Abstract
The structure of the unique bacterial tubulin BtubA/B from Prosthecobacter is very similar to eukaryotic αβ-tubulin but, strikingly, BtubA/B fold without eukaryotic chaperones. Our sequence comparisons indicate that BtubA and BtubB do not really correspond to either α- or β-tubulin but have mosaic sequences with intertwining features from both. Their nucleotide-binding loops are more conserved, and their more divergent sequences correspond to discrete surface zones of tubulin involved in microtubule assembly and binding to eukaryotic cytosolic chaperonin, which is absent from the Prosthecobacter dejongeii draft genome. BtubA/B cooperatively assembles over a wider range of conditions than αβ-tubulin, forming pairs of protofilaments that coalesce into bundles instead of microtubules, and it lacks the ability to differentially interact with divalent cations and bind typical tubulin drugs. Assembled BtubA/B contain close to one bound GTP and GDP. Both BtubA and BtubB subunits hydrolyze GTP, leading to disassembly. The mutant BtubA/B-S144G in the tubulin signature motif GGG(T/S)G(S/T)G has strongly inhibited GTPase, but BtubA-T147G/B does not, suggesting that BtubB is a more active GTPase, like β-tubulin. BtubA/B chimera bearing the β-tubulin loops M, H1-S2, and S9-S10 in BtubB fold, assemble, and have reduced GTPase activity. However, introduction of the α-tubulin loop S9-S10 with its unique eight-residue insertion impaired folding. From the sequence analyses, its primitive assembly features, and the properties of the chimeras, we propose that BtubA/B were acquired shortly after duplication of a spontaneously folding α- and β-tubulin ancestor, possibly by horizontal gene transfer from a primitive eukaryotic cell, followed by divergent evolution.
Keywords: Bacteria, Evolution, Protein Self-assembly, Protein Sequence, Tubulin, BtubA/B
Introduction
Proteins from the tubulin/FtsZ superfamily of assembling GTPases perform essential cytoskeletal, DNA segregation, and cell division functions in eukaryotes and prokaryotes (1). They share the same structural fold consisting of a N-terminal GTP-binding domain and a GTPase-activating domain, with very different C termini. The members of the family with known atomic structures include eukaryotic αβ-tubulin, the constituent of microtubules (2), prokaryotic cell division protein FtsZ (3), γ-tubulin (4), bacterial tubulin BtubA/B (5), and the recently described plasmid partition protein TubZ (6, 7). Except for γ-tubulin, which forms microtubule-nucleating ring complexes (8), they assemble forming polar cytomotive protofilaments in which the GTP-binding site is at the interface between successive subunits (1). The protofilaments coalesce in different fashions, among which microtubules have the longest range order. The contact of the GTP-binding domain with the GTPase-activating domain of the next subunit along the protofilament activates GTP hydrolysis, which eventually triggers disassembly and polymer turnover. Microtubules are central to chromosome segregation, cellular architecture, and intracellular transport in eukaryotes. Microtubules have two types of end dynamics known as dynamic instability and treadmilling (9). TubZ also treadmills in vivo (10), and the bacterial FtsZ ring has rapid assembly dynamics (11). The folding of the eukaryotic αβ-tubulin dimer is a complex process assisted by prefoldin, chaperonin, and cofactors (12). FtsZ folds spontaneously, a property associated with the absence of typical tubulin loops involved in lateral contacts in microtubule assembly and eukaryotic chaperonin CCT2 binding (13, 14). Microtubules are a well known target of antitumor drugs, which impair their dynamics by enhancing or inhibiting assembly (9), whereas FtsZ is an emerging target for new antibiotics (15).
Bacterial tubulin is the closest known prokaryotic homolog of eukaryotic tubulin. The genes coding for BtubA and BtubB, sharing ∼35% sequence identity with α- and β-tubulin, were discovered in the genome of Prosthecobacter dejongeii, a free-living member of the division Verrucomicrobia, in adjacent loci to a kinesin light chain gene fragment (16). Other Verrucomicrobia that live as ectosymbionts (epixenosomes) of ciliates in the genus Euplotium were reported to contain structures resembling microtubules (17), but no such structures were found in Prosthecobacter (16). Phylogenetic analysis indicated that BtubA and BtubB from four Prosthecobacter species are quite divergent and branch deep into the tubulin tree (16). The presence of btub genes does not exclude ftsZ, because Prosthecobacter contains coexisting genes of bacterial tubulin and cell division protein FtsZ (18). A closely related bacterium Verrucomicrobium spinosum has an ftsZ gene but no btub genes (19). This supports an FtsZ-based cell division in Verrucomicrobia rather than Btub-based division (18). On the other hand, the btubA and btubB genes failed to amplify from an isolate of the new species Prosthecobacter fluviatilis (20).
Isolated bacterial tubulins BtubA and BtubB coassemble in a 1:1 ratio into protofilament polymers that hydrolyze GTP (5, 21). The basic polymer is a twisted double filament with a tubulin-like 42 Å monomer spacing. The crystal structure of BtubA/B is strikingly similar to αβ-tubulin, including the C-terminal helices H11 and H12, supporting horizontal gene transfer. However, in contrast with αβ-tubulin, BtubA/B can fold without chaperones and weakly dimerizes (5). In the crystal structure of the BtubA/B dimer, BtubA occupies the β-tubulin position, BtubB occupies the α-tubulin position, and both contain a GTPase-activating acidic residue at the T7 loop like α-tubulin and a short S9-S10 loop as in β-tubulin. These mixed features made it preferable not to structurally assign BtubA or BtubB to α- or β-tubulin (5). Point mutations at each of the BtubA and BtubB association interfaces disrupt BtubA/B assembly (22). Bacterial tubulin lacks the highly acidic C-terminal 10–20 residue tails of α- and β-tubulin chains, which participate in interactions with microtubule associated proteins and motors (23), consistent with the absence of motor proteins in bacteria (1).
The function and subcellular structures formed by BtubA/B have not been described so far. Unfortunately, genetic methods are yet to be developed for Prosthecobacter, but given that its genes have been retained, BtubA/B may be anticipated to perform some cytoskeletal role, and one possibility would be providing a scaffold for the formation of the cell stalk. There are three other puzzling questions about bacterial tubulin that remain unanswered. First, how did bacterial tubulin originate? The answer to this question will provide essential information about the evolution of tubulins. Second, how can BtubA/B fold without chaperone assistance? This is puzzling for a close structural homolog of αβ-tubulin. Third, what are the mechanisms of bacterial tubulin assembly? This could provide insight into microtubule assembly and dynamics. In addition, bacterial tubulin is more stable and amenable to mutagenesis and heterologous expression than eukaryotic tubulin. BtubA/B is thus an attractive model protein to engineer, if it were possible, recombinant binding sites for antitumor drugs and bacterially produced microtubules. In this study, we have addressed these three questions, combining the analysis of available tubulin sequences and assembly properties of bacterial tubulin with the construction of chimeras with eukaryotic tubulin loop sequences. Our results demonstrate that BtubA and BtubB have extensively intertwining α- and β-tubulin sequence features, display primitive assembly properties, and support the insertion of several tubulin loop sequences while continuing to fold in bacteria. These results suggest that BtubA/B diverged after the transfer of ancestral α- and β-tubulin genes from a primitive eukaryote, shortly after tubulin gene duplication.
EXPERIMENTAL PROCEDURES
Sequence Analyses
Tubulin sequences were collected by running Pfam profiles (24) corresponding to N- and C-terminal domains of the FtsZ/tubulin superfamily (PF00091 and PF03953, respectively) over the whole UNIPROT database (25) by hmmpfam (26). All of the complete sequences (>300 residues) in which both domains were annotated as α- and β-tubulin were separately collected and subjected to all-against-all similarity using BLAST (27). Nonredundant sequence data sets were obtained by clustering the complete data set at either 80% identity over 80% length or 90% identity and 90% length of the sequences. Sequence alignments were produced by Muscle (28) and were compatible with structural alignments produced by Dalilite (29), except in the H1-S2 loop and the H11-H12 C-terminal region, probably because of high sequence divergence and lack of structural assignation. Tree determinants were calculated by the sequence harmony procedure (30) on sequences clustered at 80% identity and 80% length. Those alignment positions at which no single residue is shared between α- and β-tubulin families, suggesting they contribute to the specialized function of each family, were considered tree determinants. Secondary structure elements were located and named based on the previous nomenclature (31).
Protein family and domain search (26) was carried out using the Pfam 24.0 database (24) modeling 21 representative tubulin chaperones and interacting proteins (supplemental Table S1) and using all proteins encoded in the 3–4× coverage draft (∼90% complete) genome of P. dejongeii.
