcA-GlcNAc-GlcA-GlcNAc-GlcA-GlcNS-GlcA-GlcNAc-GlcA-AnMan (Fig. 5C). Taken together, our results demonstrate that the gap of the N-Ac domain for NDST-1 should be at least five saccharide residues.
FIGURE 5.
Structural characterization of dodecasaccharide 12. A, the DEAE-HPLC chromatogram of dodecasaccharide 12. The impurities were indicated. B, the MS spectrum of dodecasaccharide 12. C, the MS/MS of dodecasaccharide 12. Precursor ion selection at [M-5H]5−, m/z 461.9.
DISCUSSION
NDST is the first modification enzyme in the HS biosynthetic pathway. The actions of NDST determine the susceptibility of subsequent modifications, especially for epimerization, 2-O- and 3-O-sulfation. Therefore, understanding the substrate specificity of NDST becomes the center for investigating the control of biosynthesis of HS. However, several technical difficulties are associated with determining the substrate specificity of NDST. For example, large size oligosaccharide substrates are necessary for dissecting capability to create the N-S and N-Ac domains by the enzyme, and these substrates are typically difficult to obtain. Furthermore, to pinpoint the location of the N-sulfo group in a large oligosaccharide presents another technical challenge. Up to date, the studies of substrate specificity of NDST have been emphasized on using the polysaccharide substrate. The distribution of the N-sulfo groups was determined using nitrous acid degradation, provided that GlcNS is susceptible to the degradation by nitrous acid at pH 1.5, while the GlcNAc is resistant to nitrous acid degradation. Carlsson et al. (26) demonstrated that both NDST-1 and NDST-2 form a cluster of -GlcA-GlcNS- repeating units using this approach. However, due to the structural heterogeneity of the polysaccharide substrate, it is difficult to obtain a complete picture of the role of NDST in mapping the N-S and N-Ac domains.
In this study, we utilized structurally defined oligosaccharides and highly sensitive MS techniques to determine the substrate specificity of NDST-1 with accuracy. Our results confirm that NDST-1 generates a cluster of -GlcA-GlcNS- repeats. We also observe the enzyme sulfates a GlcNAc residue very selectively depending on the position in the oligosaccharide substrate and if a GlcNS residue is in proximity. The results from our study uncovered the mode of action of NDST-1 as illustrated in Fig. 6. In a substrate that is absent of a GlcNS residue, the enzyme can bind to any internal GlcNAc residue. Once the sulfation is initiated, NDST-1 converts the GlcNAc to GlcNS residue consecutively from nonreducing end to the reducing end until it reaches to the residue that is five units away from the reducing end. Although we cannot determine the length of the cluster of GlcNS, we clearly demonstrated that NDST-1 is fully capable of generating a domain containing four repeats of -GlcA-GlcNS-.
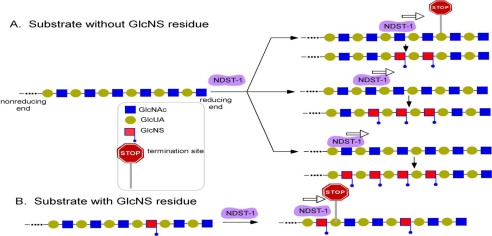
FIGURE 6.
Mode of action of NDST-1. Panel A shows the scenario of the substrate without a GlcNS residue. NDST-1 can initiate the N-sulfation at any internal GlcNAc residue, and the sulfation proceeds from nonreducing to reducing end. The N-sulfation stops at the residue that is five saccharide units away from the reducing end. Panel B shows the scenario of the substrate with a GlcNS residue. The preexisting GlcNS residues prevent the N-sulfation immediately located at the nonreducing end, leaving the gap between two GlcNS residues. The gap should be at least five saccharide units long.
