Abstract
SCF-type E3 ubiquitin ligases are crucial regulators of cell cycle progression. As the F-box protein is the substrate-specifying subunit of this family of ligases, their availability dictates the timing and the location of the ubiquitination of substrates. We report here our investigation into the regulation of the localization of F-box proteins, in particular Fbxo7, whose mislocalization is associated with human disease. We identified a motif in Fbxo7 that we have characterized as a functional leucine-rich nuclear export sequence (NES), and which allowed binding to the nuclear export protein, exportin 1 (CRM1). Unusually, the NES was embedded within the F-box domain, which is bound by Skp1 and enables the F-box protein to form part of an E3 ubiquitin ligase. The NES of Fbxo7 controlled its localization and was conserved in Fbxo7 homologues in other species. Skp1 binding prevented Fbxo7 from contacting CRM1. We propose that this competitive binding allowed Fbxo7 to accumulate within the nucleus starting at the G1/S transition. More than ten other F-box proteins also contain an NES at the same location in their F-box domains, indicating that this competitive binding mechanism may contribute to the regulation of a sixth of the known F-box proteins.
Keywords: Cancer Tumor Promoter, E3 Ubiquitin Ligase, Nuclear Transport, Parkinson Disease, Ubiquitin Ligase, CRM1, F-box Proteins, Fbxo7, NES, Skp1
Introduction
Many complex biological processes are regulated by the timely synthesis and degradation of cellular proteins. The degradation of proteins can help ensure irreversibility and thus the unidirectionality of processes like the cell cycle. Proteins are marked for degradation by the covalent attachment of polyubiquitin chains, targeting them to the proteasomes for destruction. This post-translational modification is carried out by E3 ubiquitin ligases including those of the Skp1-Cullin-F-box (SCF)2 family. The specificity of these enzymes is determined by their associated F-box protein (FBP). Functioning as adaptors, FBPs bind the Skp1 component of the SCF using a conserved 50 amino acid motif known as the F-box domain (FBD) and recruit post-translationally modified protein substrates to the holoenzyme for ubiquitination via additional protein-protein interaction domains (1). The FBP superfamily is separated into several different subclasses based on their interaction domains (2, 3). The Fbxw proteins contain WD-40 repeats, and Fbxl proteins contain leucine-rich repeats. The remaining FBPs containing other or unclassified interaction domains are designated Fbxo.
The activity of FBPs can be controlled by restricting them to a certain subcellular compartment, bringing them to or restricting them from their substrates. This can be achieved by a number of different methods. For example, the localization of Fbxw7 is controlled by differential splicing (4). The α isoform contains at least two redundant nuclear localization sequences (NLSs) and is largely nuclear, whereas the β isoform contains an N-terminal hydrophobic transmembrane domain and is permanently anchored in the cytoplasm. The γ isoform contains a nucleolar localization signal (NoLS), which is present in all three isoforms but is overridden by dominant targeting signals in the other isoforms. Only the Fbxw7γ isoform is able to counteract the growth-promoting properties of the c-Myc oncogene in the nucleolus (4).
There is also some evidence to suggest that reversible modification of FBPs can regulate their localization. For example, Skp2/Fbxl1 localization is controlled by phosphorylation. It has been shown that phosphorylation of Skp2/Fbxl1 at Ser-72 during M phase may trigger its export to the cytoplasm affecting its ability to form functional SCF complexes (5, 6). However, these observations remain controversial (7, 8).
One unusual FBP, Fbxo7 has been associated with both cancer and neurodegenerative disease (9–11). Autosomal recessive mutations in FBXO7 have been identified as causing an early-onset Parkinsonian disease (9, 11), and Fbxo7 has also been found to be highly expressed in human malignancies (10). We cloned Fbxo7 in a yeast two hybrid screen for proteins interacting with viral D-type cyclins (10). Fbxo7 is remarkable in that it operates as a conventional SCF adaptor, stimulating ubiquitination of proteins substrates (which include hepatoma up-regulated protein (HURP) and cellular inhibitor of apoptosis 1 (cIAP1) (12, 13)), and also as a selective stabilizer of other interacting proteins. Fbxo7 can enhance the assembly of cyclin D/Cdk6 complexes and augment Cdk6 activity (10, 14). It is this property of Fbxo7 that enables it to transform immortalized murine fibroblasts, which are capable of forming tumors when injected into athymic nude mice. Fbxo7 may also be a human proto-oncogene, and it has been shown to be overexpressed in various human cancers including lung and colon carcinomas (10). In addition to increases in its expression level, the mislocalization of Fbxo7 may also contribute to its role in cancer. We reported Fbxo7 as having a predominantly cytoplasmic localization in cultured cell lines; however, it was found to be enriched in nuclei of more than half of human colon carcinoma biopsies (10). This pronounced nuclear localization may enable its participation in pathogenic signaling pathways.
In this study, we sought to understand the mechanisms controlling Fbxo7 localization, and how it is regulated in cell cycle. Our results demonstrated that the subcellular localization of Fbxo7 and a subset of FBPs was regulated by a functional NES embedded within the FBD, which set up a competitive relationship between Skp1 and CRM1 binding. This also contributed to the maintenance of a net cytoplasmic localization of Fbxo7 and at least two other FBPs with this NES/FBD configuration.
EXPERIMENTAL PROCEDURES
Cell Culture, Plasmids, and Transfection
U2OS human osteosarcoma cells were obtained from Cancer Research UK (LRI Cell Production), T98G human glioblastoma cells and 293T human embryonic kidney cells were kind gifts from Dr. Koichi Ichimura and Prof. Chris Boshoff, respectively. Cell lines were maintained in DMEM supplemented with 10% fetal calf serum (FCS; PAA Laboratories, GmbH), 2 mm glutamine, 100 units/ml penicillin and streptomycin at 37 °C in a humidified 5% CO2 atmosphere. T98G cells were induced to enter G0 by contact inhibition coupled with 72 h of starvation in growth medium containing 0.1% FCS. Cells were stimulated to re-enter cell cycle by seeding cells at lower density into medium containing 10% FCS.
pcDNA3 vectors expressing N-terminal Flag or T7 epitope fusions to Fbxo7 (full-length isoform 1 (1–522), truncated, or ΔF-box) and to Skp1 have previously been described (10, 15). PCR mutagenesis was used to create Fbxo7-NESmt and Fbxo7-H1mt alleles, which were cloned into pGEX-(KG) vectors (Invitrogen). Transfections of plasmid DNA were performed using GeneJuice (Merck Biosciences) 24 h after cells were seeded onto glass or tissue culture-treated plastic.