BtubA/B Expression and Purification
Untagged P. dejongeii btubA and btubB genes were simultaneously expressed in Escherichia coli C41(DE3) cells, and BtubA/B was purified by anion exchange and size exclusion chromatography as reported (5), with modifications including an additional polymerization and depolymerization step, inspired by classical eukaryotic tubulin preparation procedures and the assembly properties of BtubA/B (see “Results”). Bacterial cell pellets from 2-liter cultures were resuspended to 15–20 ml of final volume with 50 mm Tris/HCl, pH 8, and stored frozen at −75 °C. The cell pellets were thawed, 2 mg/ml lysozyme (Sigma) and 10 μg/ml DNase I (Roche) were added, and the cells were incubated during 5 min on ice and then lysed by two or three passes through a cold French press. The lysate was centrifuged 1 h at 100,000 × g at 4 °C. The supernatant was taken to a volume of 50 ml with 50 mm Tris/HCl, pH 8, filtered through a 0.22-micron filter, and chromatographed on a 10-ml home-packed Q-Sepharose HP (Amersham Biosciences) refrigerated column (buffer A was 20 mm Tris/HCl, pH 8.5; buffer B was buffer A plus 1 m NaCl; eluted with a 0–50% gradient of B in 20 column volumes at 5 ml/min in an Amersham Biosciences FPLC system). The 10-ml fractions containing BtubA or BtubB were pooled and concentrated at 4 °C to less than 2 ml with Centriprep 30K (Amicon). The protein was chromatographed onto a home-packed Sephacryl S300 HR (Amersham Biosciences) refrigerated column (16/60 size, equilibrated in 20 mm Tris/HCl, 1 mm EDTA, 1 mm sodium azide, pH 7.5, eluted at 1 ml/min). The main fractions (4 ml each) containing BtubA and BtubB were pooled and concentrated to ∼0.5 ml at 4 °C. For BtubA/B polymerization, 1 mm EGTA, 2 mm GTP, 5 mm MgCl2, and 300 mm potassium glutamate were added to BtubA/B at 4 mg/ml final concentration, mixed, and incubated 10 min at 25 °C. The mixture was centrifuged for 30 min at 100,000 × g at 25 °C. The semitransparent polymer pellet, containing equimolar BtubA and BtubB (in a yield of ∼40% for the purified wild type protein) was thoroughly redissolved in one volume of cold 20 mm Tris/HCl, 1 mm EGTA, pH 7.5, during ∼20 min on ice, avoiding foam formation. The solution was centrifuged 30 min at 100,000 × g, 4 °C, and the supernatant, containing most BtubA/B from the previous pellet, was concentrated to ∼0.5 ml. The guanine nucleotide of BtubA/B was extracted with cold 0.5 n HClO4, measured spectrophotometrically, and analyzed by HPLC (32). The BtubA/B concentration was measured spectrophotometrically in 6 m guanidinium chloride, after subtraction of the bound nucleotide, employing an extinction coefficient 86,550 m−1 cm−1 at 280 nm (5), and BtubA/B was stored at −75 °C. These BtubA/B preparations contained 0.68 ± 0.08 GTP and 1.01 ± 0.11 GDP bound per BtubA/B, had practical values of extinction coefficient of 100,000 ± 3,000 m−1 cm−1 (280 nm) in aqueous buffer, including the bound nucleotide contribution, and were ∼100% active in polymerization above critical concentration (Cr). Alternately, the polymerization cycle was performed on the high speed supernatant of the cell lysate instead of on the purified protein. These preparations gave a somewhat higher yield of a similar BtubA/B, ∼90% active in polymerization above Cr. However, this procedure was not suited for BtubA/B constructs (below) with modified polymerization activity, and it was not employed for comparisons with them.
The BtubA and BtubB subunits were separated by performing the size exclusion chromatography in a Sephacryl S200 HR 26/60 HP column at 4 °C (using nonpolymerized BtubA/B and a factory-packed column from Amersham Biosciences/GE Healthcare) (5). Isolated BtubA contained 0.13 ± 0.06 GTP and 0.53 ± 0.11 GDP; isolated BtubB contained 0.08 ± 0.07 GTP and 0.47 ± 0.09 GDP. Eukaryotic αβ-tubulin was purified from calf brain (33).
Construction of BtubA/B Mutants and Chimera
The plasmid carrying the btubA and btubB genes (5) was used as template to generate the chimeras, using internal oligonucleotides (supplemental Table S2) carrying the minimal number of changes allowing the desired mutation/insertion by the QuikChange site-directed mutagenesis kit (Stratagene). All of the constructions were checked by complete sequencing of both BtubA and BtubB ORFs. The modified BtubA/B proteins were expressed and purified as the wild type, except for chimera AMSα, which was polymerized for 2 h at 37 °C. The secondary structure of soluble BtubA/B mosaics and mutants was compared with wild type BtubA/B with circular dichroism (13, 34), which indicated similarly folded proteins.
BtubA/B Assembly Buffers
The buffers employed were as follows: Tris-KCl or -NaCl: 20 mm Tris, 500 mm KCl or NaCl, 5 mm MgCl2, 1 mm EGTA, pH 7.5; Tris-KGlu: 20 mm Tris, 300 mm KGlu, 5 mm MgCl2, 1 mm EGTA, pH 7.5; Pipes: 80 mm Pipes2/NaOH, 5 mm MgCl2, 1 mm EGTA, pH 6.8; Pipes-D2O: 80 mm Pipes/NaOD, 6 mm MgCl2, 1 mm EGTA, made in D2O, pH meter reading 6.8; HMK: 50 mm Hepes/NaOH, 100 or 300 mm KAc, 5 mm MgAc2, 1 mm EGTA, pH 7.7; glycerol assembly buffer: 10 mm sodium phosphate, 6 mm MgCl2, 1 mm EGTA, 3.4 m glycerol, pH 6.5. EGTA was included in these buffers to suppress polymerization observed without added Mg2+, which was possibly caused by residual divalent cations in the solutions.
BtubA/B Assembly Methods
Reversible nucleotide-induced polymerization of BtubA/B was routinely performed in Tris-KCl buffer at 30 °C, to which nucleotide was added. GTP and GDP were from Sigma; GMPCPP, GMPCP, and GTPγS were from Jena Bioscience. [3H]GTP (8-[3H]GTP, ∼5 Ci/mmol) was from Amersham Biosciences. Experiments with the GTP regenerating system included 1 mm acetyl phosphate (Fluka) and 1 unit/ml acetate kinase from E. coli (Sigma).
Polymerization was monitored by right angle light scattering at 350 nm, employing a 10 × 2-mm (excitation path) cell in a Horiba Jobin-Yvon Fluromax-4 thermostatted spectrofluorometer with 0.5-nm excitation and emission slit widths. The light scattering intensity was recorded in arbitrary units of counts/s divided by the internal reference intensity. Small aliquots of BtubA/B polymer solutions were adsorbed to carbon-coated electron microscopy grids and stained with 2% uranyl acetate, and micrographs were taken in a Jeol 1239 electron microscope operating at 100 kV.
Polymer formation was quantified by centrifuging 100-μl samples at 247,000 × g (80,000 rpm) 10 min in polycarbonate tubes in a TLA100 rotor with a Beckman Optima TLX ultracentrifuge, except where indicated. Pellet and supernatant aliquots from each sample were loaded with a 20-min electrophoretic shift between them into the same lanes of 12% polyacrylamide gels with 0.1% SDS from Sigma (L 5750). The gels were stained with Coomassie Blue, scanned with a Bio-Rad CS-800 calibrated densitometer, and analyzed with Quantity One software (Bio-Rad).
Nucleotide Binding and Hydrolysis
Binding of [3H]GTP (100 μm) to BtubA/B polymers was measured by scintillation counting of supernatants and pellet aliquots, after centrifugation as above. Binding of [3H]GTP to unassembled BtubA/B or separated A and B subunits was measured from the [3H]GTP concentration in the protein-depleted top and bottom solution halves from 200-μl polycarbonate tubes after centrifugation at 386,000 × g (100,000 rpm) for 2 h in the TL100 rotor at 25 °C, correcting for the small amount of nucleotide sedimented in the absence of protein (35).
Nucleotides in polymer pellets and supernatants were extracted with cold 0.5 n HClO4, neutralized and analyzed by HPLC as described (32, 36) or with a VYDAC 3021C4.6 anion exchange column, employing a gradient of (A) 25 mm sodium phosphate (in a 1:1 ratio of mono/disodium phosphate) with acetic acid, pH 2.8, to (B) 125 mm sodium phosphate with acetic acid, pH 2.9 (0% to 100% B, 2 ml/min., detection at 260 nm). GTPase rates were measured from the HPLC analyses or with the malachite green assay of released phosphate (37).
Effects of Drugs
The effects of eukaryotic tubulin drugs on bacterial tubulin polymerization were tested by sedimentation measurements with 4 μm BtubA/B plus excess ligand in Tris-KCl buffer with 1 mm GTP) in comparison with controls (0.5–1% dimethyl sulfoxide). The compounds tested were 50 μm vinblastine, 20 μm Bis-ANS, 50 mm DAPI, 20 μm nocodazole, 50 mm podophyllotoxin, 20 μm MDL 27048 (38), 50 μm NSC 613863(R) and NSC 613862(S) (39), 50 mm colchicine, 20 μm allocolchicine, and 50 μm of their analogs MTC, TKB, TMB, TCB (40), KAC, BAC, NAC, and MAC (41), 12 μm paclitaxel, docetaxel, epothilone B, discodermolide, dyctiostatin, and laulimalide (42).
RESULTS
Mapping Bacterial Tubulin Similarities and Differences with Eukaryotic Tubulin Sequences
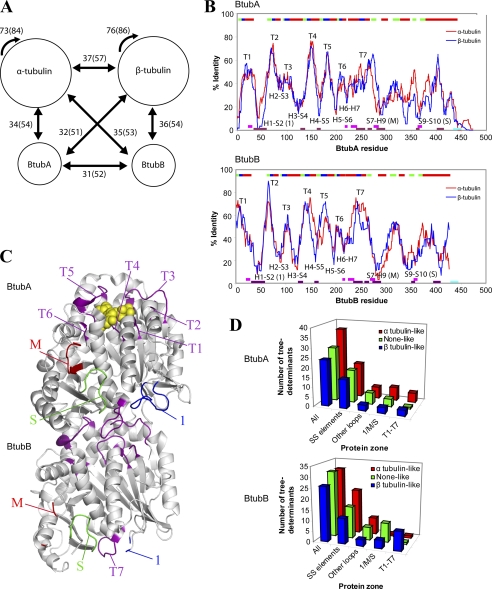
Phylogenetic analysis of the BtubA and BtubB sequences indicated that they are somewhat closer to α and β-tubulin, respectively, although the differences were small, and the position of the BtubB branch in the phylogenetic tree was unstable (16). We have examined the average identity and similarity between the BtubA and BtubB sequences and the nonredundant data sets of 41 α-tubulin and 35 β-tubulin sequences. The comparison shows that BtubA and BtubB sequences are slightly closer to both α- and β-tubulins than to each other, and there is only a marginal bias of BtubA toward α-tubulin and BtubB toward β-tubulin (Fig. 1A).
FIGURE 1.