The lack of the ability to initiate the N-sulfation at a specific GlcNAc residue results in multiple products, consequently leading to the structural heterogeneity during the biosynthesis of HS (Fig. 6A). At present time, we did not observe the preferential binding NDST-1 to the GlcNAc residue within the substrate. Thus, the initial binding process is likely to be random, allowing multiple products were produced even a single substrate was introduced. One plausible approach to reduce the product heterogeneity is to direct NDST-1 to a specific GlcNAc residue. Presto et al. (27) discovered the interaction of one HS polymerase (EXT-2) and NDST-1. Interestingly, the authors discovered that HS isolated HEK 293 cells overexpressing EXT-2 was highly sulfated and more closed to heparin than HS (Heparin is considered structurally more homogeneous than HS). Authors claimed overexpression EXT-2 elevates the expression of NDST-1, leading to more extensive N-sulfation on the polysaccharide. However, whether the interaction recruited more NDST-1 molecules to the nonreducing end is not completely ruled out. It is known that EXT2 is typically present at the nonreducing end to elongate the polysaccharide. If this is case, NDST-1 perhaps initiates the modification at the nonreducing, and thus extends the size of N-S domain to reduce the structural heterogeneity. Given the fact the modification of NDST-1 is exclusively toward the reducing end, our model suggests that the cluster of N-S domain should be longer if the N-sulfation is initiated at the nonreducing end. Indeed, structural analysis of the HS from various tissues indicated that the size of N-S domain is longer than those N-S domains identified in the middle of polysaccharide (28).
NDST-1 contains both N-deacetylase and N-sulfotransferase domains. A recombinant protein comprising of only N-sulfotransferase activity sulfates the GlcNH2 residue regardless the position in an oligosaccharide substrate or a GlcNS in the vicinity (12). Our observation suggests that the substrate specificity of NDST-1 is dominated by the deacetylase domain; however, it remains to be confirmed by studying the substrate specificity of the recombinant protein carrying only the N-deacetylase domain. Alternatively, both activities are required to exhibit the full scale substrate specificity. It is also important to note that NDST is present in multiple isoforms. Whether different isoforms have distinct capability of generating the N-S domains and N-Ac is currently unknown. In addition, the core protein of proteoglycans as well as potential molecular chaperons or the supra molecular complex involved in HS biosynthetic enzymes (27) may have direct or indirect impacts on the distribution of domain structures in vivo. Despite its limitations, using structurally defined oligosaccharide model substrates and MS technique certainly offer a new approach to delineate the substrate specificities of NDSTs to reveal the biosynthetic mechanism of HS.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants AI50050, HL094463, HL096972, and AI074775.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S7.
Y. Xu, Z. Wang, R. Liu, X. Huang, and J. Liu, submitted manuscript.
- HS
- heparan sulfate
- NDST
- glucosaminyl N-deacetylase/N-sulfotransferase.
REFERENCES
- 1. Bishop J. R., Schuksz M., Esko J. D. (2007) Nature 446, 1030–1037 [DOI] [PubMed] [Google Scholar]
- 2. Gama C. I., Tully S. E., Sotogaku N., Clark P. M., Rawat M., Vaidehi N., Goddard W. A., 3rd, Nishi A., Hsieh-Wilson L. C. (2006) Nat. Chem. Biol. 2, 467–473 [DOI] [PubMed] [Google Scholar]