Fluorescence Imaging
Plasmid constructs for the expression of FBPs as fluorescent fusions with Venus as a C-terminal tag were generated in the following way: the Venus cDNA sequence was subcloned from pCS2-Venus (a gift from Prof. Atsushi Miyawaki, RIKEN Institute, Japan), into the pEGFP-N1 vector (Clontech), replacing the EGFP gene. Sequences encoding Fbxo7 (1–522), Fbxo7 (1–522)-ΔF-box, Fbxo7 (1–522)-NESmt and Fbxo7 (1–522)-H1mt were then cloned into this pVenus-N1 plasmid. Sequences encoding full-length Fbxo2 (1–296) or (89–296) which removed the FBD from Fbxo2, were amplified by PCR from pYFP-Fbxo2 (from Chi-Yong Eom, Korea Basic Science Institute) and cloned into pVenus-N1. Sequence encoding full-length Fbxo4 (1–387) or (100–387), which removed the FBD from Fbxo4 were amplified by PCR from an Fbxo4 IMAGE clone (Source BioScience plc) and cloned into pVenus-N1. All clones generated by PCR were sequenced fully.
The FUCCI lentiviral vector pCSII-EF-MCS-mAG-geminin, (a gift from Prof. Atsushi Miyawaki, RIKEN Institute, Japan) encodes a truncation of human geminin (1–110) fused to monomeric Azami green protein (mAG). This construct was transfected into 293T cells, and the lentiviruses produced by these cells were used to infect U2OS cells. Cells expressing mAG-geminin were separated and collected by FACS and single cell cloned.
For fluorescence imaging of fixed samples, cells were grown on glass coverslips prior to fixation with 4% paraformaldehyde and permeabilization with ice-cold methanol for 5 min at −20 °C. Staining for Flag epitopes was achieved using anti-Flag antibodies (F3165, Sigma) and with anti-mouse IgG antibodies conjugated with AlexaFluor488 or AlexaFluor555 fluorescent dyes (Invitrogen). DNA was stained using Hoechst 33342 (Sigma). Fixed cells were imaged using an Axioplan 2 microscope equipped with a 20×/0.75 Plan-Apochromat objective (Carl Zeiss). Images were captured using an AxioCam MRm digital camera (Carl Zeiss) driven by AxioVision 4.6.3 software (Carl Zeiss).
For live imaging experiments, cell were grown in 35 mm coverslip dishes and visualized with a Zeiss Axiovert widefield fluorescence microscope equipped with a heated stage, humidity chamber and CO2 supply. Cells were maintained at 37 °C and 5% CO2. Images were acquired using a C4742–95 ORCA 100 digital camera (Hamamatsu) and AQM Advance software 6 (Kinetic Imaging). Single cell measurements of Hoechst 33342 and mAG fluorescence were obtained from thresholded images by delineating regions of interest around cell nuclei in Image J (NIH), recording the integrated intensity value for each cell. Fluorescence intensity is displayed as arbitrary units.
Fluorescence-activated Cell Sorting
Cells were transfected using GeneJuice (Merck Biosciences), and 48 h later were pulsed for 1 h with the nucleoside analog, EdU, prior to harvesting and fixation. EdU incorporation was detected using the Click-iT EdU Flow Cytometry Assay kit (Invitrogen) according to the manufacturer's recommendations. Samples were analyzed using a FACS CyAn ADP with Summit Software. FACS data were gated to include only single cells expressing EGFP which was used as a marker of transfection.
Cell Fractionation and Co-immunoprecipitation Assays
T98G and U2OS cells were fractionated by resuspending in 10 mm Hepes, pH 7.9, 10 mm KCl, 1.5 mm MgCl2, 0.34 m sucrose, 10% glycerol, 1 mm DTT with protease inhibitors. Triton X-100 was added at 0.1% (v/v). Cells were incubated on ice for 5 min, and lysates were centrifuged at 1,200 rcf at 4 °C for 4 min. Pelleted nuclei were lysed in RIPA buffer.
For immunoprecipitation, cells were lysed in RIPA buffer and pre-cleared with a 1:1 mix of protein A and protein G-Sepharose beads. Lysates were incubated overnight at 4 °C with rotation with anti-Flag M2 agarose (Sigma). Beads were washed four times with NET2 buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 0.05% Triton X-100). Immunoprecipitates were then boiled in 4× Laemmli buffer and resolved by SDS-PAGE.
In Vitro Protein Interaction Assays
Bi-cistronic expression vectors expressing recombinant GST-tagged alleles of human Fbxo7 and Skp1 have been previously described (15). pGEX-TEV-hCRM1 (a kind gift from Prof. Yuh Min Chook, UT Southwestern) was used to express GST fusions of human CRM1. For the protein expression, vectors were transformed into FB810 Escherichia coli. Protein expression was induced by the addition of 250 μm isopropyl-β-d-1-thiogalactopyranoside for 3 h at 30 °C. Cells were harvested by centrifugation and lysed using lysozyme and sonication in Tris-buffered saline containing 1% Triton X-100. Proteins were purified from the soluble fraction using glutathione-Sepharose beads.
T7-tagged Fbxo7 proteins were synthesized using TnT T7 Quick-coupled Transcription/Translation System® (Promega) and incubated with glutathione-Sepharose beads coated with GST fusion proteins overnight at 4 °C in binding buffer (10 mm Hepes-KOH, pH 7.6, 50 mm KCl, 2.5 mm MgCl2, 10% glycerol, 0.02% Nonidet P-40, 1 mm phenylmethanesulfonyl fluoride, and 1 mm dithiothreitol). Beads were washed four times with wash buffer (10 mm Tris-HCl, pH 7.5, 1 mm EDTA, 0.2% Nonidet P-40 and 150 mm NaCl). The beads were boiled in 4× Laemmli buffer and proteins were resolved by SDS-PAGE and visualized by immunoblotting.
Antibodies
Antibodies used for immunoblotting and immunofluorescence assays (IFA) were as follows: actin (A2066, Sigma), E2F1 (sc-193, Santa Cruz Biotechnology), Fbxo7 (previously described (10)), Flag (F3165, Sigma), GFP (3EI, Cancer Research UK), MEK-1 (sc-6250, Santa Cruz Biotechnology), p27 (sc-528, Santa Cruz Biotechnology), and T7 (69522, Novagen). HRP-conjugated secondary antibodies raised against rabbit and mouse IgG were obtained from Santa Cruz Biotechnology, Inc and Jackson ImmunoResearch Laboratories, respectively.
NES Predictive Software
Amino acid sequences were analyzed using NetNES 1.1 Server to detect putative NES motifs (16). The software uses a hidden Markov model and an artificial neural network to analyze amino acid sequences and generate an “NES score” for each residue. Amino acids with a score exceeding a given threshold are expected to participate in an NES.