Sequence comparison between BtubA/B and αβ-tubulin subunits. A, the numbers indicate the average percentages of identity between Btub and tubulin subunits (in parentheses, average similarity). Data sets of nonredundant 41 α-tubulin and 35 β-tubulin sequences, clustered at the level of 90% identity over 90% length, were employed. B, average identity along sequence between BtubA (upper graph) or BtubB (lower graph) and αβ-tubulin subunits. In all cases, a 15-residue averaging window was applied to smooth differences and analyze sequence context rather than single positions. Secondary structure is shown at the top: α-helices (red), β-strands (green), nucleotide-interacting loops (blue). CCT-interacting zones (purple) (14), taxol-interacting zones (pink) (62), and the acidic C-terminal tails (cyan) of tubulin are shown at the bottom. Lateral interactions in microtubules involve loops H1-S2 and M and loop S9-S10 of α-tubulin (54). C, loops mapped in the BtubA/B structure (Protein Data Bank entry 2BTQ; GDP, yellow spheres): nucleotide interacting loops T1–T7 (purple), loops H1-S2 (marked 1, blue), S7-H9 (M, red), and S9-S10 (S, green). Note that BtubA loop M and BtubB loops 1 and M were incomplete in the BtubA/B structure. D, conservation of eukaryotic tubulin tree determinant residues in several zones of bacterial tubulin structure: nucleotide-binding loops T1–T7, loops H1-S2 (marked 1), S7-H9 (M), and S9-S10 (S), all other loops and secondary structure elements (see supplemental Tables S3 and S4).
To clarify the relationship between the subunits of bacterial and eukaryotic tubulin, we carried out a detailed sequence analysis. The local closeness of BtubA and BtubB to either α- or β-tubulin was analyzed by plotting identity values along their sequences. The similar peak and valley patterns (Fig. 1B) show that the BtubA and BtubB sequences locally resemble and differ from both α- and β-tubulin in a similar way, and it is not clear whether they are more related to one or to the other. Several specific zones in the BtubA and BtubB profiles switch from being closer to α-tubulin than to β-tubulin or vice versa. This is consistent with having intertwined α-tubulin and β-tubulin residues (see below). Comparing these identity profiles indicates that the N-terminal nucleotide-binding domain and the core helix H7 (the first ∼280 residues) are generally more conserved than the C-terminal domain. The first six sharp peaks (>50% identity) match the strand-to-helix loops T1 to T6 at the apical face of the N-domain (Fig. 1C), which together with loop T7 from the next subunit form the nucleotide-binding site (31). Interestingly, the identity minima (<20% in BtubA or BtubB) in this domain correspond to the helix-to-strand loops H1-S2, H2-S3, H3-S4, H4-S5, H5-S6, and H6-H7, which alternate with the conserved loops T1–T7 and localize to the bottom and the side of the nucleotide-binding domain. In the C-terminal domain, the minima map in loop S7-H9 (the M-loop) (23), loop S9-S10, and the loop between helices H11 and H12. Microtubule assembly lateral contacts, cytoplasmic chaperonin CCT-binding zones, and several taxol-binding residues of tubulin conspicuously map in these loop sequences that are more divergent in BtubA/B (Fig. 1B), several of which are absent in the more distant relative FtsZ (14). Thus, the tubulin nucleotide-binding site is conserved in BtubA/B, but the loop sequences involved in microtubule-forming contacts and CCT binding are not, presumably because they diverged when the btubA and btubB genes left the eukaryotic cell. This is compatible with the absence of any eukaryotic genes encoding chaperonin CCT, prefoldin, tubulin cofactors A, B, C, and D, or any other 14 tubulin-interacting proteins in the ∼90% complete genome of P. dejongeii, scanned employing sensitive protein family (Pfam) profiles (supplemental Table S1). The only significant hits were bacterial GroEL chaperone (with the TCP-1/cpn60 Pfam profile) and the tetratricopeptide repeat in the reported kinesin light chain fragment (16). These results suggest that BtubA/B may fold in vivo without the assistance of the tubulin chaperones.
We next analyzed the conservation of key αβ-tubulin residues in BtubA/B. Tree determinant residues are typically responsible for subtle functional differences in homolog protein families (43). Using the sequence harmony method (30), we detected a total of 88 tree determinants, with nonoverlapping residue distributions, in α- and β-tubulin, which spread along their sequences (supplemental Table S3). When mapped onto the BtubA/B structure, α and β-tubulin key residues intertwine in BtubA and BtubB (supplemental Fig. S1). Globally, BtubA has more α-tubulin than none-like or β-tubulin tree determinant residues, whereas BtubB has more α-tubulin or none-like than β-tubulin tree determinants (supplemental Table S3 and Fig. 1D). However, careful inspection of the conserved nucleotide-binding loops T1–T7 revealed a correlation pattern between BtubA and α-tubulin and between BtubB and β-tubulin (Fig. 1D and supplemental Table S4). BtubA has more α-tubulin than β-tubulin tree determinant residues (five α, three β, and one none), and BtubB has mostly β-tubulin tree determinants (one α, seven β, and one none) in these positions. This includes the residues at the top monomer interaction interface αT73/βG71 (at loop T2), αE97/βS95 and αY103/βW101 (at loop T3), and αT179/βD177 (at loop T5) (31), which follow a perfect BtubA-α-tubulin and BtubB-β-tubulin correlation. One important exception is the cocatalytic Glu-254 in loop T7 of α-tubulin, essential for hydrolysis of the GTP bound to the β-tubulin monomer below in microtubule protofilaments. This residue is Lys-252 in β-tubulin, which does not activate GTPase in the contacting α-subunit in αβ-tubulin dimers (31). However, both BtubA and BtubB contain a corresponding Glu residue in their T7 loops (16), suggesting that, unlike tubulin, both may hydrolyze GTP upon protofilament formation. Note that a single base change is enough to mutate Glu into Lys. The T7 loop sequence is slightly more similar to tubulin in BtubB than in BtubA. On the other hand, the eight residue insertion in loop S9-S10 of α-tubulin (2) is absent from both BtubA and BtubB. The rest of the loops and secondary structure elements of both BtubA and BtubB have more α-tubulin than β-tubulin tree determinants (Fig. 1D). This bias may be partially due to random noise in the tree determinant analysis, because many of these positions are occupied by amino acids with a high natural occurrence (Ala, Leu, and Glu).
In summary, our sequence analyses indicate that although BtubA and BtubB have mosaic sequences with mixed α- and β-tubulin features, their conserved nucleotide-binding loops mainly have tree determinant residues from either α-tubulin or β-tubulin, and their most divergent loops correspond to tubulin loops involved in microtubule assembly and CCT binding.
Analysis of BtubA/B Polymer Structures and Reversible Assembly
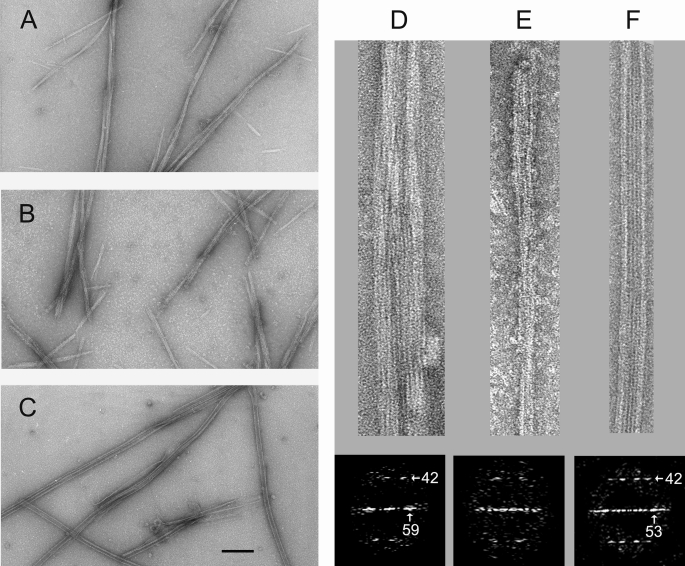
In view of the similarities and differences with eukaryotic tubulin sequences, we first screened conditions for GTP-dependent assembly of tag-free bacterial tubulin BtubA/B (5). Initial electron microscopy tests indicated that BtubA/B (20 μm) did not polymerize in two typical microtubule assembly buffers (5), Pipes with 0.1 mm GMPCPP or glycerol assembly buffer with 1 mm GTP, at 37 °C. However, further enhancing the protein self-association by addition of 200 g/liter Ficoll 70, a macromolecular crowding agent (44), or the cosolute 1 m glutamate (45) induced the formation of filamentous bundles and cables of BtubA/B (Fig. 2, A and B). These BtubA/B polymers with additives did not assemble with GDP, although they hardly disassembled with time. Comparison of the BtubA/B polymers with microtubules assembled from αβ-tubulin under identical conditions (Fig. 2C) showed that they are structurally different, because bacterial tubulin never formed tubes. Diffraction patterns from BtubA/B polymers show a tubulin-like 42 Å spacing between subunits along the protofilaments (5) and a 59 Å equatorial reflection, indicating a slightly larger average lateral spacing than the 53 Å between microtubule protofilaments (Fig. 2, D–F).
FIGURE 2.
Bacterial tubulin polymers formed under enhanced microtubule assembly conditions. A, electron micrograph of negatively stained BtubA/B polymers assembled in Pipes buffer with 200 g/liter Ficoll 70 and 0.1 mm GMPCPP at 37 °C. B, BtubA/B polymers assembled in Pipes buffer with 1 m sodium glutamate and 0.1 mm GMPCPP. C, microtubules assembled from calf brain αβ-tubulin in Pipes buffer with 200 g/liter Ficoll 70 and 0.1 mm GMPCPP. The bar corresponds to 100 nm for A–C. D, enlarged view (top panel) and computed diffractogram (bottom panel) of one BtubA/B polymer assembled in Ficoll. E, BtubA/B polymer in sodium glutamate and its diffractogram. F, enlarged αβ-tubulin microtubule assembled in Ficoll and its diffractogram. The spots indicated by the arrows corresponding to a characteristic 42-Å axial spacing between tubulin monomers along protofilaments and a 53-Å lateral spacing.