- 3. Murphy K. J., Merry C. L., Lyon M., Thompson J. E., Roberts I. S., Gallagher J. T. (2004) J. Biol. Chem. 279, 27239–27245 [DOI] [PubMed] [Google Scholar]
- 4. Kreuger J., Spillmann D., Li J. P., Lindahl U. (2006) J. Cell Biol. 174, 323–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Esko J. D., Selleck S. B. (2002) Annu. Rev. Biochem. 71, 435–471 [DOI] [PubMed] [Google Scholar]
- 6. Peterson S. P., Frick A., Liu J. (2009) Nat. Prod Rep. 26, 61–627 [DOI] [PubMed] [Google Scholar]
- 7. Aikawa J., Esko J. D. (1999) J. Biol. Chem. 274, 2690–2695 [DOI] [PubMed] [Google Scholar]
- 8. Aikawa J., Grobe K., Tsujimoto M., Esko J. D. (2001) J. Biol. Chem. 276, 5876–5882 [DOI] [PubMed] [Google Scholar]
- 9. Rong J., Habuchi H., Kimata K., Lindahl U., Kusche-Gullberg M. (2001) Biochemistry 40, 5548–5555 [DOI] [PubMed] [Google Scholar]
- 10. Li J. P., Gong F., El Darwish K., Jalkanen M., Lindahl U. (2001) J. Biol. Chem. 276, 20069–20077 [DOI] [PubMed] [Google Scholar]
- 11. Domon B., Costello C. E. (1988) Glycoconj J. 5, 397–409 [Google Scholar]
- 12. Liu R., Xu Y., Chen M., Weïwer M., Zhou X., Bridges A. S., DeAngelis P. L., Zhang Q., Linhardt R. J., Liu J. (2010) J. Biol. Chem. 285, 34240–34249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Forsberg E., Kjellen L. (2001) J. Clin. Invest. 108, 175–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pallerla S. R., Lawrence R., Lewejohann L., Pan Y., Fischer T., Scholomann U., Zhang X., Esko J. D., Grobe K. (2008) J. Biol. Chem. 283, 16885–16894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fan G., Xiao L., Cheng L., Wang X., Sun B., Hu G. (2000) FEBS Lett. 467, 7–11 [DOI] [PubMed] [Google Scholar]
- 16. MacArthur J. M., Bishop J. R., Stanford K. I., Wang L., Bensadoun A., Witztum J. L., Esko J. D. (2007) J. Clin. Invest. 117, 153–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Crawford B. E., Garner O. B., B. J. R., Zhang D. Y., B. K. T., Nigam S. K., Esko J. D. (2010) PLOS One 5, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang L., Fuster M., Sriramarao P., Esko J. D. (2005) Nat. Immunol. 6, 902–910 [DOI] [PubMed] [Google Scholar]
- 19. Fuster M. M., Wang L., Castagnola J., Sikora L., Reddi K., Lee P. H. A., Radek K. A., Schuksz M., Bishop J. R., Gallo R. L., Sriramarao P., Esko J. D. (2007) J. Cell Biol. 177, 539–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Saribas A. S., Mobasseri A., Pristatsky P., Chen X., Barthelson R., Hakes D., Wang J. (2004) Glycobiology 14, 1217–1228 [DOI] [PubMed] [Google Scholar]
- 21. Duncan M. B., Liu M., Fox C., Liu J. (2006) Biochem. Biophys. Res. Commun. 339, 1232–1237 [DOI] [PubMed] [Google Scholar]
- 22. Sigulinsky C., Babu P., Victor X. V., Kuberan B. (2010) Carbohydr Res. 345, 250–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen J., Jones C. L., Liu J. (2007) Chem. Biol. 14, 986–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen M., Bridges A., Liu J. (2006) Biochemistry 45, 12358–12365 [DOI] [PubMed] [Google Scholar]
- 25. Sismey-Ragatz A. E., G. D. E., Otto N. J., Rejzek M., Field R. A., Deangelis P. L. (2007) J. Biol. Chem. 282, 28321–28327 [DOI] [PubMed] [Google Scholar]
- 26. Carlsson P., Presto J., Spillmann D., Lindahl U., Kjellen L. (2008) J. Biol. Chem. 283, 20008–20014 [DOI] [PubMed] [Google Scholar]
- 27. Presto J., Thuveson M., Carlsson P., Busse M., Willen M., Eriksson I., Kusche-Gullberg M., Kjellen L. (2008) Proc. Natl. Acad. Sci. 105, 4751–4756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Staples G. O., Shi X., Zaia J. (2010) J. Biol. Chem. 285, 18336–18343 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.