RESULTS
Fbxo7 Subcellular Distribution Varies Over Time in Growing Cells
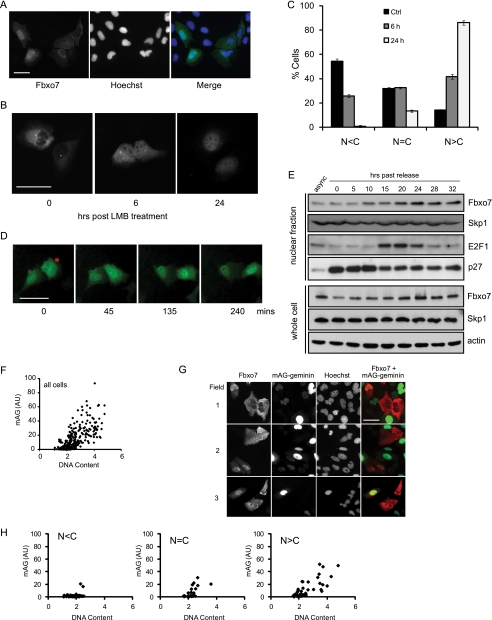
We have previously reported in experiments utilizing a fluorescent fusion protein of dsRED to the C terminus of Fbxo7 that in an asynchronous population of live cells, its localization is predominantly cytoplasmic and that it can shuttle between the nucleus and the cytoplasm (10). Fbxo7 localization was further investigated using an N-terminal Flag epitope-tagged Fbxo7 construct, which was transfected into U2OS cells, which were fixed and subjected to indirect IFAs. We found that a small number of cells had Fbxo7 localized predominantly in the nucleus (Fig. 1A), indicating that Fbxo7 can accumulate in the nucleus. To investigate the dynamics of Fbxo7 subcellular localization, U2OS cells expressing a fluorescent Fbxo7-Venus fusion protein were treated with the nuclear export inhibitor, leptomycin B (LMB) and the localization of the fusion protein was monitored in live cells over a 24 h period. Surprisingly, the rate of LMB-induced nuclear accumulation of Fbxo7-Venus was slow, taking up to 24 h for nuclear fluorescence to exceed that of cytoplasmic fluorescence in most cells (Fig. 1, B and C). This was much slower than seen for other proteins such as NF-κB which is also exported from the nucleus in a CRM1-dependent manner (17).
FIGURE 1.
Fbxo7 shuttles between the nucleus and the cytoplasm and accumulates in the nucleus at the G1/S transition. A, images of fixed U2OS cells expressing Flag-Fbxo7 visualized by immunofluorescence assays using anti-Flag antibodies (green) and Hoechst 33342 (blue). B, images of live U2OS cells expressing Fbxo7-Venus treated with 20 ng/ml of LMB for the indicated times. C, graph of the subcellular distribution of Fbxo7 was determined for a minimum of 167 cells per condition and categorized as being predominantly cytoplasmic (N < C), evenly distributed (N = C), or predominantly nuclear (N > C) at the indicated times after LMB treatment. Error is represented as the S.D. D, images of a time course tracking live cell fluorescence in individual U2OS cells expressing Fbxo7-Venus (green). A typical cell that demonstrated a change in localization from cytoplasmic to nuclear is indicated by an asterisk. E, immunoblots of endogenous Fbxo7, Skp1, E2F1, and p27 proteins in nuclear extracts and Fbxo7, Skp1, and actin in whole-cell lysates made from T98G cells synchronously released from quiescence and harvested at the indicated time points. F, graph of quantification of the intensity of Hoechst staining (DNA content) and mAG fluorescence. Data presented were obtained from over 400 asynchronously growing, clonal U2OS cells expressing mAG-geminin which were fixed and stained with Hoechst 33342, and quantified as described under “Experimental Procedures.” G, fluorescence microscopy images of mAG-geminin (green) expressing cells stained with Hoechst 33342 and for Flag-Fbxo7 expression using anti-Flag mAb (red). H, graphs of cells shown in G, analyzed as described for F. Single cell data were sorted into categories based on the Fbxo7 distribution; N < C, N = C, or N>C. Data from 125 cells expressing Flag-Fbxo7 were collected. Scale bars represent 50 μm.
This slow nuclear accumulation of Fbxo7 after LMB treatment might indicate that its translocation was influenced by the cell cycle. To see whether the rate of Fbxo7 nuclear import to export changed as cells progressed through the cell cycle, live cell imaging was used to monitor individual Fbxo7-Venus-expressing cells growing normally in culture. In some cells, a clear shift in bulk Fbxo7 localization from the cytoplasm to the nucleus took place over a period of a few hours (Fig. 1D), suggesting that at a particular point in the cell cycle the rate of export had decreased, or the rate of import increased, allowing nuclear accumulation.
We then investigated whether the shift in Fbxo7 localization was correlated with a particular phase of the cell cycle. Human glioblastoma cells (T98G) were made quiescent by contact inhibition and serum starvation and then stimulated to synchronously re-enter the cell cycle by seeding at lower density into medium containing full serum (18). Cells were harvested at different time points post release from quiescence and used to produce either whole-cell protein lysates or nuclear- and cytoplasmic-enriched fractions, which were analyzed by immunoblotting (Fig. 1E). While we saw only a very subtle increase in the total levels of Fbxo7 as cells re-entered the cell cycle, a clear increase in its nuclear concentration was observed, starting at 10 h post release. This coincided with other markers of the G1/S phase transition, including a decrease in nuclear p27 levels and a marked increase in E2F1 levels at 15 h post release. Together these data indicate that the increase in nuclear Fbxo7 occurred around the G1/S phase transition. However, no changes in the total levels or in the nuclear concentrations of Skp1, the SCF-recruiting component, at the G1/S transition were observed, indicating that there was no net change in the levels or localization of the total pool of SCF complexes during the cell cycle.
To further investigate when this shift in Fbxo7 localization occurred, an S/G2 phase marker, mAG-geminin, from the fluorescence ubiquitin cell cycle indicator system (FUCCI) (19) was utilized. The fusion protein, mAG-geminin, is unstable in early G1 but accumulates throughout the remainder of the cell cycle after the G1/S transition. In clonal U2OS cells stably expressing mAG-geminin, mAG fluorescence intensity correlated with total cellular DNA content, as measured by quantification of Hoechst staining. This indicated that the combination of the mAG-geminin and Hoechst staining could be used to distinguish between cells in G1 or in S/G2 phase (Fig. 1F). The mAG-geminin-expressing U2OS cells were transiently transfected with Flag-Fbxo7 (1–522) and assayed by IFA staining for the Flag epitope to track Fbxo7 localization during the cell cycle (Fig. 1G). In cells where staining for Flag-Fbxo7 was predominantly cytoplasmic, Hoechst, and mAG fluorescence was low, consistent with cells being in early G1 phase (Fig. 1H). In those cells were Fbxo7 staining was evenly distributed or predominantly nuclear, cells expressed higher levels of mAG-geminin and had increased Hoechst staining, consistent with being in S/G2 phase. Together these data strongly suggest that Fbxo7 was cytoplasmic during G0/G1 and accumulated in the nucleus as cells transited into S/G2 phase.