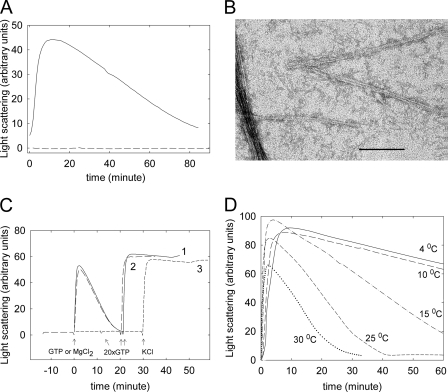
We then analyzed conditions for reversible assembly of bacterial tubulin by monitoring the time course of polymerization with light scattering and with electron microscopy. Polymer formation required both BtubA and BtubB (Fig. 3A), in agreement with previous pelleting results that showed an optimal equimolar A:B ratio for polymerization (5, 21) and was followed by disassembly upon GTP consumption. Under our conditions, we observed ∼8 nm-wide double filaments (Fig. 3B) similar to the BtubA/B double filaments (5) and looser bundles than with Ficoll, but not BtubB rings (21, 22). The properties of bacterial tubulin prompted us to include a polymerization/depolymerization cycle in the purification procedure, which yielded highly purified BtubA/B with a 1:1 A:B ratio, containing 0.7 GTP and 1.0 GDP per A/B. The cycled BtubA/B preparation undergoes fully reversible assembly and disassembly in our standard of Tris–0.5 m KCl buffer, in a practical time scale at 30 °C (Fig. 3C). This works better in our hands than with 0.1 m Pipes-NaOH, 0–200 mm KCl (5), or HMK buffer (22).
FIGURE 3.
Reversible GTP-induced assembly of BtubA/B in Tris-KCl monitored by 90° light scattering. A, solid line, polymerization time course of 5 μm purified BtubA plus 5 μm purified BtubB at 30 °C upon the addition of 100 μm GTP at time 0; dashed line, 5 μm separated BtubA or BtubB. B, electron micrograph of double filaments and bundle formed by 5 μm BtubA + BtubB. C, line 1, solid, polymerization of 5 μm BtubA/B upon addition of 50 μm GTP (arrow at time 0) and subsequent addition of 1 mm GTP (arrow at 20 min); line 2, long dashes, reaction started by 5 mm MgCl2 addition to a sample containing 0.1 mm GTP in buffer without MgCl2; line 3, short dashes, reaction started by the addition of 0.5 m KCl to a sample containing 1 mm GTP and MgCl2 in buffer without KCl. D, time course of 10 μm BtubA/B polymerization induced by 100 μm GTP at different temperatures.
BtubA/B polymerization required GTP, Mg2+, and K+; assembly can be triggered by the final addition of any of these three inducers (Fig. 3C). Qualitative examination of the effects of solution conditions on BtubA/B polymerization indicated optimal [KCl] of ∼0.5 m and showed weak differences with [MgCl2] of 2–10 mm and pH 6.8–7.8. This is in contrast with eukaryotic tubulin assembly, which is inhibited by high ionic strength and at the higher pH and is more markedly Mg2+-dependent (46). BtubA/B assembly was slower with NaCl than with KCl. Organic salts (acetate, glutamate, and Pipes) enhanced polymerization, similar to tubulin (45). In marked contrast with mammalian tubulin (46), BtubA/B polymerizes at low temperatures. Maximal light scattering values were observed in a 5–25 °C temperature range with an excess of GTP (not shown). With a limiting GTP concentration BtubA/B reached maximal scattering values at 4–15 °C, forming the same type of polymers as at 30 °C and depolymerized more slowly upon GTP consumption (Fig. 3D).
Cooperative Polymerization of Bacterial Tubulin: Nucleotide, Divalent Cation, and Drug Effects in Comparison with Eukaryotic Tubulin
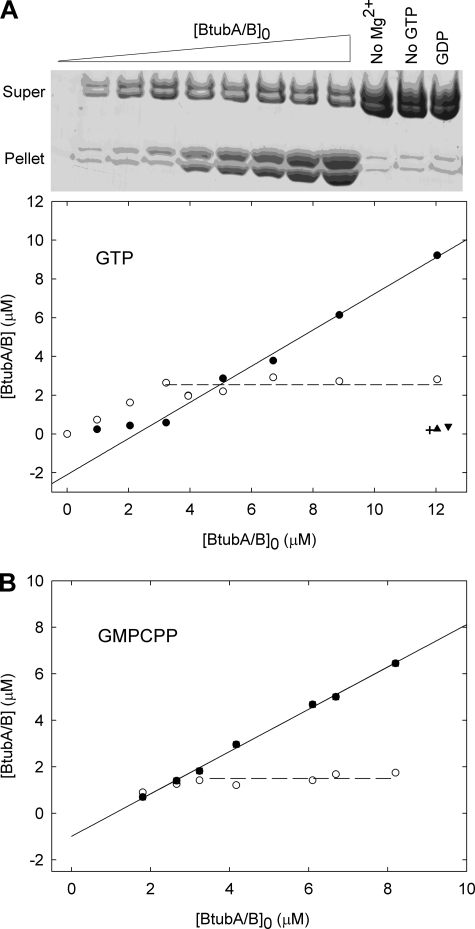
We quantitatively determined BtubA/B assembly and the effects of ligands, measuring polymer formation by sedimentation. Previous observations by scattering and GTPase measurements had indicated a ∼1.0 μm BtubA/B critical concentration for polymerization (21). In our standard buffer with GTP at 25 °C, BtubA/B polymers do not significantly pellet below a critical protein concentration, Cr, 2.05 ± 0.32 μm, above which practically all the protein added is incorporated into the polymers (Fig. 4A). This behavior supports a nucleated polymerization mechanism (47, 48) consistent with the multi-stranded nature of the protein filaments observed (Fig. 3B). Cr values and the corresponding free energy changes of polymer elongation were determined under different solution conditions (Table 1). BtubA/B assembly was enhanced in Tris-KGlu buffer (supplemental Fig. S2A) or by D2O (Table 1). D2O is a known stabilizer of microtubules which reduces their GTPase activity (49). Interestingly, these two buffers allow for the reversible assembly of BtubA/B and αβ-tubulin under identical conditions, with GTP at 30 °C.
FIGURE 4.
Cooperative assembly of BtubA/B measured by sedimentation. A, polymer formation at varying concentrations of BtubA/B with 1 mm GTP in Tris-KCl at 25 °C, analyzed by SDS-PAGE (top panel) and quantified (bottom panel); the filled and open circles are pelleted and supernatant concentrations, respectively. The cross-hair is a control without Mg2+, the triangle is without GTP, and the inverted triangle is with 1 mm GDP. B, a similar experiment made with 0.1 mm GMPCPP.
TABLE 1.
Buffer and nucleotide effects on the critical concentration for polymerization of bacterial tubulin
Nucleotide concentrations were 1 mm for GTP and GDP and 0.1 mm for GMPCPP, GMPCP, and GTPγS. ND, not determined.
| Buffer | Nucleotide | Cr | ΔGapp0a |
|---|---|---|---|
| μm BtubA/B | kcal mol−1 | ||
| Tris-KCl | GTP | 2.05 ± 0.32b | −7.76 ± 0.10 |
| GDP | >20b | >−6.41 | |
| GMPCPP | 0.99 ± 0.20b | −8.19 ± 0.12 | |
| GMPCP | >10b | >−6.82 | |
| GTPγS | 2.11 ± 0.26b | −7.74 ± 0.07 | |
| Tris-NaCl | GTP | 10 < Cr < 20c | ND |
| Tris-KGlu | GTP | 0.94 ± 0.10b0.76 ± 0.10c | −8.22 ± 0.06 |
| Tris-KGlu | GDP | >10c | ND |
| Tris-0.2 m KCl | GTP | 2.32 ± 0.30b | −7.69 ± 0.08 |
| Pipes | GTP | 10 < Cr < 20c | ND |
| GMPCPP | 10 < Cr < 30c | ND | |
| Pipes-D2O | GTP | 6 ± 1b,c | −7.2 ± 0.2 |
| Pipes-0.2 m KCl | GTP | <14c | ND |
| Pipes-0.2 m NaCl | GTP | 20 < Cr < 40c | ND |
| HMK | GTP | <10, aggregatesc | ND |
a Apparent elongation free energy changes calculated from the sedimentation Cr values (ΔGapp0 = RT lnCr−1).
b Determined by sedimentation at 25 °C.
c Determined by light scattering at 30 °C.
GMPCPP, a widely employed slowly hydrolyzable GTP analog, enhanced BtubA/B assembly (Fig. 4B and Table 1). GTPγS also induced BtubA/B polymerization (5), with a Cr similar to GTP (supplemental Fig. S2B and Table 1) giving stable polymers morphologically similar to the GTP and GMPCPP polymers.
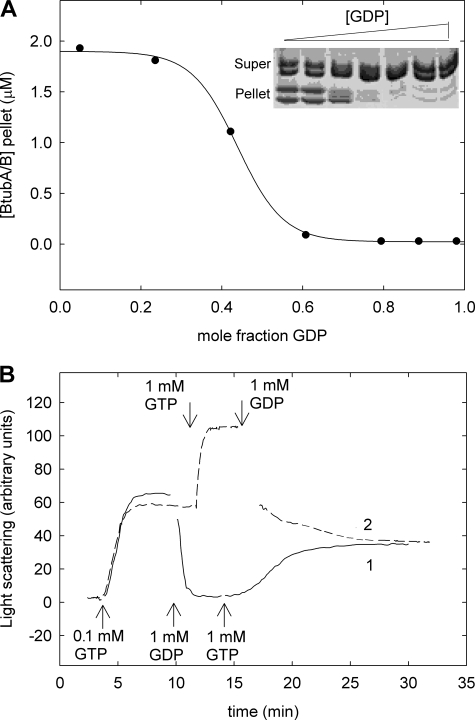
GDP was an effective inhibitor of GTP-induced assembly. Employing different ratios of GTP to GDP at constant total nucleotide concentration (1 mm), the half-inhibition of polymerization took place at ∼0.44 GDP mol fraction (Fig. 5A). Complementary experiments showed that the addition of 1 mm GDP to BtubA/B polymers preassembled with 0.1 mm GTP resulted in rapid and complete disassembly (Fig. 5B, solid line; rate constant, 3.18 ± 0.06 min−1). Further addition of 1 mm GTP induced reassembly (Fig. 5B, dashed line). BtubA/B assembly behaved as a nucleotide sensor that reversibly responded to the GTP/GDP ratio in the solution (Fig. 5B), more readily in our hands than αβ-tubulin assembly (not shown). These results would be compatible with a reversible inactivation of BtubA/B by GDP binding with an affinity similar to that of GTP.