The FBD of Fbxo7 Contains a Functional NES
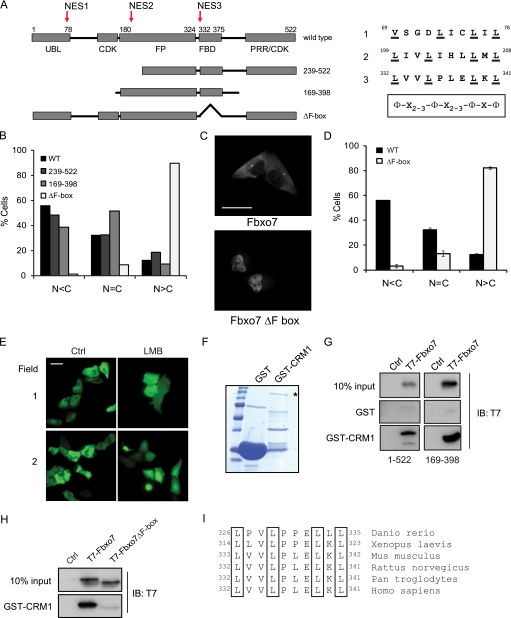
To identify sequence motifs that regulated Fbxo7 localization, we undertook a systematic analysis of the primary sequence of Fbxo7. Although no canonical nuclear localization sequences could be identified, three candidate motifs were identified that closely conformed to an NES consensus: Φ-x2–3-Φ-x2–3-Φ-x-Φ, where Φ is Leu, Ile, Val, Phe, or Met and x is any amino acid (Fig. 2A, indicated with arrows). These were located within the ubiquitin-like (Ubl) domain at amino acids 69–78, within the FP dimerization domain at amino acids 199–208, and within the FBD at amino acid 332–341. As well as scrutinizing the primary sequence, the NES recognition software, NetNES 1.1 Server (16), was used to analyze and predict whether any of these putative NESs was likely to be functional. Only NES3 contained amino acids exceeding the “NES score” threshold, indicating that it was highly likely to be functional.
FIGURE 2.
Fbxo7 contains a functional NES within its FBD. A, schematic representation showing functional domains and truncation mutants of Fbxo7. Arrows indicate motifs within Fbxo7 that conform to the consensus sequence for a leucine-rich NES, and corresponding NES sequences are listed on the right. B, graphs of the percentage of cells showing predominantly cytoplasmic (N < C), equivalent (N = C), or predominantly nuclear (N > C) distribution of exogenous Fbxo7 proteins. Localization of Flag-tagged Fbxo7 truncation mutants was determined by immunofluorescent staining with anti-Flag antibodies. C, images of WT and Fbxo7 (ΔF-box) Venus fusions expressed in U2OS cells visualized by fluorescence microscopy. D, quantification of the distribution of the localization of WT and Fbxo7 (ΔF-box) Venus fusions from a minimum of 415 cells per condition. Error is represented as the S.E. E, fluorescence images of U2OS cells expressing a Venus-NES-Venus reporter fusion protein. Cells imaged in the right-hand side panels were treated with 20 ng/ml LMB for 15 h. F, image of the Coomassie-stained SDS-PAGE gel of GST and GST-CRM1 fusion proteins. Asterisk denotes the full-length fusion protein. G, images of immunoblots of in vitro binding assays for T7-tagged Fbxo7 (1–522) and (169–398) bound to GST-CRM1. H, immunoblots of in vitro binding assay for T7-tagged Fbxo7 (169–398) and the corresponding ΔF-box mutant binding to GST-CRM1. I, alignment of the NES amino acids located within the FBDs from Fbxo7 homologues in other species. Scale bars represent 50 μm.
To begin to test the activity of these NESs in vivo, Flag-tagged truncation mutants of Fbxo7 that removed one or more of the putative NESs (239–522, 169–398, and ΔF-box) were transfected in U2OS cells, which were then fixed and assayed for the presence of the Flag epitope. Transfected cells were visualized by microscopy and at least 400 cells were scored as having predominantly cytoplasmic (C>N), predominantly nuclear (C<N), or an equivalent (C = N) signal intensity. This experiment revealed that the mutant lacking the FBD showed a dramatic change in its localization, becoming predominantly nuclear in 90% of cells, as compared with WT (Fig. 2B). On the other hand, the Fbxo7 mutants lacking either NES1 or NES1 and NES2, showed a subcellular distribution similar to WT Fbxo7 (Fig. 2B). The fact that the deletion of the FBD alone resulted in such a dramatic change to protein localization suggested that NES3 was the only functional NES and largely responsible for the export of Fbxo7 from the nucleus. To verify this finding in live cells, fusions of WT or ΔF-box mutant Fbxo7 to Venus fluorescent protein were generated and transfected into U2OS cells. Consistent with the results in fixed cells, the Fbxo7 (ΔF-box)-Venus protein was found to be predominantly nuclear in the majority of cells as compared with Fbxo7-Venus, which was predominantly cytoplasmic (Fig. 2, C and D). These data suggest that the mostly cytoplasmic localization of WT Fbxo7 was due to sequences within the FBD, which bring about its export from the nucleus.
To determine the capacity of NES3 to bring about nuclear export, we tested its ability to influence the localization of a protein other than Fbxo7. To achieve this, a fluorescent reporter protein, consisting of two Venus proteins separated by NES3 was created. This reporter protein was of sufficient size to prevent its diffusion across the nuclear membrane, and the synthesis of the full-length fusion protein was confirmed by immunoblotting (data not shown). The Venus-NES3-Venus protein was expressed in U2OS cells and visualized in live cells. As can be seen in Fig. 2E, it was excluded from the nucleus; moreover, the cytoplasmic localization of this protein was not due to its inability to enter the nucleus, as evidenced by the fact that LMB treatment induced its accumulation in the nucleus. These data argue that NES3 alone was sufficient to mediate nuclear export.
NES sequences function by direct interaction with the nuclear export receptor, CRM1. The ability of Fbxo7 to bind to CRM1 was therefore tested using in vitro binding assays. GST alone and a GST fusion protein to CRM1 were produced in bacteria (Fig. 2F) and tested for their ability to interact with full-length Fbxo7 (1–522) and Fbxo7 (169–398), which lacked the other putative NESs. Both Fbxo7 proteins interacted specifically with GST-CRM1, but not GST alone, in vitro (Fig. 2G). Moreover, this interaction was dependent on the presence of the FBD, because when it was deleted, the interaction with GST-CRM1 was lost (Fig. 2H). Together, these experiments indicate that Fbxo7 contained a functional NES sequence within its FBD, which imposed its predominantly cytoplasmic localization. In addition, an alignment of multiple Fbxo7 homologues from different species from zebrafish to higher mammals showed that the defining leucine residues of the NES were conserved (Fig. 2I), suggesting that it is important for Fbxo7 localization across multiple species.
A Subset of F Box Proteins Contains an NES Embedded within the F-box Domain
In a study by Cenciarelli et al. (3) it was suggested that neither the FBD nor Skp1 binding influenced the localization of FBPs. However, because the NES was evolutionarily conserved and the loss of the FBD clearly altered Fbxo7 localization, we were interested in determining if other FBPs also contained an NES within their FBD and whether this affected their localization. In 2004, Jin et al. (20) reported the identification of 68 human FBPs. The subcellular localization of 23 out of 68 FBPs has been reported, and of these, 8 had a consensus NES at the start of the FBD (Table 1) as identified by sequence alignment. This included Fbxo2/NFB42, Fbxo4, Fbxo6, Fbxo7, Fbxo8, Fbxl5, Fbxw2, and Fbxw7. Most (5 out of 8) of these NESs also scored above the threshold for a functional NES by NetNES analysis (Table 1); however, all 8 FBPs have been reported to have a cytoplasmic localization. Fbxw7 has additionally been found in the nucleus and nucleolus (4).
TABLE 1.
F-box proteins with experimentally tested localizations
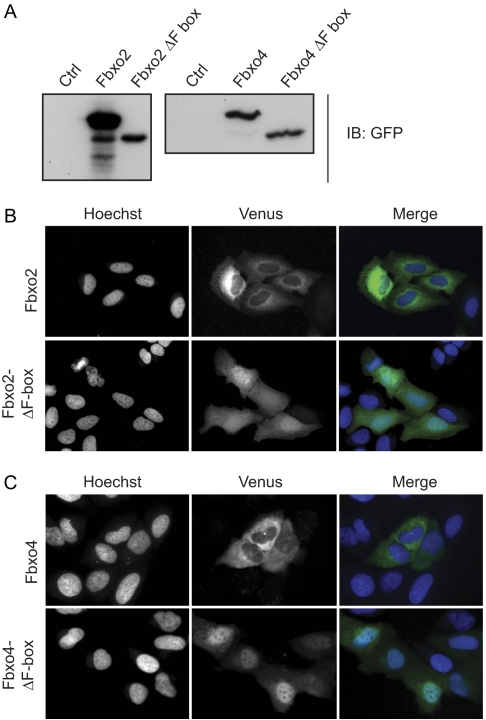
The fact that all of the FBPs which contained this NES were cytoplasmic, suggested that it might contribute to their exclusion from the nucleus. To test this, Fbxo2, Fbxo4, and F-box deletion mutants of both proteins were fused to Venus and expressed in U2OS cells. Immunoblotting of lysates from transfected cells using anti-GFP antibodies demonstrated that full-length fusion proteins corresponding to the predicted molecular weights were expressed (Fig. 3A). The localization of the fluorescent fusion proteins was then visualized in live cells. The cytoplasmic localization of full-length Fbxo2 and Fbxo4 was verified; moreover, this was dependent on the presence of the FBD, as the mutants which removed this motif resulted in their re-localization to the nucleus (Fig. 3, B and C). These data indicate that the presence of an NES within the FBD was not unique to Fbxo7, and furthermore, suggest that the NES in the FBD contributed to the localization of these proteins.
FIGURE 3.
The FBDs of Fbxo2/NFB42 and Fbxo4 contain NESs. A, immunoblots of lysates from cells expressing Fbxo2 and Fbxo4 fusion proteins to Venus with anti-GFP antibodies. Images of the localization of the Venus fusions (green) of the WT and ΔF-box deletion mutants of Fbxo2 (B) and Fbxo4 (C) transfected into U2OS cells and visualized in live cells by fluorescence microscopy. Nuclei are stained by Hoechst 33342 (blue). The size bar in these images is 50 μm.
Residues of the Fbxo7 NES Are Required for Interaction with Skp1
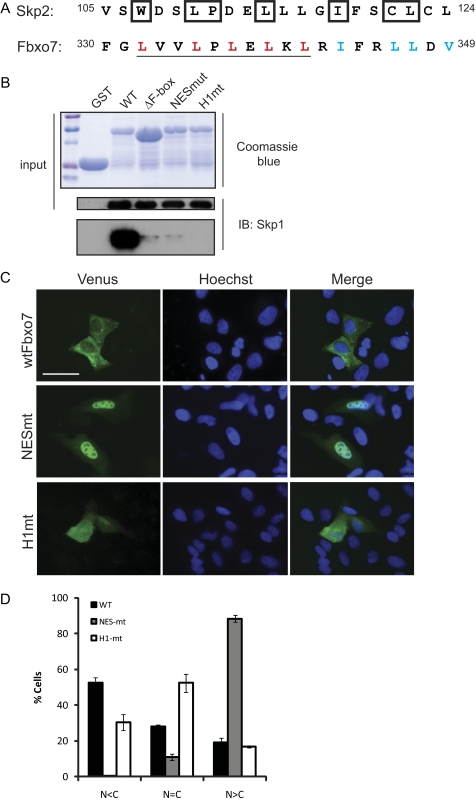
The juxtaposition of Skp1 and CRM1 binding residues within an FBP implied a potential regulatory relationship. To investigate this further, we examined the solved structure of Skp2/Fbxl1 bound to Skp1 and noted the residues in the FBD of Skp2/Fbxl1 that directly contacted Skp1 (Fig. 4A, boxed residues) (21). By aligning the FBD of Skp2/Fbxl1 with that of Fbxo7, residues that were predicted to be essential for interaction with Skp1 were identified. This comparison suggested that at least three of the leucine residues comprising the NES were likely to be involved in direct contacts with Skp1 (Fig. 4A, NES is underlined). Because of the apparent overlap of key interacting residues within the FBD, we wanted to determine whether it was possible to separate CRM1 and Skp1 binding and what effect this would have on the localization of Fbxo7. To test this, site-specific mutagenesis was used to engineer Fbxo7-NESmt, an allele in which all of the leucine residues in the NES motif were changed to alanine, and which was predicted to ablate NES function (Fig. 4A, mutated NES residues marked in red). There were additional residues in the FBD that form an α-helix that also directly contacted Skp1 as deduced from the crystal structure (21). This helix was not part of the NES. Therefore, a second mutant, called Fbxo7-H1mt, was created in which four hydrophobic residues in helix 1 were changed to alanine (Fig. 4A, mutated residues marked in blue).
FIGURE 4.
Residues in the Fbxo7 FBD are required for nuclear export and Skp1 binding. A, alignment of the amino acids of the FBDs of human Skp2/Fbxl1 and Fbxo7. The residues in Skp2/Fbxl1 that directly interact with Skp1 are boxed. The NES residues in Fbxo7 are underlined. Residues in Fbxo7 that were mutated to alanine to produce the NESmt allele are marked in red. Residues mutated to alanine to produce the H1mt allele are marked in blue. B, in vitro binding assay for Skp1 binding to GST-Fbxo7 WT and mutant alleles. Coomassie-stained SDS-PAGE gel of GST and GST-Fbxo7 (169–398) alleles (top) and immunoblots for Skp1 bound to GST-Fbxo7 alleles (bottom). C, images of the localization of the Venus fusions (green) of WT, NESmt and H1mt Fbxo7 expressed in U2OS cells. Nuclei are stained by Hoechst 33342 (blue). Live cells were visualized by fluorescence imaging. D, quantification of the distribution of the localization of WT and mutant Venus fusions from a minimum of 260 single cells per condition. Error is represented as the S.E. Scale bar represents 50 μm.