FIGURE 5.
Effects of GDP of BtubA/B assembly. A, polymerization of 5 μm BtubA/B with 1 mm GDP/GTP mixtures in Tris-KCl buffer at 25 °C, quantified by pelleting; the line is drawn solely to show the trend of the data. B, polymerization-depolymerization time course of 5 μm BtubA/B with GTP and GDP additions. Polymerization of sample 1 (solid line) was initiated by 0.1 mm GTP, and 1 mm GDP and 1 mm GTP were subsequently added as indicated by the arrows at the bottom. Assembly was equally initiated in sample 2 (dashed line), but then 1 mm GTP and 1 mm GDP were added as indicated by the arrows at the top.
Bacterial tubulin polymerized with different divalent cations, 10 mm MgCl2, CaCl2, MnCl2, CoCl2, or NiCl2 and to a lesser extent with ZnCl2, in electron microscopy and light scattering tests. The polymers formed in each case were similar, in contrast with the known polymorphic assembly of tubulin with divalent cations (50) and its inhibition by low Ca2+ concentrations (46). Finally, bacterial tubulin polymerization was insensitive, in our sedimentation assays, to 24 compounds containing 12 chemically different types of eukaryotic tubulin modulators, including well known microtubule assembly inhibitors (colchicine and vinblastine) and inducers (paclitaxel and epothilone) (see “Experimental Procedures”).
GTP Binding and Hydrolysis by BtubA/B: GTPase Mutants
To gain insight into the role of the nucleotide in BtubA/B assembly dynamics, we studied the GTP incorporation, hydrolysis, and nucleotide content in bacterial tubulin polymers. Purified BtubA/B contains close to one GTP and one GDP bound (see above). Upon the addition of [3H]GTP to trigger assembly of 8–10 μm BtubA/B at 25 °C, the polymers incorporated 2.06 ± 0.41 (n = 8) 3H-GXP per BtubA/B. Steady-state polymers with a GTP regenerating system incorporated 1.86 ± 0.42 (n = 4) 3H-GXP per BtubA/B. These measurements indicated that BtubA/B has two GTP exchangeable sites, which are the structurally identified nucleotide sites in BtubA and BtubB (5). This is compatible with BtubA/B being a weak dimer, in contrast with the tight αβ-tubulin dimer in which the α-subunit contains nonexchangeable GTP (23, 51). Increasing the BtubA/B concentration to 15–20 μm reduced the nucleotide incorporation by steady-state polymers to 0.95 ± 0.22 (n = 4) 3H-GXP, which might be related to BtubA/B polymers bundling at higher concentrations. In contrast to polymerizing BtubA/B, the unassembled protein (20 μm) exchanged in only 0.42 ± 0.24 (n = 14) [3H]GTP in buffer without magnesium. These measurements suggested that either Mg2+ ions or polymerization were required for effective nucleotide exchange. Separated BtubA and BtubB subunits (12 μm), respectively, incorporated 0.16 ± 0.07 (n = 4) and 0.16 ± 0.03 (n = 4) [3H]GTP with 5 mm MgCl2. This accounts for the exchange by the unassembled protein, within experimental error, and would suggest that BtubA/B polymerization favors GTP exchange.
Bacterial tubulin polymers hydrolyzed GTP from the solution (Fig. 6A and supplemental Fig. S3). The GTP turnover rate was ∼0.3 min−1 (∼2 min−1 in Tris-KGlu buffer; referred to total BtubA/B, with 0.1 mm GTP at 30 °C, from HPLC measurements). The nucleotide bound to the polymers was 35 ± 5% GTP and 65 ± 5% GDP (30 ± 11% GTP in Tris-KGlu buffer). This GTP content is similar to the 40% GTP, 60% GDP content in our BtubA/B preparations made with a polymerization cycle and indicated that both subunits can hydrolyze GTP. Interestingly, steady-state populations of BtubA/B polymers with a GTP regenerating system contained 47 ± 3% GTP (51 ± 1% GTP in Tris-KGlu buffer; Fig. 6B and supplemental Fig. S3). These equal proportions of GTP and GDP suggested the possibility that only one of the subunits had hydrolyzed the nucleotide in the steady-state BtubA/B polymers.
FIGURE 6.
GTP hydrolysis by BtubA/B polymers and effects of mutations. A, filled circles and squares, percentages of nonhydrolyzed GTP in the supernatants and polymer pellets, respectively, during assembly of 7 μm BtubA/B with 125 μm GTP in Tris-KCl buffer at 30 °C; the line is the light scattering, and the open squares show the pelleted polymer, both relative to the maxima reached shortly after starting the reaction. B, a similar experiment performed with GTP regenerating system, 10 μm BtubA/B, and 125 μm GTP. C, assembly time courses of 15 μm BtubA/B GTPase mutants with 100 μm GTP. Solid line, B-S144G mutant; dashed line, A-147G mutant; dotted line, wild type. D, electron micrograph of the BtubA//B-S144G polymers. The bar indicates 100 nm.
To reveal the role of each subunit in GTP hydrolysis, we constructed the mutants BtubA-T147G/BtubB and BtubA/BtubB-S144G in the T4 phosphate-binding loops, designed to selectively impair their GTPase activity by removing the first hydroxyl residue (T/S) from the tubulin signature motif GGG(T/S)G(S/T)G (31). Analogous mutations inhibit the GTPase activity in E. coli FtsZ (52) and in TubZ (53). The BtubA-T147G mutation had modest effects, consisting of ∼2-fold slower assembly and disassembly and a reduction of the GTPase rate to 65% of the wild type. By contrast, the BtubB-S144G mutation resulted in much slower assembly and disassembly (Fig. 6C), accompanied by a reduction of the GTPase rate to 10% of the wild type, which means that the more actively hydrolyzing subunit is BtubB. This suggests that steady-state BtubA/B polymers contain predominantly GTP at their A subunits and GDP at their B subunits, similar to α- and β-tubulin in microtubules. BtubA-T147G/BtubB polymers were similar to wild type polymers (Fig. 3B), and BtubA/BtubB-S144G formed more extensive bundles (Fig. 6D).
Folding and Assembly of Bacterial Tubulin Chimera with Eukaryotic Loop Sequences
We observed an inhibition of eukaryotic tubulin assembly by bacterial tubulin under buffer conditions in which both proteins polymerize, without an extensive copolymer formation (supplemental Fig. S4), indicating that BtubA/B has a limited capacity to recognize tubulin and inhibit microtubule assembly (possibly by incorporation of BtubA/B capping microtubules or distorting the microtubule lattice). This is compatible with their divergent loop sequences (Fig. 1), presumably implied in the exclusive αβ-tubulin ability of assembling into microtubules, its requirement of eukaryotic chaperonin CCT for folding and in the binding of drugs. To address these questions, we constructed chimerical proteins in which one or more loop sequences of bacterial tubulin were replaced by the corresponding eukaryotic tubulin sequences. This approach would allow us to assess the contribution of these sequences to the different properties of eukaryotic and bacterial tubulin.
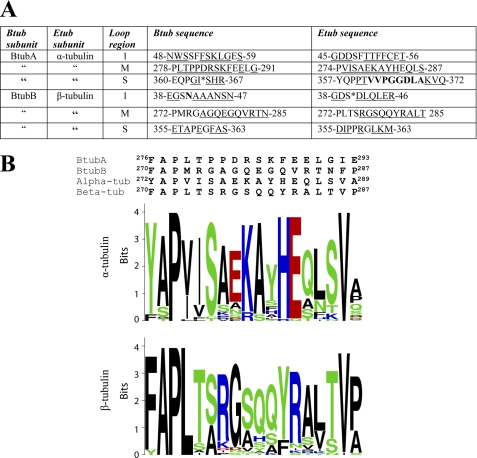
The eukaryotic sections that were exchanged into BtubA/B were the more divergent part of H1-S2 loop (named loop 1 for this work) and the S7-H9 loop (known as the M-loop), which are the main partners involved in the lateral interactions between protofilaments in microtubules (54), as well as the S9-S10 loop (named S-loop for this work). The α-tubulin S-loop contains an eight-amino acid extension that occupies a position corresponding to the microtubule stabilizing drug taxol in β-tubulin (2, 54). These sections from α and β-tubulin were exchanged into BtubA, BtubB, or both. Fig. 7A shows these sequences and Fig. 1C their position in the BtubA/B structure. To design the chimera, we first inspected the local alignments and conservation pattern in each zone (see LOGO schemes in Fig. 7B and supplemental Fig. S5). The amino acid residues substituted were from calf (Bos taurus) brain tubulin sequences, the tubulin source employed in this study. They are identical to the human (Homo sapiens) α-1 and β-2 tubulin sequences.
FIGURE 7.
Sequences from eukaryotic tubulin exchanged into bacterial tubulin. A, sequence differences between loops in bacterial tubulin (Btub) and bovine brain tubulin (Etub). Exchanged residues are underlined, and extra residues from tubulin are in bold type. Insertions in Btub are marked with asterisks. B, alignment of the M-loops of BtubA and BtubB with bovine αβ-tubulin and sequence logos for M-loop from nonredundant eukaryotic tubulin data sets clustered at 90% identity and 90% protein length. Logos were produced using the WebLogo server and colored according to amino acid biochemical group. Analogous information for loops 1 and S is available in supplemental Fig. S5.