The NESmt and H1mt both altered residues within the FBD that were predicted to contact Skp1. We first tested whether these proteins were capable of interacting with Skp1 using an in vitro binding assay. A GST fusion protein of Fbxo7 (169–398) was mutated to incorporate either the NESmt or the H1mt changes. These constructs also co-expressed Skp1 from an IRES in the plasmid. Proteins were expressed in bacteria and immobilized on a glutathione column, and assayed for the presence of Skp1 on the column. However, despite robust expression of Skp1 and all of the GST-Fbxo7 proteins, Skp1 associated with neither the NESmt nor the H1mt in vitro (Fig. 4B). These data strongly indicate that mutation of the predicted Skp1 contact residues in helix 1 or in the NES abrogated binding to Skp1.
The NESmt and H1mt Fbxo7 alleles were next engineered as fusions to Venus, expressed in U2OS cells, and observed by live cell fluorescence microscopy. As expected, Fbxo7-NESmt-Venus was almost exclusively nuclear in >90% of cells scored (Fig. 4, C and D), which was similar to the distribution seen with the ΔF-box mutant (Fig. 2, C and D). On the other hand, the localization of the H1mt-Venus protein within cells was distributed across the three scoring categories: cytoplasmic, nuclear, or equal distribution within the nucleus and cytoplasm (Fig. 4, C and D), with fewer cytoplasmic cells and an even distribution across the cell being the most prevalent. This distribution suggested that the H1 mutations did not ablate NES function and that the rate of import and export were equivalent. In sum, these data indicate that residues in the NES imparted a predominantly cytoplasmic localization to Fbxo7.
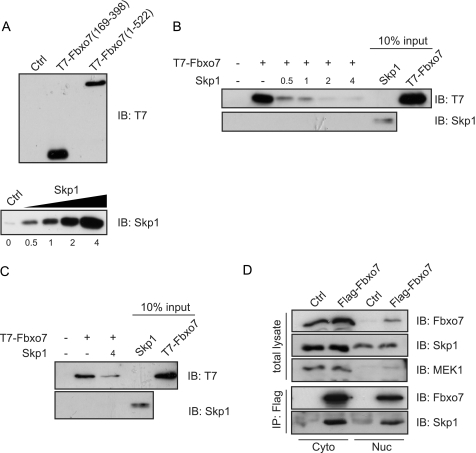
Skp1 Competes with CRM1 for Binding to Fbxo7
The data showed that the CRM1 and Skp1 interacting residues of the FBD overlapped. To test whether CRM1 and Skp1 bound simultaneously or in a mutually exclusive manner to Fbxo7, an in vitro competition assay was devised. GST-CRM1 was produced in and purified from bacteria, immobilized on a glutathione column, and incubated with in vitro synthesized T7-tagged Fbxo7 (169–398), and Skp1, which was titrated at increasing concentrations into this binding reaction (Fig. 5, A and B). The amount of Fbxo7 bound to the beads was assayed by immunoblotting for the T7 epitope. As previously observed in Fig. 2G, Fbxo7 interacted with GST-CRM1; however, the addition of Skp1 into the binding assay decreased substantially the amount of Fbxo7 bound to GST-CRM1, and this effect was dose-dependent (Fig. 5B). The addition of Skp1 also inhibited binding in the context of full-length Fbxo7 (1–522) to GST-CRM1 (Fig. 5C). These data indicate that Skp1 competed with CRM1 for interaction with Fbxo7, suggesting mutually exclusive binding. These data further imply that if Fbxo7 interacted with Skp1 within the nucleus, it would be unable to interact with the nuclear export machinery and would lead to Fbxo7 accumulation in the nucleus.
FIGURE 5.
Binding of Fbxo7 to Skp1 and CRM1 is mutually exclusive. A, immunoblots for in vitro synthesized T7-Fbxo7 (1–522), T7-Fbxo7 (169–398), and Skp1 (1–146) used as inputs for in vitro binding assays. Immunoblots of T7 and Skp1 for proteins present on GST and GST-CRM1 columns. In vitro synthesized Skp1 was titrated into a binding reaction of immobilized GST-CRM1 and in vitro synthesized T7-Fbxo7 (169–398) (B) or T7-Fbxo7 (1–522) (C) proteins. (D) Images of immunoblots for the indicated proteins on input and anti-Flag immunoprecipitated proteins from nuclear (Nuc) or cytoplasmic (Cyto) extracts of U2OS cells expressing Flag-Fbxo7 (1–522).
To test whether Fbxo7 interacted with Skp1 in the nucleus in vivo, U2OS cells were transfected with Flag-tagged Fbxo7. Cells were fractionated into nuclear and cytoplasmic lysates, which were immunoprecipitated with antibodies to the Flag epitope, and the immunoprecipitates were then assayed for the presence of endogenous Skp1. Skp1 co-immunoprecipitated with Flag-Fbxo7 from both nuclear and cytoplasmic cellular extracts (Fig. 5D), indicating that Skp1 interacted with Fbxo7 in either subcellular compartment. These data demonstrate that Fbxo7 was recruited as part of an E3 ubiquitin ligase in both the nucleus and the cytoplasm.
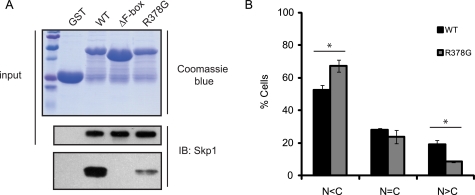
A Pathogenic Point Mutation in Fbxo7 Promoted a More Cytoplasmic Localization
A naturally occurring autosomal recessive mutation in Fbxo7 has been reported to cause an atypical, early-onset Parkinson disease (PD). The R378G mutation is located in an unstructured region of the protein. However, as this mutation is next to the FBD, the ability of Fbxo7 (R378G) to interact with Skp1 was tested in an in vitro binding assay as described above. Surprisingly, the ability of this allele of Fbxo7 to interact with Skp1 was significantly compromised in comparison to WT Fbxo7 (Fig. 6A). A lack of binding to Skp1 might be expected to prevent the retention and accumulation of Fbxo7 (R378G) in the nucleus. To test this, an Fbxo7 (R378G)-Venus fusion protein was expressed in U2OS cells and its localization scored as in Fig. 2. The R378G mutant was found to be more abundant in the cytoplasm than the WT protein (p value 0.020; Student's t test) (Fig. 6B). In addition, the number of cells where nuclear Fbxo7 exceeded cytoplasmic Fbxo7 was significantly decreased (p value 0.022; Student's t test). These data indicate that Skp1 binding was important for nuclear accumulation of Fbxo7, and suggest that the decrease in Skp1 binding and the resulting lack of nuclear retention or increased cytoplasmic levels of Fbxo7 contribute to its pathogenicity in PD.
FIGURE 6.
A Parkinson disease-associated allele of Fbxo7 is impaired for Skp1 binding and its subcellular localization is altered. A, immunoblots from an in vitro binding assay of WT and R378G Fbxo7 to Skp1 were conducted as in 4B. B, quantification of the distribution of the localization of WT and R378G Fbxo7 Venus fusions for a minimum of 168 cells per condition. Error is represented as the S.E. * denotes statistical significance, p value < 0.05.
DISCUSSION
Many cellular proteins have activities which must be controlled both spatially and temporally so that appropriate and timely interactions occur. This can be regulated transcriptionally or post-translationally, and the latter almost certainly allows for more rapid cellular responses. As the substrate receptors for E3 ubiquitin ligases, the ability of FBPs to interact with their substrates is crucial and thus heavily regulated, both by modification of the substrate and the availability of the FBP (22). One common method of controlling availability is by regulating localization. Here we report our studies conducted on the localization of FBPs, using in particular Fbxo7. In this and previous studies (10, 15), we have shown that Fbxo7 is a predominantly cytoplasmic protein, and we have demonstrated this using a number of different techniques: by fractionation into nuclear and cytoplasmic protein extracts and detection of endogenous Fbxo7 protein; by direct visualization in live, single cells using fluorescent fusions to Venus (this report) or dsRed at the C terminus of Fbxo7 (10, 15); and by indirect IFA staining of fixed cells containing N-terminal Flag-tagged Fbxo7. Previous studies of Fbxo7 localization performed using fixed cells containing N-terminal HA-epitope-tagged Fbxo7 have also demonstrated a mostly cytoplasmic localization (12). Here we show that although Fbxo7 is generally localized in the cytoplasm, it can accumulate in the nucleus, and we demonstrated this occurs at the G1/S transition. Several putative leucine-rich NESs were identified in the primary sequence of Fbxo7, and one of these was shown to be functional. Unusually it was located at the start of the FBD, the binding site for Skp1. This juxtaposition of the two binding sites was not unique to Fbxo7 and an examination of the sequences of FBPs for which a subcellular localization has been reported showed that there were several other FBPs that had this motif at the start of the FBD, suggesting that they would also potentially be subject to this mode of regulation. Indeed deletion of the FBD in two other FBPs, Fbxo2, and Fbxo4, caused their re-localization to the nucleus. However, the presence of an embedded NES/FBD was not required for or did not always enforce an entirely cytoplasmic localization. We noted, for example, that Fbxw7 contains other motifs allowing its localization to other compartments (4), and which may be dominant to the NES. In addition, the NES was not required for the cytoplasmic localization of other FBPs. For example, Fbxw2 also has an embedded NES within its FBD, yet its deletion did not alter localization: both WT and an FBD deletion mutant were cytoplasmic (3). It is possible that there may be other NESs in Fbxw2, or alternatively the deletion of the FBD may have also removed other localization signals, like an NLS. Nonetheless, this finding led the authors to suggest that the FBD had no effect on the localization of FBPs, but this is clearly not the case for some FBPs, including Fbxo2, Fbxo4, and Fbxo7. These examples illustrate that the identification of localization motifs in an FBP and an understanding of their relationships and dominance must be evaluated for each FBP. Although we limited the FBPs in Table 1 to those with a reported localization, a consensus NES at the start of the FBD was also evident in several other FBPs, for example, Fbxw3, Fbxo9, Fbxo31, Fbxo32. Our studies therefore suggest that the NES may participate in the localization of 12 out of 68, approximately one-sixth of FBPs.
Our data indicate that this type of NES-containing FBD can be a key regulatory domain in the protein, by virtue of its interactions with CRM1 and Skp1, which dictated both the localization of the FBP as well as enabling its ability to act as part of an E3 ubiquitin ligase. We showed that Fbxo7 can interact with Skp1 and therefore be part of an SCF-type E3 ubiquitin ligase in both the cytoplasm and in the nucleus. The net change in its localization, beginning at the onset of S-phase, might allow Fbxo7 to interact with different sets of substrates at different times in the cell cycle. Thus, its activity would be spatially regulated in time with the cell cycle, for example, by acting as an assembly scaffold for cyclin D/Cdk6 complexes in the cytoplasm during G1 and then later participating in the ubiquitination of as yet unidentified nuclear substrates.
Mechanistically, we show that the Fbxo7 NES directly binds CRM1, and that CRM1 and Skp1 binding were mutually exclusive. This supports a model whereby binding to Skp1 in the nucleus inhibits the export of Fbxo7 to the cytoplasm, and allows its accumulation. There are several scenarios for how nuclear accumulation might occur. For example, the rates of import and export are similar until the G1/S transition when export is inhibited due to Skp1 binding. This would suggest that Skp1 binding to Fbxo7 is a regulated event, specifically enhanced at the G1/S transition, perhaps due to other signals. Although we did not observe any change to the bulk Skp1 levels or to the nuclear levels of Skp1 at the G1/S transition, it is still possible that Fbxo7 binding to Skp1 may rely on an S phase-dependent modification to either protein. Precedence for a changeable interaction of an FBP with Skp1 includes the enhanced interaction of the Saccharomyces cerevisiae FBP, Grr1 for Skp1 under high glucose conditions (23). An alternative possibility is that nuclear import and export are similar until the rate of import significantly increases at the G1/S phase transition. Although we did not find a canonical NLS in Fbxo7, it is possible that it gains entry to the nucleus by interacting with other proteins that contain these sequences, like p21 or p27, which are proposed import factors for cyclin D/Cdk complexes (24, 25), and to which Fbxo7 is known to bind directly (10). The possibility that Fbxo7 import might be due to Skp1 is unlikely as its net nuclear protein levels did not change at the G1/S transition. Finally, we did note that although these studies focused on the most abundant isoform of Fbxo7 (isoform 1), endogenous isoform 2 was also readily detected in nuclear-enriched cell fractions (data not shown). These two isoforms differ at their N termini, where isoform 2 lacks the Ubl motif, present only in isoform 1. As endogenous isoform 2 also accumulated in the nucleus, this suggests that nuclear localization is not dependent at least on the Ubl motif.
The localization of Fbxo7 appears to be deregulated in two different human diseases. Fbxo7 had a nuclear localization in a percentage of biopsies from human colon cancer samples, and yet, interestingly, data presented here indicated a different localization defect in PD. Homozygous inheritance of an R378G allele of Fbxo7 causes an atypical PD (9, 11), and finding that this pathogenic point mutation, located outside of the FBD, severely compromised Skp1 binding was surprising. Moreover, in agreement with our model for the role of Skp1 binding in nuclear retention, R378G did not accumulate in the nucleus and was more cytoplasmic than the WT protein. This suggests that one route to developing early-onset PD would be due to the lack of ubiquitination of nuclear or cytoplasmic targets of Fbxo7, or alternatively, cytoplasmic Fbxo7 substrates are stabilized by interaction with increased levels of free Fbxo7. We are currently investigating additional nuclear and cytoplasmic Fbxo7 substrates, as this should shed light on the deregulated pathways that cause PD and cancer. In sum, our data show a novel mechanism of regulation of both localization and E3 ubiquitin ligase activity through the juxtaposition of an NES within the FBD, which occurs for a number of FBPs including Fbxo7, whose mislocalization is linked to both neurodegenerative disease and cancer.