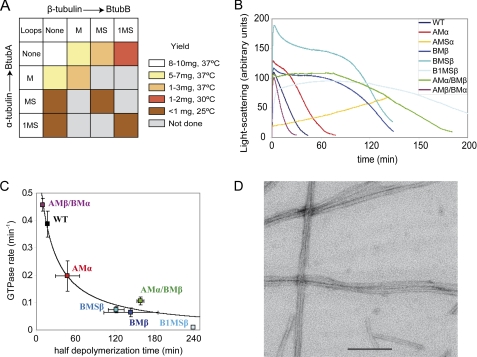
Bacterial tubulin chimeras containing the eukaryotic tubulin M loop sequences were expressed as soluble proteins in E. coli and purified in typical final yields of ∼65% with respect to the wild type. However, the yield decreased to ∼20% upon successive introduction of β-tubulin loop S and loop 1 (Fig. 8A). This effect was more marked for chimeras containing the α-tubulin S loop; despite good expression, only one-third of the protein was soluble, and only a part of that passed the polymerization step during purification, resulting in less than 10% final yield. The purified chimeras had circular dichroism spectra similar to wild type BtubA/B (supplemental Figs. S6 and S7). These results indicate that the bacterial tubulin framework supports these human tubulin loops, because the chimeras fold in the bacterial cytosol without the assistance of eukaryotic chaperones and acquire average secondary structures similar to BtubA/B, although the yield severely decreases with the introduction of loop S from α-tubulin.
FIGURE 8.
Expression, assembly, and GTPase of bacterial tubulin chimera with eukaryotic tubulin loops. A, color-ranked recovery of purified BtubA/B chimera following expression in E. coli. B, light scattering assembly time courses of the different chimera in Tris-KCl buffer, 0.1 mm GTP, at 25 °C. C, GTPase activity of the chimeras plotted versus the time of half-depolymerization. The line is a least squares reciprocal fit to the data. D, electron micrographs of negatively stained polymers BtubA/B1MSβ chimera (10 μm) in Pipes microtubule assembly buffer with 0.1 mm GMPCPP at 30 °C. The bar indicates 200 nm.
The M-loop chimeras polymerized with GTP under standard conditions for BtubA/B assembly but showed different GTPase activity (Fig. 8, B and C). Introducing the M-loop of α-tubulin in BtubA (in Btub chimera AMα/Β) induced a 2-fold reduction in GTPase compared with the wild type. In contrast, introduction of β-tubulin M-loop in the chimera A/BMβ and in the double chimera AMα/BMβ resulted in an ∼6-fold reduced GTPase rate and a correspondingly longer half-disassembly time. However, the inverted double chimera AMβ/BMα assembled, hydrolyzed GTP, and disassembled similar to wild type BtubA/B (Fig. 8, B and C). This is compatible with a reported lack of effect of inserting the β-tubulin M-loop into BtubA (22).
In view of the sequence analysis results and the properties of the BtubA/B GTPase mutants and of the M-loop chimeras, we chose to insert loops from α into A and from β into B in the construction of further chimeras. All of these chimeras, except those containing the α-tubulin S-loop (Fig. 8A), assembled with GTP into filamentous polymers similar to the wild type. We found an inverse correlation between the GTPase rate and the half-disassembly time (Fig. 8C). The AMSα/B chimera did not polymerize at 25 °C and slowly polymerized at 37 °C with GTP (Fig. 8B) into morphologically aberrant heteropolymers containing both AMSα and B subunits, which did not form with GDP (not shown). Chimeras BMβ and B1MSβ were also tested for polymerization under microtubule assembly conditions in Pipes with 0.1 mm GMPCPP at 30 °C. Chimera BMβ was able to polymerize into BtubA/B-like filaments and bundles (similar to Fig. 3B). These results showed that the substitution of the eukaryotic β-tubulin M-loop into BtubA/B allowed polymerization under microtubule assembly conditions. Under these conditions, the triple chimera B1MSβ assembled cooperatively into more ordered bundles (Fig. 8D), still different from microtubules assembled from αβ-tubulin in the same experiment (not shown). We concluded that the ability to assemble into microtubules possibly requires an accumulation of eukaryotic tubulin sequences, which becomes incompatible with CCT-independent folding.
DISCUSSION
Sequence Comparisons Suggest a Model for the Evolution of Bacterial and Eukaryotic Tubulins
BtubA and BtubB have mixed α- and β-tubulin features. Our sequence analysis has shown that they are like mosaics with intermingling residues found in α- and β-tubulin, and their most divergent zones correspond to the eukaryotic tubulin loops involved in microtubule assembly and CCT binding. The latter is consistent with BtubA/B lacking the ability to assemble into microtubules and the apparent absence of CCT genes in P. dejongeii. The question is how bacterial tubulin was originated during the evolution of the tubulin superfamily. When the btubA and btubB genes were discovered in Prosthecobacter, phylogenetic analysis superficially favored the hypothesis that bacterial tubulin genes are ancestral to eukaryotic tubulin genes rather than a recent horizontal gene transfer from a eukaryote (16). Bacteria from the Plantomycetes-Verrucomicrobia-Chlamydiae superphylum display several features typical of eukaryotes and archaea, which may suggest that they descend from intermediate steps in the prokaryote-eukaryote transition (55). However, the presence of BtubA/B in a single genus of Verrucomicrobia, Prosthecobacter, and the low number of eukaryotic signature proteins in its genome raise doubts that Prosthecobacter is ancestral to the eukaryotes (56). The striking similarity of the bacterial and eukaryotic tubulin structures (5) supports horizontal transfer of tubulin genes from an eukaryotic host to Prosthecobacter (5, 57) (transfer of the tubulin genes from a prokaryote to the common ancestor of eukaryotes seems much less probable, because of the very rare occurrence of tubulin genes in the prokaryotic world). Following horizontal transfer of eukaryotic tubulin genes, it could be possible that in the absence of CCT and a selective pressure to maintain cellular microtubule assembly, the sequences of the CCT-binding loops of both tubulin subunits relaxed in the Prosthecobacter lineage so that bacterial tubulin can fold without chaperones. However, this model does not explain how tubulin could initially fold (22) or how tubulin genes were maintained in Prosthecobacter, silently mutating until the protein could fold (unless there was an eukaryotic symbiont that provided the chaperones for tubulin folding). In addition, such a process would be expected to originate BtubA and BtubB sequences resembling either α- or β-tubulin, instead of having mixed features. Another possibility would be that bacterial tubulin derives from a single common ancestor of α- and β-tubulin that was transferred to a prokaryote and subsequently gave origin to BtubA and BtubB by gene duplication, but the fact that the BtubA and BtubB sequences are slightly less similar than both are to the eukaryotic tubulin sequences (Fig. 1A) argues against this possibility. Moreover, in this case, BtubA and BtubB sequences would contain only the features common to α- and β-tubulin. Each of these hypotheses is contrary to the fact that BtubA and BtubB each have features separately found in both α- and β-tubulin sequences.
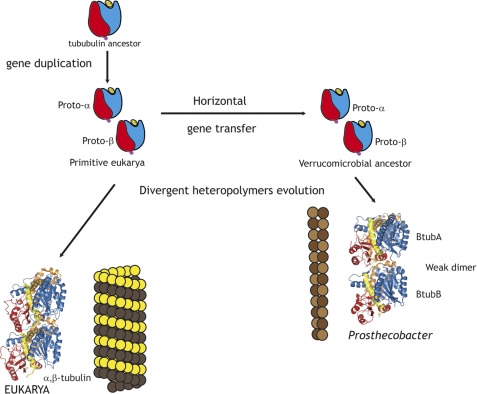
We favor a model in which the primitive α and β-tubulin genes were acquired shortly after duplication by a Prosthecobacter ancestor from a primitive eukaryotic cell, where proto-tubulins assembled into filaments (see scheme in Fig. 9). In this model, BtubA and BtubB each contain sequence features found both in α- and in β-tubulin. This hypothesis is supported by the primitive biochemical properties of bacterial tubulin and the behavior of chimera with inserted eukaryotic tubulin sequences, as will be discussed later. The tubulin subunits subsequently evolved to assemble into microtubules, which required CCT-assisted folding, and acquired their precise relationship in the tight αβ-tubulin heterodimer. The combination of one active GTP hydrolyzing β-subunit and a passive α-subunit was successfully selected for the crucial tasks of microtubules during the evolution of the eukaryotic cell. Tubulin subunits and isotypes are among the most conserved proteins (57, 58), and microtubules from different organisms have closely related structures. On the other hand, the BtubA/B ancestor possibly evolved in a simpler manner after it was transferred to prokaryotic life. This ancestor may have been either shared with αβ-tubulin or a side branch of tubulin heteropolymers that did not succeed in early eukaryotic life. Horizontal transfer could take place to an epibiontic bacterium, which either transferred the bacterial tubulin genes to a free living relative or later switched to free life. No eukaryotic donor is currently identifiable, because both BtubA and BtubB show comparable sequence identities to successful tubulins from protozoa, plants, and metazoans. How BtubA and BtubB ended up with the intermingling α- and β-tubulin residues that we have detected remains to be explained, one possibility being convergent amino acid usage related to heteropolymer formation.
FIGURE 9.
Model scheme for the evolution of bacterial tubulin. The α- and β-tubulin proto-proteins appeared in a primitive eukaryotic cell by gene duplication from a tubulin ancestor, which had a GTP-binding domain (blue) and a GTPase-activating domain (red). Shortly after duplication, both genes were transferred to a bacterium, one possibility being a Verrucomicrobal ancestor of Prosthecobacter. Primitive tubulins coassembled into filamentous heteropolymers that underwent divergent more extensive evolution in eukarya than in bacteria.
Primitive Biochemical Properties of Bacterial Tubulin
We have carried out the first systematic study of the assembly properties of BtubA/B in comparison with αβ-tubulin. BtubA and BtubB coassemble into double filaments and bundles that show the 42 Å monomer spacing characteristic of the tubulin/FtsZ superfamily, but the protofilaments have a different lateral arrangement than in microtubules. BtubA/B polymerizes over a wider range of pH, ionic strength and temperature than αβ-tubulin and with different divalent cations. The cold stability of BtubA/B polymers agrees with Prosthecobacter species being able to grow at 1–10 °C, despite being considered mesophilic organisms (59).