Acknowledgments
We thank Nigel Miller, Jürgen Scheer, and Suzanne Randle for their technical assistance. We appreciate the gift of these reagents: pCS2-Venus and the FUCCI lentiviral vector pCSII-EF-MCS-mAG-geminin, both from A Miyawaki, RIKEN Institute, Japan; pYFP-Fbxo2 from Chi-Yong EOM, Korea Basic Science Institute; T98G human glioblastoma cells from Dr. Koichi Ichimura; and 293T human embryonic kidney cells from Prof. Chris Boshoff.
This work was supported by the BBSRC (BB/F012764/1).
- SCF
- Skp1-Cullin-F-box
- FBP
- F-box protein
- FBD
- F-box domain
- NES
- nuclear export sequence
- NLS
- nuclear localization sequence
- Ubl
- ubiquitin-like domain
- FP domain
- Fbxo7-PI31 domain
- LMB
- leptomycin B
- N
- nuclear
- C
- cytoplasmic
- PD
- Parkinson disease
- EdU
- 5-ethynyl-2′-deoxyuridine.
REFERENCES
- 1. Skowyra D., Craig K. L., Tyers M., Elledge S. J., Harper J. W. (1997) Cell 91, 209–219 [DOI] [PubMed] [Google Scholar]
- 2. Bai C., Sen P., Hofmann K., Ma L., Goebl M., Harper J. W., Elledge S. J. (1996) Cell 86, 263–274 [DOI] [PubMed] [Google Scholar]
- 3. Cenciarelli C., Chiaur D. S., Guardavaccaro D., Parks W., Vidal M., Pagano M. (1999) Curr. Biol. 9, 1177–1179 [DOI] [PubMed] [Google Scholar]
- 4. Welcker M., Orian A., Grim J. E., Eisenman R. N., Clurman B. E. (2004) Curr. Biol. 14, 1852–1857 [DOI] [PubMed] [Google Scholar]
- 5. Gao D., Inuzuka H., Tseng A., Chin R. Y., Toker A., Wei W. (2009) Nat. Cell Biol. 11, 397–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lin H. K., Wang G., Chen Z., Teruya-Feldstein J., Liu Y., Chan C. H., Yang W. L., Erdjument-Bromage H., Nakayama K. I., Nimer S., Tempst P., Pandolfi P. P. (2009) Nat. Cell Biol. 11, 420–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bashir T., Pagan J. K., Busino L., Pagano M. (2010) Cell Cycle 9, 971–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boutonnet C., Tanguay P. L., Julien C., Rodier G., Coulombe P., Meloche S. (2010) Cell Cycle 9, 975–979 [DOI] [PubMed] [Google Scholar]
- 9. Di Fonzo A., Dekker M. C., Montagna P., Baruzzi A., Yonova E. H., Correia Guedes L., Szczerbinska A., Zhao T., Dubbel-Hulsman L. O., Wouters C. H., de Graaff E., Oyen W. J., Simons E. J., Breedveld G. J., Oostra B. A., Horstink M. W., Bonifati V. (2009) Neurology 72, 240–245 [DOI] [PubMed] [Google Scholar]
- 10. Laman H., Funes J. M., Ye H., Henderson S., Galinanes-Garcia L., Hara E., Knowles P., McDonald N., Boshoff C. (2005) EMBO J. 24, 3104–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shojaee S., Sina F., Banihosseini S. S., Kazemi M. H., Kalhor R., Shahidi G. A., Fakhrai-Rad H., Ronaghi M., Elahi E. (2008) Am. J. Hum. Genet. 82, 1375–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chang Y. F., Cheng C. M., Chang L. K., Jong Y. J., Yuo C. Y. (2006) Biochem. Biophys. Res. Commun. 342, 1022–1026 [DOI] [PubMed] [Google Scholar]
- 13. Hsu J. M., Lee Y. C., Yu C. T., Huang C. Y. (2004) J. Biol. Chem. 279, 32592–32602 [DOI] [PubMed] [Google Scholar]
- 14. Laman H. (2006) Cell Cycle 5, 279–282 [DOI] [PubMed] [Google Scholar]
- 15. Kirk R., Laman H., Knowles P. P., Murray-Rust J., Lomonosov M., Meziane el K., McDonald N. Q. (2008) J. Biol. Chem. 283, 22325–22335 [DOI] [PubMed] [Google Scholar]
- 16. la Cour T., Kiemer L., Mølgaard A., Gupta R., Skriver K., Brunak S. (2004) Protein Eng. Des. Sel. 17, 527–536 [DOI] [PubMed] [Google Scholar]
- 17. Nelson G., Paraoan L., Spiller D. G., Wilde G. J., Browne M. A., Djali P. K., Unitt J. F., Sullivan E., Floettmann E., White M. R. (2002) J. Cell Sci. 115, 1137–1148 [DOI] [PubMed] [Google Scholar]
- 18. Stein G. H. (1979) J. Cell. Physiol. 99, 43–54 [DOI] [PubMed] [Google Scholar]
- 19. Sakaue-Sawano A., Kurokawa H., Morimura T., Hanyu A., Hama H., Osawa H., Kashiwagi S., Fukami K., Miyata T., Miyoshi H., Imamura T., Ogawa M., Masai H., Miyawaki A. (2008) Cell 132, 487–498 [DOI] [PubMed] [Google Scholar]
- 20. Jin J., Cardozo T., Lovering R. C., Elledge S. J., Pagano M., Harper J. W. (2004) Genes Dev. 18, 2573–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schulman B. A., Carrano A. C., Jeffrey P. D., Bowen Z., Kinnucan E. R., Finnin M. S., Elledge S. J., Harper J. W., Pagano M., Pavletich N. P. (2000) Nature 408, 381–386 [DOI] [PubMed] [Google Scholar]
- 22. Cardozo T., Pagano M. (2004) Nat. Rev. Mol. Cell Biol. 5, 739–751 [DOI] [PubMed] [Google Scholar]
- 23. Li F. N., Johnston M. (1997) EMBO J. 16, 5629–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bagui T. K., Mohapatra S., Haura E., Pledger W. J. (2003) Mol. Cell. Biol. 23, 7285–7290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cheng M., Olivier P., Diehl J. A., Fero M., Roussel M. F., Roberts J. M., Sherr C. J. (1999) EMBO J. 18, 1571–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]