Eukaryotic αβ-tubulin tight dimers are the functional assembly units of microtubules, but BtubA/B only forms weak dimers in solution, whose role in the assembly of BtubA/B heteropolymers is to be further studied. BtubA/B cooperatively polymerizes with apparent protomer dissociation constant Kd values of ∼1 μm (Table 1 and Ref. 21). Eukaryotic αβ-tubulin assembles with typical Kd values of ∼10 μm (46). Tubulin dimer Kd values range between ∼10 pm (60) and ∼100 nm (61). In comparison, BtubA/B incipiently dimerizes in solution with Kd values ranging between ∼10 and 100 μm (5, 22), which are above the observed Kd of its polymers. It is clear that BtubA/B has acquired the capacity to form heteropolymers, but none of the two different association interfaces forming the protofilament (A-B and B-A) is differentiated enough to form a permanent BtubA/B dimer. We do not know whether the solution Btub dimers (5) are A-B dimers or B-A dimers. In the BtubA/B dimer structure (5), BtubA is at the plus end with the nucleotide site exposed like β-tubulin. This has been employed by Sontag et al. (22) to suggest that BtubA corresponds to β-tubulin and BtubB to α-tubulin. However, the BtubA/B crystal structure does not determine what the dimer is, because it contains a continuous …ABABAB… filament (5). The bacterial tubulin dimer needs not correspond to and αβ-tubulin dimer. One possibility is that the A-B association is very weak and BtubA/B forms a B-A dimer in solution. Note that in this case the point mutants designed to disable the association interfaces within dimers and between dimers (22) would work the other way around. In the simplest case, BtubA/B heteropolymers could grow via alternating monomer addition and annealing reactions.
We conclude that BtubA and BtubB, although they form heteropolymers, are clearly less specialized than α- and β-tubulin. In addition, BtubA/B is insensitive to 12 types of eukaryotic tubulin drugs that we have tested so far. This is initially discouraging for the potential use of bacterial tubulin for antitumor drug binding studies. Has bacterial tubulin lost the ability to assemble into microtubules and bind tubulin drugs? Or did it never acquire these properties? The bacterial tubulin sequence features discussed above support the second possibility. In this case, bacterial tubulin would have originated before plant and marine small molecules acting on microtubules such as colchicine, vinblastine, taxol, discodermolide, and laulimalide (9, 42), poisons that presumably appeared as defensive mechanisms against eukaryotic predators.
BtubA/B GTPase and Implications for Polymer Dynamics
The interactions of bacterial tubulin with the interfacial nucleotide indicate a weakly differentiated role of A and B subunits in the BtubA/B dimer, in comparison with the clearly differentiated properties of α- and β-tubulin (23). BtubA/B hydrolyzes GTP at a slow rate during assembly. BtubA/B polymers rapidly disassemble with GDP (half-life ≈ 13 s), behaving like a sensor of the GTP/GDP ratio in the solution. We do not know whether the nucleotide exchange takes place in the polymers or in recycling subunits.
Both BtubA and BtubB in A/B polymers can exchange bound nucleotide. Steady-state polymers of BtubA/B with a GTP regenerating system contain one GTP and one GDP bound (50% GTP), similar to microtubules. However, both BtubA and BtubB subunits can hydrolyze GTP, as predicted (5, 16). These features are less specialized than in αβ-tubulin, in which the α-subunit has nonexchangeable GTP bound and only the β-subunit can exchange the nucleotide in unassembled heterodimers (23, 51). Intriguingly, [3H]GTP nucleotide exchange in unassembled BtubA/B was less efficient than in polymerizing BtubA/B, suggesting that the complete nucleotide site contributed by the A and B subunits might be required for nucleotide dissociation.
The nucleotide contents of BtubA/B polymers suggest the possibility that one of the subunits hydrolyzes GTP immediately after assembly and the other does it later, which triggers disassembly. The effects of the mutations BtubA-T147G and BtubB-S144G in the tubulin signature motif indicate that BtubB is a faster GTPase than BtubA and that its inhibition leads to polymer stabilization. This is supported by preliminary results indicating that BtubA/BtubB-S144G polymers with the GTP regenerating system contained close to 100% GTP, whereas BtubA-T147G/BtubB polymers contain ∼50% GTP, similar to the wild type. Thus, like in microtubules, hydrolysis of the BtubB GTP is a necessary step, but in contrast with microtubules, polymer disassembly seems to require hydrolysis of part of the GTP molecules bound to the Btub A subunits. The BtubA/B functionality seems somewhat more concentrated in BtubB than in BtubA, like a predifferentiated β-tubulin. The nucleotide exchange and hydrolysis properties of BtubA/B polymers, even more primitive than tubulin, might be compatible with microtubule-related polymer dynamics, including dynamic instability and treadmilling.
Insights into Tubulin Evolution from Bacterial Tubulin Chimera with Eukaryotic Loop Sequences
We proceeded to replace in BtubA/B selected αβ-tubulin loop sequences involved in microtubule assembly contacts and CCT binding (Figs. 7 and 8). Related to our previous results introducing tubulin loops into FtsZ (14), we found that bacterial tubulin chimera can still fold in acceptable yield without CCT and assemble after introduction of substantial eukaryotic portions. However, we have not observed binding of the BtubA/B chimera to CCT (not shown), perhaps because of steric impediments in the more complex BtubA/B molecule. Polymer stabilization and a progressive reduction of GTPase were observed (Fig. 8), particularly when substituting the α M-loop in A and β M-loop in B. If these properties are considered microtubule-like, this may be taken as additional evidence in favor of a certain α-A/β-B relatedness over α-B/β-A. On the practical side, spontaneously folding BtubA/B-tubulin chimera could be useful to engineer eukaryotic tubulin functions into these bacterially expressed proteins. Introducing in BtubB the β-tubulin loop S9-S10 sequence still permitted folding, as did further introduction of β-tubulin loop H1-S2 residues. The combination of these loops increased the filament bundling but did not result in microtubule-like polymers. However, we found that the α-tubulin S9-S10 sequence, with its unique eight-residue insertion, impaired BtubA/B folding, giving mostly insoluble chimera. These results with BtubA/B chimera reinforce the idea that assembly of the first microtubules required an accumulation of eukaryotic tubulin surface sequences, which necessitated CCT chaperonin for folding (13, 14). From these observations, we hypothesize that bacterial tubulin originated before α-tubulin acquired its loop S9-S10 insertion and evolved into the passive subunit of the αβ-dimer.
In conclusion, the sequence analyses, the biochemical properties of bacterial tubulin, and the behavior of the chimera with eukaryotic loop sequences, taken together, support the notion that BtubA/B is a weakly differentiated form of αβ-tubulin. BtubA bears some resemblance to α-tubulin, and BtubB bears some resemblance to the more active β-tubulin. This does not imply that α and A or β and B are orthologs, but analogs forming differentiated heterodimers. A possible scenario for αβ-tubulin evolution is as follows. All members of the tubulin/FtsZ family have a N-terminal GTP-binding domain and a GTPase-activating domain, hypothesized to come from the fusion of two previously separate proteins that copolymerized forming protofilaments, thus linking nucleotide hydrolysis with polymerization (63). The common features of tubulin and FtsZ depolymerization suggest an ancient protofilament bending motor (64, 65). At some point, gene duplication gave rise to a primitive α- and β-tubulin that assembled into heteropolymers with the GTP-binding site between monomers (Fig. 9), that performed some cytomotive function (1). Protofilaments with alternating subunits have two distinct association interfaces. It is probable that the top end of the α-subunit coevolved with the bottom end of the β-subunit, and the bottom end of the α-subunit with the top end of the β-subunit, abolishing homopolymerization. Each of the two interfaces may have evolved separately, but both of them are under pressure to conserve nucleotide binding. A heteropolymer with alternating subunits could be thought to facilitate different dynamic properties at each polymer end, such as in microtubule dynamic instability (66). Bacterial tubulin could have originated from incipiently differentiated tubulin genes that were transferred from a primitive eukaryotic cell. This would explain why the Btub sequences are like α/β-tubulin mosaics and no existing eukaryotic donor is identifiable. In eukaryotes, primitive tubulin substantially evolved into the current αβ-tubulin tight dimer to accurately assemble into microtubules, performing their complex chromosome segregation and cytoskeletal functions. In the bacterial world, tubulin found little continuation, except in the Prosthecobacter lineage, where it is possibly an evolutionary remnant with a less demanding cytoskeletal role.
Supplementary Material
Acknowledgments
We thank J. Martín-Benito for computing the diffractograms in Fig. 2; J. F. Díaz for testing several microtubule stabilizing agents with BtubA/B; C. Schaffner for discussions; and D. Juan, M. Cabezas, and M. Gómez de Cedrón for technical assistance.
This work was supported in part by Ministerio de Ciencia e Innovación Grants BFU2008-00013 (to J. M. A.) and BFU2010-15703 (to J. M. V.) and contracts Juan de la Cierva (to A. J. M.-G. and M. A. O.), a contract from the Consejo Superior de Investigaciones Científicas Junta de Ampliación de Estudios-Doctores (to A. J. M.-G.), a Formacion de Personal Universitario fellowship (to L. S.), and a fellowship from the Madrid Regional Government (to M. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S4 and Figs. S1–S7.
- CCT
- chaperonin containing TCP-1, also termed TRiC
- Cr
- critical concentration
- GMPCPP
- guanosine-5-[(α,β)-methyleno]triphosphate
- GMPCP
- guanosine-5-[(α,β)-methyleno]diphosphate
- GTPγS
- guanosine-5-(γ-thio)triphosphate
- Pipes
- piperazine-N,N′-bis(2-ethanesulfonic acid)
- KGlu
- potassium glutamate.
REFERENCES
- 1. Löwe J., Amos L. A. (2009) Int. J. Biochem. Cell Biol. 41, 323–329 [DOI] [PubMed] [Google Scholar]
- 2. Nogales E., Wolf S. G., Downing K. H. (1998) Nature 391, 199–203 [DOI] [PubMed] [Google Scholar]
- 3. Löwe J., Amos L. A. (1998) Nature 391, 203–206 [DOI] [PubMed] [Google Scholar]
- 4. Aldaz H., Rice L. M., Stearns T., Agard D. A. (2005) Nature 435, 523–527 [DOI] [PubMed] [Google Scholar]
- 5. Schlieper D., Oliva M. A., Andreu J. M., Löwe J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 9170–9175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ni L., Xu W., Kumaraswami M., Schumacher M. A. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 11763–11768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Aylett C. H., Wang Q., Michie K. A., Amos L. A., Löwe J. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 19766–19771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kollman J. M., Polka J. K., Zelter A., Davis T. N., Agard D. A. (2010) Nature 466, 879–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jordan M. A., Wilson L. (2004) Nat. Rev. Cancer. 4, 253–265 [DOI] [PubMed] [Google Scholar]
- 10. Larsen R. A., Cusumano C., Fujioka A., Lim-Fong G., Patterson P., Pogliano J. (2007) Genes. Dev. 21, 1340–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stricker J., Maddox P., Salmon E. D., Erickson H. P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 3171–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lewis S. A., Cowan N. J. (2002) Nat. Med. 8, 1202–1203 [DOI] [PubMed] [Google Scholar]
- 13. Andreu J. M., Oliva M. A., Monasterio O. (2002) J. Biol. Chem. 277, 43262–43270 [DOI] [PubMed] [Google Scholar]
- 14. Bertrand S., Barthelemy I., Oliva M. A., Carrascosa J. L., Andreu J. M., Valpuesta J. M. (2005) J. Mol. Biol. 346, 319–330 [DOI] [PubMed] [Google Scholar]
- 15. Haydon D. J., Stokes N. R., Ure R., Galbraith G., Bennett J. M., Brown D. R., Baker P. J., Barynin V. V., Rice D. W., Sedelnikova S. E., Heal J. R., Sheridan J. M., Aiwale S. T., Chauhan P. K., Srivastava A., Taneja A., Collins I., Errington J., Czaplewski L. G. (2008) Science 321, 1673–1675 [DOI] [PubMed] [Google Scholar]
- 16. Jenkins C., Samudrala R., Anderson I., Hedlund B. P., Petroni G., Michailova N., Pinel N., Overbeek R., Rosati G., Staley J. T. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 17049–17054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Petroni G., Spring S., Schleifer K. H., Verni F., Rosati G. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 1813–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pilhofer M., Rosati G., Ludwig W., Schleifer K. H., Petroni G. (2007) Mol. Biol. Evol. 24, 1439–1442 [DOI] [PubMed] [Google Scholar]
- 19. Yee B., Lafi F. F., Oakley B., Staley J. T., Fuerst J. A. (2007) BMC Evol. Biol. 7, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Takeda M., Yoneya A., Miyazaki Y., Kondo K., Makita H., Kondoh M., Suzuki I., Koizumi J. (2008) Int. J. Syst. Evol. Microbiol. 58, 1561–1565 [DOI] [PubMed] [Google Scholar]
- 21. Sontag C. A., Staley J. T., Erickson H. P. (2005) J. Cell Biol. 169, 233–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sontag C. A., Sage H., Erickson H. P. (2009) PLoS One 4, e7253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nogales E. (2000) Annu. Rev. Biochem. 69, 277–302 [DOI] [PubMed] [Google Scholar]
- 24. Finn R. D., Mistry J., Tate J., Coggill P., Heger A., Pollington J. E., Gavin O. L., Gunasekaran P., Ceric G., Forslund K., Holm L., Sonnhammer E. L., Eddy S. R., Bateman A. (2010) Nucleic Acids Res. 38, D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. UniProt Consortium (2011) Nucleic Acids Res. 39, D214–D219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Eddy S. R. (1998) Bioinformatics 14, 755–763 [DOI] [PubMed] [Google Scholar]
- 27. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 28. Edgar R. C. (2004) BMC Bioinformatics 5, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Holm L., Park J. (2000) Bioinformatics 16, 566–567 [DOI] [PubMed] [Google Scholar]
- 30. Pirovano W., Feenstra K. A., Heringa J. (2006) Nucleic Acids Res. 34, 6540–6548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nogales E., Downing K. H., Amos L. A., Löwe J. (1998) Nat. Struct. Biol. 5, 451–458 [DOI] [PubMed] [Google Scholar]
- 32. Huecas S., Andreu J. M. (2003) J. Biol. Chem. 278, 46146–46154 [DOI] [PubMed] [Google Scholar]
- 33. Andreu J. M. (2007) Methods Mol. Med. 137, 17–28 [DOI] [PubMed] [Google Scholar]
- 34. Greenfield N. J. (2006) Nat. Protoc. 1, 2876–2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huecas S., Schaffner-Barbero C., García W., Yébenes H., Palacios J. M., Díaz J. F., Menéndez M., Andreu J. M. (2007) J. Biol. Chem. 282, 37515–37528 [DOI] [PubMed] [Google Scholar]
- 36. Oliva M. A., Huecas S., Palacios J. M., Martín-Benito J., Valpuesta J. M., Andreu J. M. (2003) J. Biol. Chem. 278, 33562–33570 [DOI] [PubMed] [Google Scholar]
- 37. Kodama T., Fukui K., Kometani K. (1986) J. Biochem. 99, 1465–1472 [DOI] [PubMed] [Google Scholar]
- 38. Peyrot V., Leynadier D., Sarrazin M., Briand C., Rodriquez A., Nieto J. M., Andreu J. M. (1989) J. Biol. Chem. 264, 21296–21301 [PubMed] [Google Scholar]
- 39. Barbier P., Peyrot V., Leynadier D., Andreu J. M. (1998) Biochemistry 37, 758–768 [DOI] [PubMed] [Google Scholar]
- 40. Andreu J. M., Perez-Ramirez B., Gorbunoff M. J., Ayala D., Timasheff S. N. (1998) Biochemistry 37, 8356–8368 [DOI] [PubMed] [Google Scholar]
- 41. Pérez-Ramírez B., Gorbunoff M. J., Timasheff S. N. (1998) Biochemistry 37, 1646–1661 [DOI] [PubMed] [Google Scholar]
- 42. Buey R. M., Barasoain I., Jackson E., Meyer A., Giannakakou P., Paterson I., Mooberry S., Andreu J. M., Díaz J. F. (2005) Chem. Biol. 12, 1269–1279 [DOI] [PubMed] [Google Scholar]
- 43. Casari G., Sander C., Valencia A. (1995) Nat. Struct. Biol. 2, 171–178 [DOI] [PubMed] [Google Scholar]
- 44. Zhou H. X., Rivas G., Minton A. P. (2008) Annu. Rev. Biophys. 37, 375–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hamel E., del Campo A. A., Lowe M. C., Waxman P. G., Lin C. M. (1982) Biochemistry 21, 503–509 [DOI] [PubMed] [Google Scholar]
- 46. Lee J. C., Timasheff S. N. (1977) Biochemistry 16, 1754–1764 [DOI] [PubMed] [Google Scholar]
- 47. Oosawa F., Asakura S. (1975) Thermodynamics of the Polymerization of Protein, pp. 25–40, Academic Press, London [Google Scholar]
- 48. Huecas S., Llorca O., Boskovic J., Martín-Benito J., Valpuesta J. M., Andreu J. M. (2008) Biophys. J. 94, 1796–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Panda D., Chakrabarti G., Hudson J., Pigg K., Miller H. P., Wilson L., Himes R. H. (2000) Biochemistry 39, 5075–5081 [DOI] [PubMed] [Google Scholar]
- 50. Gaskin F. (1981) Biochemistry 20, 1318–1322 [DOI] [PubMed] [Google Scholar]
- 51. Spiegelman B. M., Penningroth S. M., Kirschner M. W. (1977) Cell 12, 587–600 [DOI] [PubMed] [Google Scholar]
- 52. Mukherjee A., Saez C., Lutkenhaus J. (2001) J. Bacteriol. 183, 7190–7197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tinsley E., Khan S. A. (2006) J. Bacteriol. 188, 2829–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li H., DeRosier D. J., Nicholson W. V., Nogales E., Downing K. H. (2002) Structure 10, 1317–1328 [DOI] [PubMed] [Google Scholar]
- 55. Devos D. P., Reynaud E. G. (2010) Science 330, 1187–1188 [DOI] [PubMed] [Google Scholar]
- 56. Staley J. T., Bouzek H., Jenkins C. (2005) FEMS Microbiol. Lett. 243, 9–14 [DOI] [PubMed] [Google Scholar]
- 57. Erickson H. P. (2007) Bioessays 29, 668–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ludueña R. F. (1998) Int. Rev. Cytol. 178, 207–275 [DOI] [PubMed] [Google Scholar]
- 59. Staley J. T., Bont J. A., Jonge K. (1976) Antonie Van Leeuwenhoek J. Microbiol. 42, 333–342 [DOI] [PubMed] [Google Scholar]
- 60. Caplow M., Fee L. (2002) Mol. Biol. Cell. 13, 2120–2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Menéndez M., Rivas G., Díaz J. F., Andreu J. M. (1998) J. Biol. Chem. 273, 167–176 [DOI] [PubMed] [Google Scholar]
- 62. Löwe J., Li H., Downing K. H., Nogales E. (2001) J. Mol. Biol. 313, 1045–1057 [DOI] [PubMed] [Google Scholar]
- 63. Oliva M. A., Cordell S. C., Löwe J. (2004) Nat. Struct. Mol. Biol. 11, 1243–1250 [DOI] [PubMed] [Google Scholar]
- 64. Ludueña R. F., McIntosh J. (2009) ASCB 49th Annual Meeting, San Diego, CA, December 5–9, 2009, Abstract 239/B186, p. 116, American Society for Cell Biology, Bethesda, MD [Google Scholar]
- 65. McIntosh J. R., Volkov V., Ataullakhanov F. I., Grishchuk E. L. (2010) J. Cell Sci. 123, 3425–3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Nogales E., Whittaker M., Milligan R. A., Downing K. H. (1999) Cell 96, 79–88 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.